Association between single nucleotide polymorphisms of nLvALF1 and PEN2-1 genes and resistance to Vibrio parahaemolyticus in the Pacific white shrimp Litopenaeus vannamei
Pwpol Kongchum, Suphvdee Chimtong, Nprt Prpiwong
aFaculty of Animal Sciences and Agricultural Technology, Silpakorn University, Phetchaburi Campus, Cha-am, Phetchaburi, 76120, Thailand
bSamut Sakhon Coastal Aquaculture Research and Development Center, Mueang, Samut Sakhon, 74000, Thailand
A B S T R A C T
Vibrio parahaemolyticus carrying a plasmid encoding the PirAvp and PirBvp toxins is the causative agent of acute hepatopancreatic necrosis disease (AHPND) in penaeid shrimps. In the Pacific white shrimp farming industry,one possible strategy to reduce economic loss due to AHPND is the development of a shrimp line resistant to the disease. In this study, we identified single nucleotide polymorphisms (SNPs) in the Litopenaeus vannamei antilipopolysaccharide factor 1 (nLvALF1) and penaeidin 2-1 (PEN2-1) genes, and we analyzed the associations between these SNPs and resistance/susceptibility to V. parahaemolyticus infection in the Pacific white shrimp.Postlarvae (PL20) shrimp from a local hatchery in Prachuap Khiri Khan Province were challenged with an isolate of VPAHPND and mortality was observed for 14 days. DNA was extracted from susceptible (died within 6 days) and resistant (survived the challenge) shrimp (45 individuals/group) and used for PCR amplifications of nLvALF1(397 bp) and PEN2-1 (637 bp) gene fragments. PCR products were sequenced by direct sequencing and SNPs were identified from sequencing chromatograms. Nine and seven SNPs were identified in nLvALF1 and PEN2-1 gene fragments, respectively. Analyses of allele frequencies in susceptible and resistant samples using Chi-square tests revealed that four and six SNPs in nLvALF1 and PEN2-1, respectively, were associated with resistance/susceptibility to V. parahaemolyticus infection (P <0.05). The SNPs in the candidate genes identified here are potential DNA markers for breeding V. parahaemolyticus-resistant Pacific white shrimp in the study population;however, further validation will be required if these SNPs are to be used across populations.
Keywords:
Single nucleotide polymorphisms
Anti-lipopolysaccharide factor
Penaeidin
Early mortality syndrome
Acute hepatopancreatic necrosis disease
1.Introduction
The Pacific white shrimp (Litopenaeus vannamei, Boone, 1931) is the most widely cultivated penaeid shrimp. It is native to Central and South America but was introduced to Asia in 1978; in Taiwan and China,commercial culture ofL. vannameibegan in 1996 (Liao & Chien, 2011),after which commercial farming of the species expanded to South and Southeast Asian countries making it an important aquaculture produce of the region. Global aquaculture production of the Pacific white shrimp increased from 2,648,541 mt in 2010 to 4,966,241 mt in 2018 (FAO,2020) due to increased global demand; thus, intensive farming systems have been implemented in most shrimp-producing countries to boost yields. However, intensification of stock density and feeding has led to adverse pond and water quality conditions and negative effects on the health of shrimps with increased risk of disease outbreak. Emerging diseases caused by viruses, rickettsia-like bacteria, bacteria, protozoa,and fungi include acute hepatopancreatic necrosis disease (AHPND),hepatopancreatic microsporidiosis, hepatopancreatic haplosporidiosis,aggregated transformed microvilli, covert mortality disease, white spot disease, yellow head disease, infectious myonecrosis, Taura syndrome,and infectious hypodermal and hematopoietic necrosis; such diseases have led to reduced production of the Pacific white shrimp in both Asia and the Americas (Lightner, 2011; Thitamadee et al., 2016).
Penaeid shrimp disease, known as “early mortality syndrome” or more specifically AHPND, is a serious disease that affects shrimp production and causes significant economic loss in the shrimp farming industries of many countries. The first outbreak was reported in China in 2009 (Flegel, 2012); thereafter, outbreaks were reported in many other countries and regions including Vietnam, Malaysia, Thailand, Mexico,South America, and the USA (de la Peña et al., 2015; Dhar et al., 2019;Kondo et al., 2014; Kua et al., 2016; Nunan et al., 2014; Restrepo et al.,2016; Tran et al., 2013). Pacific white shrimp postlarvae with AHPND usually show mortality rates of 40%-100% within 35 days of stocking in culture ponds (Hong et al., 2016). Tran et al. (2013) identified the causative agent associated with the disease as the bacteriumVibrio parahaemolyticus. It was discovered thatV. parahaemolyticuscausing AHPND carries a plasmid encoding the toxins PirAvpand PirBvp; these secreted toxins are the primary virulence factors that cause the disease and the acquisition of PirABvpby horizontal gene transfer between microorganisms is possible (Lee et al., 2015). Recently,V. parahaemolyticusandVibrio harveyi, which cause AHPND with PirA and PirB toxin genes,were isolated from Pacific white shrimp ponds in Malaysia (Muthukrishnan et al., 2019); the isolatedV. harveyistrain was found to be more virulent to the Pacific white shrimp than was theV. parahaemolyticusstrain.
Prevention and treatment approaches for AHPND have been attempted including the use of plant extracts as feed additives to inhibit bio film formation (Soowannayan et al., 2019), biocontrol using a microalgal-bacterial consortium (Chang et al., 2020), degradation of PirA/BVPtoxins byBacillus subtilis(Nguyen et al., 2021), and dietary supplementation with the probiotic bacteriumRhodobacter sphaeroides(Torpee et al., 2021). Nevertheless, the disease continues to occur prevalently in Pacific white shrimp. One potential strategy for dealing with AHPND besides the existing prevention and treatment methods is the use of selective breeding to develop a shrimp stock resistant toV. parahaemolyticusAHPND infection. In general, selection of broodstock based on genetic information helps accelerate genetic gain; however, this approach requires genetic markers that are associated with the traits of interest.
Antimicrobial peptides (AMPs) are a diverse class of natural molecules produced by all multicellular organisms; they act as the first line of defense against bacteria, yeast, fungi, viruses, and cancer cells (Zhang &Gallo, 2016). These multifunctional peptides are the major component of the shrimp immune system represented by three cationic peptide families, namely penaeidins, crustins, and anti-lipopolysaccharide factors (ALFs) (Hou et al., 2014). ALFs are AMPs with broad-spectrum antimicrobial activity; they were isolated for the first time from horseshoe crab haemocytes (Tanaka et al., 1982) and various ALF isoforms have subsequently been identified in many shrimp and other crustaceans(Liu et al., 2014). Crustacean ALFs exhibit activities against bacteria(Beale et al., 2008; Somboonwiwat et al., 2005) and viruses (Li et al.,2015; Ponprateep et al., 2012; Tharntada et al., 2009). Penaeidins are AMPs that were discovered in shrimp; they are synthesized and stored in haemocytes to be released upon microbial challenge (Destoumieux et al., 2000), and they exhibit antimicrobial activities against bacteria,fungi (Destoumieux et al., 1997, 1999), and viruses (Woramongkolchai et al., 2011; Xiao et al., 2020).
In general, gene mutations can lead to changes in protein structure and expression (Lodish et al., 2000); thus, mutations in genes encodingL. vannameipenaeidins and ALFs may result in increased susceptibility or resistance to microbial infections. SNPs inALFgenes are reportedly associated with resistance to white spot syndrome virus (WSSV) and VPAHPNDinfections inL. vannamei(Liu et al., 2014; Zhang et al., 2019) as well as toVibrio alginolyticus-resistance/susceptibility in the swimming crab,Portunus trituberculatus(Li et al., 2013). In addition, variousL. vannameipenaeidins (BigPEN,PEN2,PEN3, andPEN4) were found to restrict WSSV infection, whereas the downregulation of penaeidins rendered shrimp more susceptible to WSSV (Xiao et al., 2020). However,SNPs in genes that encode penaeidins, as well as the association of these SNPs with the resistance/susceptibility ofL. vannameito viral and/or bacterial infections, have yet to be reported. In the present study, we identified SNPs in two genes,nLvALF1andPEN2-1, and investigated whether these SNPs were associated with resistance toV. parahaemolyticus(AHPND strain) infection inL. vannamei.
2.Materials and methods
2.1.Experimental shrimp
TheL. vannameilarvae used in this study were obtained from Sailom Farm, a shrimp hatchery in Kuiburi District, Prachuap Khiri Khan Province. Postlarvae 12 were collected from several nursery tanks to ensure that they were from different pairs of broodstock. Shrimp larvae were transferred to the wet lab and reared to allow acclimatization to laboratory conditions for 1 week prior to the challenge tests: they were kept in artificial seawater (salinity 20) and fed four times a day with an artificial diet (35% protein). Feces and uneaten food were siphoned out and ~30%-50% of rearing water was exchanged daily.
2.2.Preparation of V. parahaemolyticus
The pure culture stock of VPAHPNDused in this study was isolated from the hepatopancreas of diseased Pacific white shrimp. Bacterial culture was streaked onto thiosulfate citrate bile salts (TCBS) agar plates and incubated at 37°C for 24 h. Single green colonies were then subcultured in tryptic soy broth (TSB) +2% NaCl and incubated at 37°C for 24 h. Subsequent cultures were performed in 15-mL conical tubes followed by 1-L fasks with TSB +2% NaCl medium; a large volume of bacteria culture was obtained for the challenge tests by incubation overnight. After incubation, bacterial density was determined using a spectrophotometer at an optical density (OD) of 600 nm. The culture suspension was then serially diluted, and spread onto TCBS agar plates to determine CFU/mL. Bacterial cells in the TSB culture medium were collected by centrifugation (4000 rpm for 30 min at 4°C), washed once with 1% NaCl, and resuspended in 1% NaCl. The OD of the bacterial suspension was measured using a spectrophotometer at 600 nm, and the density was adjusted to 109CFU/mL using the CFU/mL concentration predetermined by plate counts and used in the challenge test.
2.3.Con firmation of VPAHPND identification
The VPAHPNDused for the challenge test was con firmed by multiplex PCR and real-time PCR assays. For multiplex PCR, bacterial cultures were analyzed for the presence of the AHPND molecular markers using two primer pairs (TUMSAT-Vp3F 5′-GTGTTGCATAAT TTTGTGCA-3′and TUMSAT-Vp3R 5′-TTGTAGAGAAACCACGACTA-3’; TUMSAT-Vp1F 5′-CGCAGATTTGC TTTTGTGAA-3′and TUMSAT-Vp1R 5′-AGAAGCTGGCCGAAGTGATA-3′); a pair of primers for the 16S rRNA gene (Vp-faE-79F 5′-GCAGCTGATCAAAACGTT GAGT-3′and Vp-faE-34R 5′-ATTATCGATCGTGCCACTCAC-3′) was used as the internal control (Tinwongger et al., 2014). Bacterial DNA was extracted by the boiling method (Koch et al., 2001). Briefy, single bacterial colonies from TCBS plates were suspended in distilled water and boiled at 95°C for 5 min. They were then centrifuged at 12,000 rpm for 5 min and the clear supernatant was used as a DNA template. PCR was performed in a 20 μL reaction comprising 10 μL of AccuStart II GelTrack PCR Supermix(QuantaBio, USA), 0.2 μM of each primer, 25-50 ng of DNA template,and ddH2O up to 20 μL. PCR reactions were run on the ProFlex 3 ×32-Well PCR System (Applied Biosystems, USA). Cycling conditions were as follows: 94°C for 3 min; 30 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 15 s; followed by a final extension step at 72°C for 3 min.The PCR products were electrophoresed on a 1.5% agarose gel, stained by Invitrogen SYBR Safe DNA Gel Stain, and visualized under an Azure c200 Gel documentation system (Azure Biosystems, USA).
The identification of VPAHPNDwas also verified molecularly by realtime PCR with the primers VpPirA-F 5′-TTGGACTGTCGAACCAAACG-3′,VpPirA-R 5′-GCACCCC ATTGGTATTGAA TG-3′, and VpPirA TaqMan probe 5′-FAM-AGA-CAG-CAA-ACA-TAC-ACC-TAT-CAT-CCC-GGABHQ1-3’ (Han et al., 2015). The reaction mixture constituted 10 μL of PerfeCTa Fastmix II (QuantaBio, USA), 0.3 μM of each primer, 0.1 μM of TaqMan probe, and ddH2O to a final volume of 20 μL. Real-time PCR was performed on a QuanStudioTM5 real-time PCR system (Applied Biosystems, USA). The cycling protocol was 95°C for 3 min before 45 cycles of 95°C for 10 s and 60°C for 15 s. Amplification results for VPAHPNDwere compared with those from the positive control.
2.4.Determination of median lethal dose
The median lethal dose (LD50) of VPAHPNDinfection was determined by a challenge test of postlarvae 20 (PL20). Groups of 10 PL20 shrimp were each stocked in one of 15 plastic containers filled with 5 L of artificial seawater (salinity 20) with proper aeration. The next day,VPAHPND(109CFU/mL) suspension in 1% NaCl was added to the containers at concentrations of 104, 105, 106, and 107CFU/mL, whereas 1% NaCl was added to the control group. Each concentration was tested in triplicate. Shrimp were fed an artificial diet three times per day and 20% of the water was exchanged daily from day 2 to day 10. Shrimp mortality was observed and recorded for 10 days. Cumulative mortality was used to calculate the LD50 according to the method of Reed and Muench(1938).
2.5.VPAHPND challenge test
For the challenge test, the PL20 shrimp, which were acclimated to wet lab conditions, were allocated into three plastic containers filled with 20 L of artificial seawater and aerated by an air-pump at a density of 5 larvae/L (i.e., 100 larvae per container). VPAHPNDsuspension (109CFU/mL) in 1% NaCl was added to two containers (challenge groups) at 107CFU/mL (the predetermined LD50 was 106.74CFU/mL) and the remaining container was used as the control group. Shrimp were fed an artificial diet three times per day, their waste matter was removed, and~30% of rearing water was exchanged daily from the second day onward. Shrimp survival was observed for 14 days. Dead and moribund shrimp were collected daily and preserved in 95% ethanol, whereas shrimp that survived the challenge were collected at day 14 and preserved in 95% ethanol prior to DNA extraction.
2.6.DNA extraction and PCR amplification
Samples of 45 resistant (survived the challenge) and 45 susceptible(died within 6 days) individuals were used for DNA extraction. DNA was extracted from the muscle tissue using a QIAGEN spin column (QIAGEN DNeasy Blood & Tissue Kit), and the purity and concentration of genomic DNA samples was estimated with an Eppendorf Bio-Spectrometer basic spectrophotometer (Eppendorf, Germany).
PCR amplifications were performed in a 25-μL reaction mixture comprising 50 ng of template DNA, 10 × PCR buffer, 0.5 μM of each primer, 0.2-mM (each) dNTPs, and 0.625 units ofTaqDNA Polymerase(New England Biolabs). The primers used for thenLvALF1gene were those previously published (Liu et al., 2014) and the specific primers for thePEN2-1gene were designed with NCBI’s Primer-BLAST based on thePEN2-1gene sequence (DQ206401). All primer sequences are listed in Table 1. Thermal cycling conditions were as follows: initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 95°C for 15 s, annealing at a specific temperature for each gene for 20 s, and extension at 68°C for 30 s; and a final extension at 68°C for 7 min. The PCR products were electrophoresed on 2% agarose gel, stained with ethidium bromide, and the gel was visualized and photographed with a gel documentation system (Syngene, UK). The PCR products of resistant and susceptible samples were then sent to Bio Basic (Canada) for purification and sequencing.

Table 1Information on the primers used in this study.
2.7.SNP identification and analysis of the association between nLvALF1/PEN2-1 SNPs and V. parahaemolyticus resistance
SNPs innLvALF1andPEN2-1genes and the genotypes of SNP loci of resistant and susceptible individuals were identified from sequencing chromatograms using Sequencher software version 5.4.6 (Gene Codes Corporation, USA). Statistical analysis for significance tests of allele frequencies and genotype frequencies was performed with Chi-square tests of independence in R statistical software (R Core Team, 2020).Associations were considered significant whenP <0.05.
3.Results
3.1.Con firmation of VPAHPND
A multiplex PCR assay using the primer set TUMSAT-Vp3, TUMSATVp1, and Vp-faE con firmed that theV. parahaemolyticusisolate used in the challenge test was the AHPND strain. The PCR product showed three bands (Fig. A1): TUMSAT-Vp3 amplified thepirAvptoxin gene (360 bp)found in the VPAHPNDstrain; TUMSAT-Vp1 (500 bp) was positive for VPAHPND, but in a rare case with a non-AHPND strain (Tinwongger et al.,2014); Vp-faE amplified an endogenous normalizer, theflaEgene, that was positive in both AHPND and non-AHPNDV. parahaemolyticus.Real-time PCR results con firmed that theV. parahaemolyticusisolate possessed thepirAvptoxin gene (see the real-time amplification plot in Fig. A2).
3.2.Shrimp mortality
The mortality rate of shrimp challenged with 107CFU/mL VPAHPNDduring the 14-day trial was 37% (Fig. 1). Mortality occurred from day 2 to day 9 and the highest mortality (7.5% per day) was observed on days 5 and 6. Mortality was not observed from day 10 onward. Cumulative death was 74 individuals; shrimp that died within 6 days (49 individuals) were assigned to the susceptible group and used for SNP analysis. Mortality was not observed in the control group.
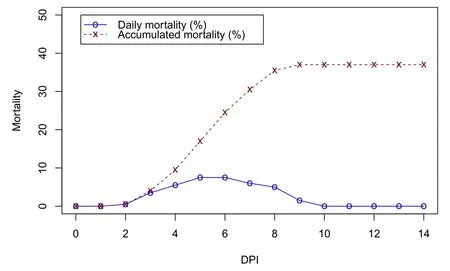
Fig. 1.Mortality rate (%) of experimental shrimp during the 14-day challenge trial with Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain. DPI:day post infection.
3.3.SNP identification
3.3.1.nLvALF1 gene
PCR amplification of shrimp DNA samples using a primer pair specific to theALFconserved domain ofnLvALF1(CX-ALF1) (Liu et al.,2014) yielded amplified products of approximately 400-bp in length(Fig. A3). Sequencing results revealed the exact fragment to be 397-bp long, which is consistent with the study of Liu et al. (2014). Based on the sequencing chromatograms of 12 random samples from the study population, the 397-bpnLvALF1gene fragment contained nine SNPs, i.e., 189G/A, 226A/T, 230T/G, 236A/G, 257C/T, 271T/A, 275G/A,284G/A, and 297G/A (Fig. 2). The minor allele frequencies at SNP loci 189G/A, 226A/T, 230T/G, 236A/G, 257C/T, 271T/A, 275G/A,284G/A, and 297G/A were 16.67%, 8.33%, 16.67%, 8.33%, 25%,18.18, 25%, 16.67%, and 9.09%, respectively.3.3.2.PEN2-1 gene

Fig. 2.The SNP loci (189G/A, 226A/T, 230T/G, 236A/G, 257C/T, 271T/A, 275G/A, 284G/A, and 297G/A) identified in a 397-bp fragment of the nLvALF1 gene.Primer sequences are underlined and SNP positions are indicated in red boldface. The SNP locus numbers are assigned according to the nucleotide numbers from the forward primer CX-ALF1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The PCR reaction amplified using PEN2-1F and PEN2-1R primers yielded an amplicon of the expected size (~600 bp) on agarose gel electrophoresis (Fig. A.4). The precise length of the target fragment from the DNA sequence data was 637 bp, and nucleotide BLAST showed 100% identity with theL. vannamei PEN2-1gene (g.473-1109; accession no. DQ206401.1). SNP analysis from sequencing chromatograms of random samples from the study population (n =21) revealed seven polymorphic loci including g.582A/G, g.657G/T, g.664T/G, g.708C/T,g.798G/A, g.869G/T, and g.1013C/T (Fig. 3), with all SNPs located in the intronic region. The minor allele frequencies at each SNP locus were 21.43%, 21.43%, 30.95%, 47.62%, 11.90%, 14.29%, and 21.43%,respectively.

Fig. 3.SNPs identified in a 637-bp fragment of the PEN2-1 gene. Coding regions are highlighted in gray, primer binding sites are underlined and SNP positions are in red boldface. The locus numbers (g.582A/G, g.657G/T, g.664T/G, g.708C/T, g.798G/A, g.869G/T, and g.1013C/T) are assigned according to the nucleotide sequence of Litopenaeus vannamei PEN2-1 (GenBank accession no.: DQ206401.1). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4.SNP association analysis
3.4.1.nLvALF1 gene
Association analysis between genotypes and resistance/susceptibility toV. parahaemolyticusinfection at nine SNP loci (susceptible group,n=25; resistant group,n=18) showed that no significant difference (Chi-square test;P >0.05) existed between the genotype distributions of the susceptible group and resistant group (Table 2).
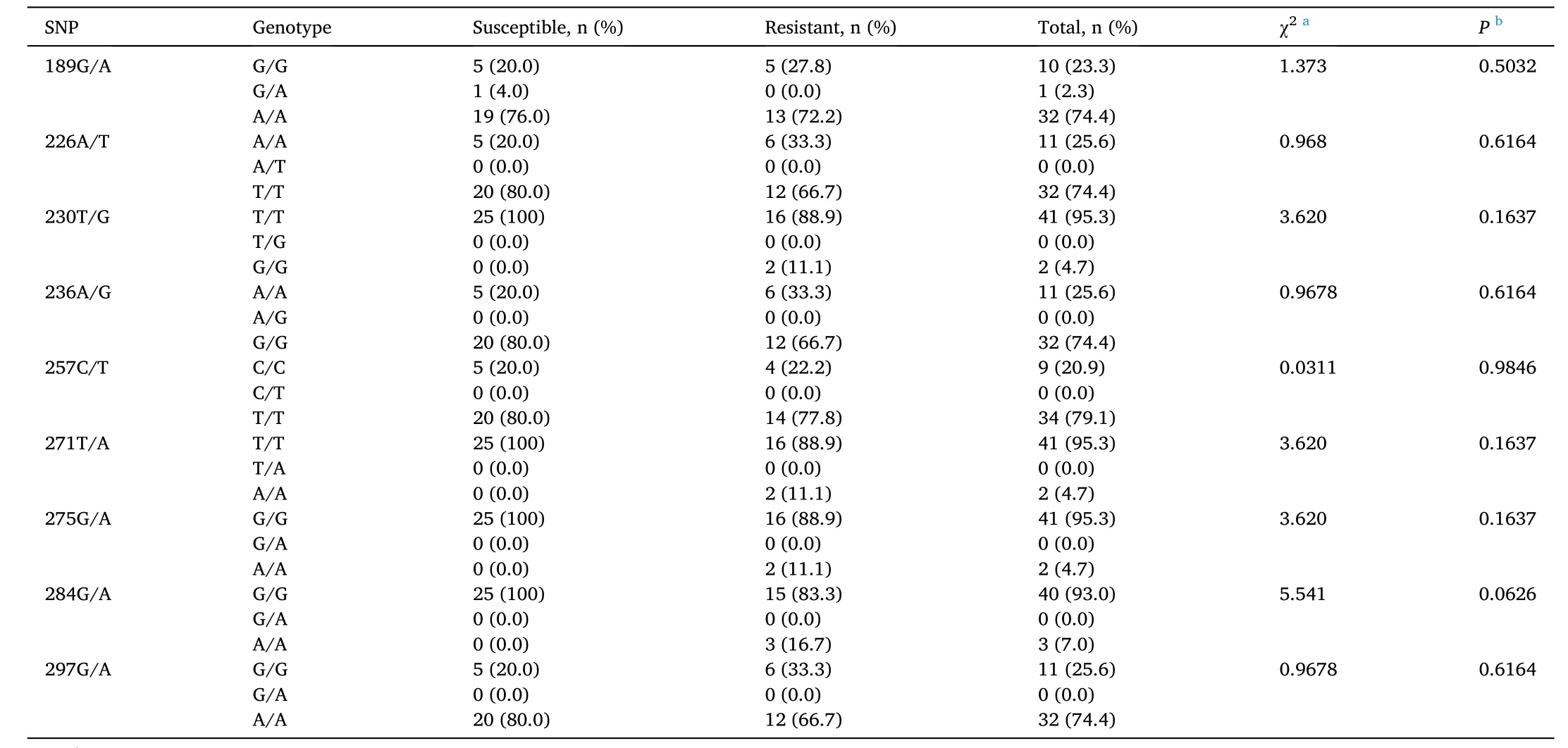
Table 2Genotype distributions of shrimp susceptible and resistant to Vibrio parahaemolyticus at different SNP loci in the nLvALF1 gene.
The allele frequencies in the susceptible and resistant groups at nine SNP loci are listed in Table 3. Association analysis using maximum likelihood ratio Chi-square tests indicated that significant differences existed between the allele frequencies of susceptible and resistant shrimps at SNP loci 230T/G, 271T/A, 275G/A, and 284G/A. Based on sequence data, the haplotype G-A-A-A at SNPs 230T/G, 271T/A, 275G/A, and 284G/A was found only in the resistant shrimp.
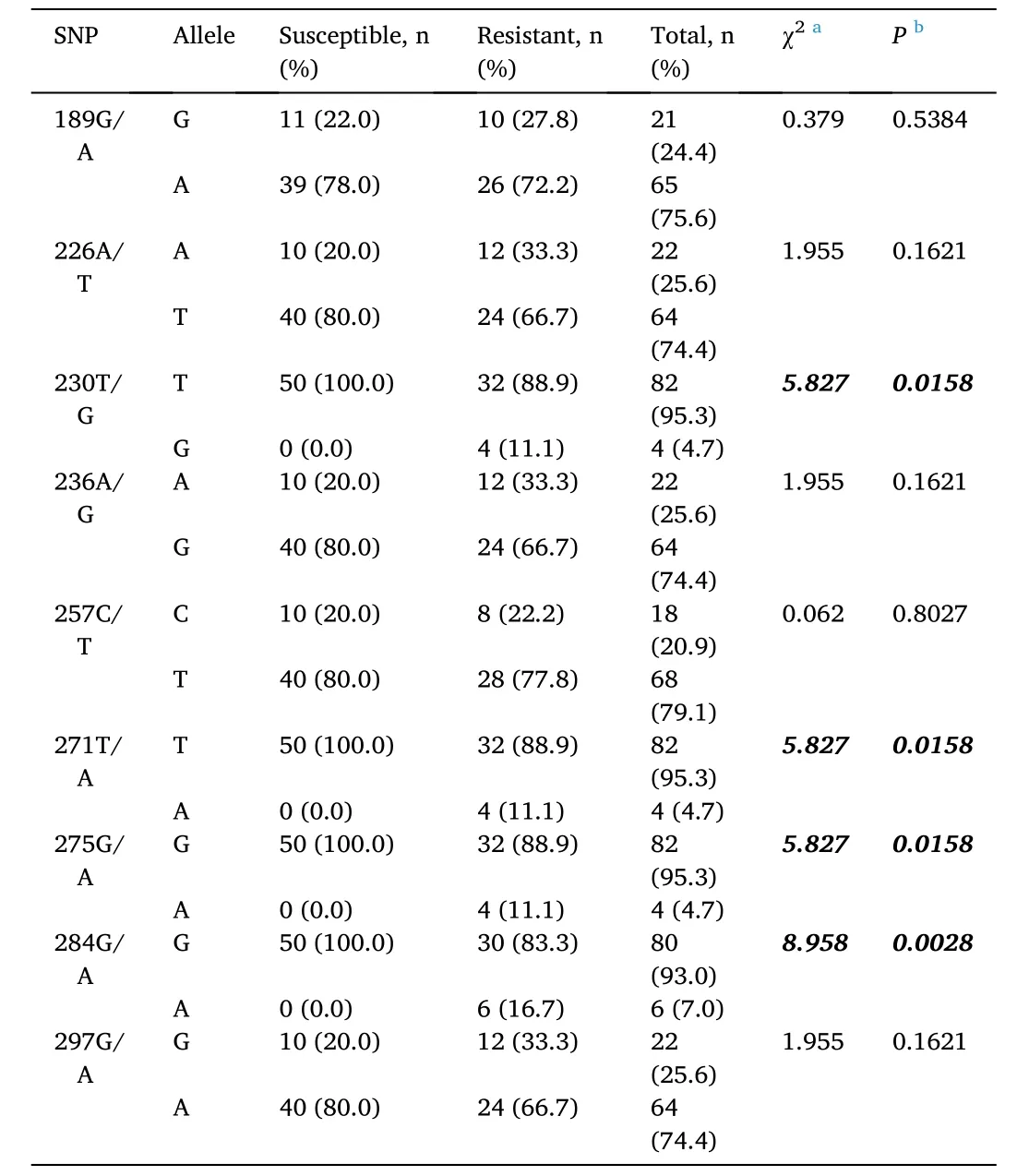
Table 3SNP allele frequencies at different loci of shrimp susceptible and resistant to Vibrio parahaemolyticus.
3.4.2.PEN2-1 gene
The genotype distributions between susceptible shrimp (n =36) and resistant shrimp (n =39) at each of the SNP loci were tested using Chisquare tests; significant associations (P <0.05) between genotype and resistance/susceptibility toV. parahaemolyticuswere found at SNP loci g.657G/T, g.664T/G, and g.708C/T. Cramer’s V statistics for each locus were 0.291, 0.311, and 0.315, respectively, indicating moderate level relationships between genotypes and resistance/susceptibility. Signi ficant associations were not observed at other SNP loci. Genotype frequencies, Chi-square statistics, andPvalues for each SNP locus are shown in Table 4.
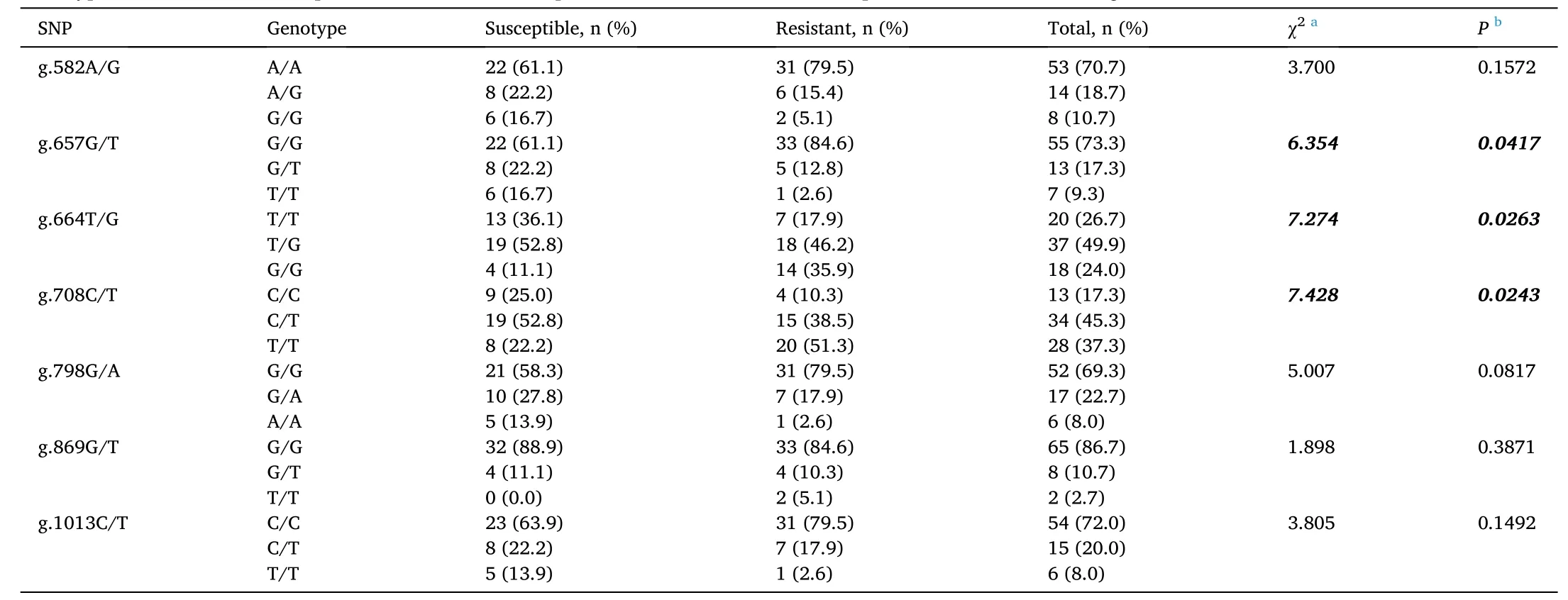
Table 4Genotype distributions of susceptible and resistant shrimp at different SNP loci of the Litopenaeus vannamei PEN2-1 gene.
Association analysis of the allele frequency at each SNP locus in theL.vannameiPEN2-1gene and resistance/susceptibility toV. parahaemolyticusinfection showed that significant associations (P <0.05) existed at six SNP loci, i.e., g.582A/G, g.657G/T, g.664T/G,g.708C/T, g.798G/A, and g.1013C/T (Table 5). In the resistant group,the frequencies of A (g.582A/G), G (g.657G/T), G (g664 T/G), T(g.708C/T), G (g.798G/A), and C (g.1013C/T) alleles were higher than the expected values, whereas in the susceptible group, the frequencies of G (g.582A/G), T (g.657G/T), T (g.664T/G), C (g.708C/T), A (g.798G/A), and T (g.1013C/T) alleles were higher than the expected values.Based on DNA sequences, the haplotype A-G-G-C (g.582A/G - g.657G/T- g.798G/A - g.1013C/T) was found in the resistant group, whereas the haplotype G-T-A-T (g.582A/G - g.657G/T - g.798G/A - g.1013C/T) was found in the susceptible group.
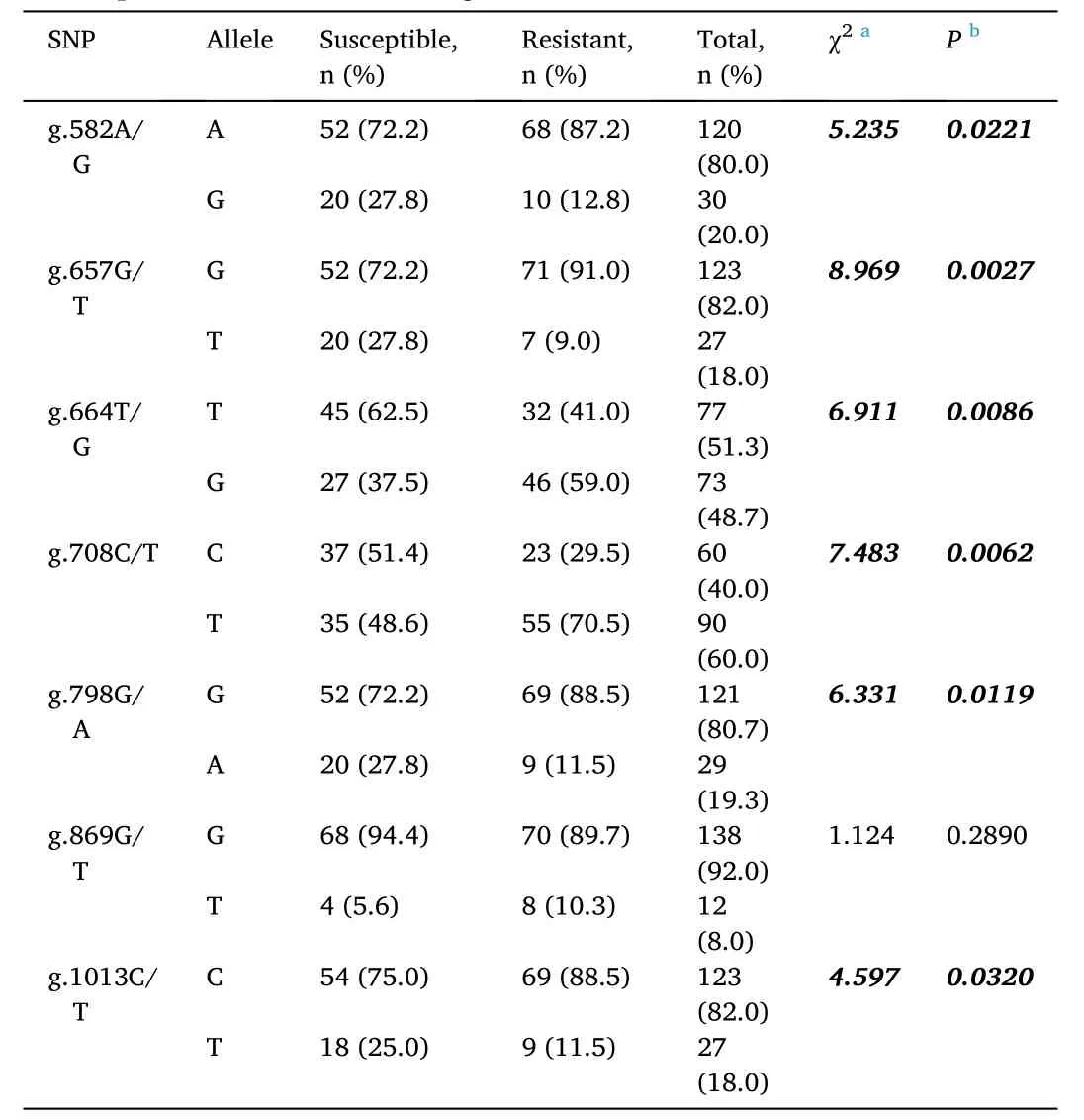
Table 5SNP allele frequencies of susceptible and resistant shrimp at different SNP loci of the Litopenaeus vannamei PEN2-1 gene.
4.Discussion
The effectiveness of disease prevention and control is one of the most important components of successful shrimp farming; thus, revealing the genetic basis of disease resistance is crucial to the aquaculture production of Pacific white shrimp. In the current study, we identified SNPs and investigated their associations with the resistance toV. parahaemolyticusAHPND strain with the aim of revealing potential genetic markers for the breeding disease-resistantL. vannamei.
We detected nine and seven SNPs from the sequence chromatograms of 397-bp and 637-bp fragments ofnLvALF1andPEN2-1genes,respectively. The minor allele frequencies were 8.33%-25% innLvALF1and 11.9%-47.62% inPEN2-1. The published primer pair for thenLvALF1gene amplified a fragment from the Intron II/Exon II region containing a conservedALFdomain (Liu et al., 2014); however, we were unable to identify the type of SNPs (intronic/exonic) innLvALF1due to limited sequence information. In thePEN2-1gene, all SNPs were in the intronic region. Although intronic SNPs have unknown functional significance, those that occur within intron branchpoint sites, especially at adenine (A), presumably affect splicing (Chiang et al., 2017). Alternative splicing is considered one of the major sources of functional diversity in proteins from multicellular organisms; alternative splicing events may affect the conserved regions of a protein structure, resulting in an isoform with unique functional features (Birzele et al., 2008). Thus,SNPs or point mutations that occur in the intronic regions of AMP-encoding genes could affect the structure and function of synthesized antimicrobial peptides, and consequently alter the resistance of shrimps to pathogens. Several studies have reported the relationship between intronic SNPs and disease resistance; for example, intronic SNPs in theLECT2gene are associated with resistance toV. harveyiinfection in the Asian seabass,Lates calcarifer(Fu, Bai, et al., 2014); an intronic SNP in theMCP-8gene is linked to resistance toStreptococcus agalactiaein tilapia (Fu et al., 2014); and intronic SNPs in theGAB3gene are related to resistance to viral nervous necrosis in the Asian seabass(Yang et al., 2020).
ShrimpALFandPENare immune-related genes in the AMP families;therefore, SNPs innLvALF1andPEN2-1genes are useful for genetic association studies. We conducted a challenge test in a hatchery population ofL. vannameiusing an isolate of VPAHPND. The alleles and genotypes at different SNP loci of the susceptible and resistant groups were determined from sequence chromatograms and then used in association analysis. Association analysis of nine SNP loci in thenLvALF1gene revealed no difference in genotype distributions between the susceptible and resistant groups, whereas allele frequencies at four SNP loci(230T/G, 271T/A, 275G/A, and 284G/A) did differ significantly between the two groups. Similarly, Liu et al. (2014) analyzed the association betweennLvALF1gene SNPs and WSSV resistance in two independentL. vannameipopulations, and they found no difference in genotype frequencies between WSSV-susceptible and -resistant groups,but found that allele distributions between the two groups differed significantly. The genotypes GG, AA, AA, and AA at the SNP loci 230T/G, 271T/A, 275G/A, and 284G/A were only found in the resistant group; the alleles at these SNP loci, of which the frequencies between the two groups differed significantly, were in the same haplotype (G-A-A-A).This result suggests that shrimp possessing these genotypes, alleles, and haplotype are likely to be resistant toV. parahaemolyticusinfection.
Penaeidin is an AMP found only in penaeids and plays a significant role in protection against bacteria and fungi (Destoumieux et al., 1997,1999) as well as viruses (Woramongkolchai et al., 2011; Xiao et al.,2020). In the present study, we found an association between SNPs in thePEN2-1gene and resistance/susceptibility toV. parahaemolyticusinfection. Genotype distributions of the resistant group and susceptible group at three SNP loci (g.657G/T, g.664T/G, and g.708C/T) were significantly different, whereas allele frequencies at six SNP loci(g.582A/G, g.657G/T, g.664T/G, g.708C/T, g.798G/A, and g.1013C/T)in the two groups were significantly different. The alleles A (g.582A/G),G (g.657G/T), G (g664 T/G), T (g.708C/T), G (g.798G/A), and C(g.1013C/T) are possibly associated with resistance toV. parahaemolyticusinfection. Moreover, at the SNP loci g.582A/G,g.657G/T, g.798G/A, and g.1013C/T, the haplotype A-G-G-C was more common in the resistant group, whereas the haplotype G-T-A-T was more common in the susceptible group. As mentioned earlier,PEN2-1SNPs were detected in the intron region so will not affect amino acid change. Thus, the observed significant associations might be due to the linkage of these SNPs and exonic SNPs inPEN2-1or other closely located genes that control resistance to VPAHPNDinfection. In addition, intronic SNPs may affect splicing in mRNA synthesis (Chiang et al., 2017) and thereby result in disease resistance/susceptibility. Splicing could affect disease as a direct cause, as a modifier of severity, and as a determinant of disease susceptibility (Wang & Cooper, 2007).
Identifying the genetic polymorphisms in candidate genes involved in the immune response to microbial infection is vital to improving the genetics of aquaculture stock. The SNP polymorphisms identified here,and their association with resistance toV. parahaemolyticusinfection inL. vannamei, provide evidence of genetic resistance at the DNA level in this aquaculture species. SNPs innLvALF1andPEN2-1genes related toV. parahaemolyticusresistance could be potential DNA markers for improving the disease resistance traits of Pacific white shrimp. However only one population was used in the current study; therefore, more research is necessary to verify these results in a larger sample size and in different populations. In addition, asALFandPENare genes that encode AMPs, i.e., key components of the innate immune system and the only protective mechanism of crustaceans (Wang, 2014), it will be worth studying the association between the SNPs innLvALF1andPEN2-1genes and resistance to other pathogens that cause disease in these shrimp. If the results obtained are similar to those reported in our study, then these SNPs could be used as universal markers for breeding a disease-resistantL. vannameiline.
5.Conclusions
In this study, we identified SNPs in thenLvALF1andPEN2-1genes ofL. vannamei, and we analyzed their associations with resistance/susceptibility toV. parahaemolyticusinfection. Nine SNPs in annLvALF1-gene fragment (397 bp) and seven SNPs in aPEN2-1-gene fragment (637 bp) were detected. Four SNPs (230T/G, 271T/A, 275G/A, and 284G/A)innLvALF1and six SNPs (g.582A/G, g.657G/T, g.664T/G, g.708C/T,g.798G/A, and g.1013C/T) inPEN2-1are associated with the resistance/susceptibility toV. parahaemolyticusinfection, and thus might be potential genetic markers for breedingV. parahaemolyticus-resistantL. vannamei.
CRediT authorship contribution statement
Pawapol Kongchum: Conceptualization, Methodology, Investigation, Formal analysis, Writing - original draft, Funding acquisition.Suphavadee Chimtong: Methodology, Investigation. Naparat Prapaiwong: Methodology, Investigation.
Declaration of competing interest
All authors have no af filiations with or involvement in any organization or entity with any financial interest, or non- financial interest in the subject matter or materials discussed in this manuscript.
Acknowledgment
Funding for this research was provided by the National Science and Technology Development Agency, Ministry of Higher Education, Science, Research and Innovation. The authors wish to thank Saichol Preanjit (Sailom Farm) for providing shrimp postlarvae, Khomkrit Leamnak and Yodchat Katthamrong for their assistance during the experiment.The authors would also like to thank the editor and reviewers for the valuable suggestions made to improve this manuscript.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2021.08.003.
 Aquaculture and Fisheries2022年4期
Aquaculture and Fisheries2022年4期
- Aquaculture and Fisheries的其它文章
- A review on fishing gear in China: Selectivity and application
- atg7 and beclin1 are essential for energy metabolism and survival during the larval-to-juvenile transition stage of zebra fish
- Comparative analysis of the morphology, karyotypes and biochemical composition of muscle in Siniperca chuatsi, Siniperca scherzeri and the F1 hybrid (S. chuatsi ♀ × S. scherzeri ♂)
- Effects of salinity and alkalinity on fatty acids, free amino acids and related substance anabolic metabolism of Nile tilapia
- Differences in postembryonic dorsal fin development resulted in phenotypic divergence in two gold fish strains, Red Cap Oranda and Ranchu
- Chemical characteristics changes associated with extracted protein and salt addition and effect of different additives in the curing process on sensory acceptance of liquid fermented fish from tilapia frame
