A novel peptide-based probe 99mTc-PEG6-RD-PDP2 for the molecular imaging of tumor PD-L2 expression
Qi Luo, Yunwei Zhang, Zihua Wang, Yining Sun, Linqing Shi, Yue Yu,Jiyun Shi,*, Zhiyuan Hu, Fan Wanga,,,*
a Guangzhou Laboratory, Guangzhou 510005, China
b Medical Isotopes Research Center and Department of Radiation Medicine, State Key Laboratory of Natural and Biomimetic Drugs, School of Basic Medical Sciences, Peking University, Beijing 100191, China
c Fujian Provincial Key Laboratory of Brain Aging and Neurodegenerative Diseases, School of Basic Medical Sciences, Fujian Medical University, Fuzhou 350122, China
d Key Laboratory of Protein and Peptide Pharmaceuticals, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of
Sciences, Beijing 100101, China
e CAS Key Laboratory of Standardization and Measurement for Nanotechnology, CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety,CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology of China, Beijing 100190, China
ABSTRACT Tumor-related PD-L2 expression is associated with the clinical efficacy of PD-1/PD-L1 blockade therapy.PD-L2-specific imaging can help selecting patients for appropriate immunotherapy.In this study, a PD-L2-targeting peptide (PDP2) was screened by the one-bead one-compound combinatorial library approach.Using the retro-inverso D-peptide of PDP2 (RD-PDP2) and PEGylation strategies, we developed a novel Tc-99m-labeled PD-L2-targeting peptide as a SPECT tracer (99mTc-PEG6-RD-PDP2) for imaging of tumor PD-L2 expression.The radiolabeling yield of 99mTc-PEG6-RD-PDP2 was greater than 95% by the standard HYNIC/tricine/TPPTS labeling procedure.99mTc-PEG6-RD-PDP2 displayed high PD-L2-binding specificity both in vitro and in vivo.SPECT/CT imaging with 99mTc-PEG6-RD-PDP2 showed that the A549-PD-L2 tumors were clearly visualized, whereas the signals in PD-L2-negative A549 tumors were much lower.In vivo blocking study suggested that the tumor uptake of 99mTc-PEG6-RD-PDP2 was PD-L2 specifically mediated.99mTc-PEG6-RD-PDP2 is a promising SPECT probe for the non-invasive imaging of tumor PD-L2 expression and has a great potential in guiding the anti-PD-1 or anti-PD-L1 immunotherapy of cancer.
Keywords:PD-L2 Peptide probe SPECT imaging Immunotherapy
Immune checkpoint blockade (ICB) therapy, including programmed cell death protein-1 (PD-1)/programmed cell deathligand 1 (PD-L1) blockade, has been approved to treat various cancers, leading to a paradigm shift in cancer therapy [1,2].However,due to the limited clinical efficacy, prevalent immune-related adverse events, and expensive treatment costs, more effective therapies and predictive biomarkers are needed to distinguish responders from non-responders [3,4].
Tumor-associated PD-L1 expression has been reported to be an adopted predictive biomarker for responding to anti-PD-1/PDL1 immunotherapy [5,6].In some clinical trials, cancer patients were treated with monoclonal anti-PD-1 or anti-PD-L1 antibodies (KeytrudaR○, OPDIVOR○, or TECENTRIQR○), and the high PD-L1 expression was related to the increased objective response rate and clinical benefit [7–9].Nonetheless, not all PD-L1-positive patients showed clinical responses to such therapies, and some PDL1-negative patients also had responses [10,11].In another clinical trial, no significant difference in objective response rate was observed between the PD-L1-positive subgroup and PD-L1-negative subgroup [12].These researches suggested that other molecular interactions with PD-1 should also be considered.
Programmed cell death ligand-2 (PD-L2), another ligand of PD-1, is also detected in various tumors and inversely correlated with the overall survival of patients (Fig.S1 in Supporting information) [13–17].PD-L2 is expressed in immune, stromal, and tumor cells, and has a higher affinity to PD-1 compared with PD-L1 (sixto ten-fold) [18].Similar to PD-L1, the interaction of PD-L2 and PD-1 inhibits T cell proliferation, cytokine production and T cell cytolysis function, which also regulate T cell-mediated immune response and play a role in tumor immune escape [19–21].Yearleyet al.’s study suggested that PD-L2 expression might provide the information in predicting clinical response to anti-PD-1 therapy [22].Besides, when PD-L2-positive patients were treated with anti-PD-L1 therapy, the interaction of PD-L2 and PD-1 made the blockade of PD-L1 ineffective [21,22], thus, PD-L2-positive patients should choose the anti-PD-1 treatment instead of anti-PD-L1 therapy.The detection of PD-L2 is significant for the precision treatment of cancer.
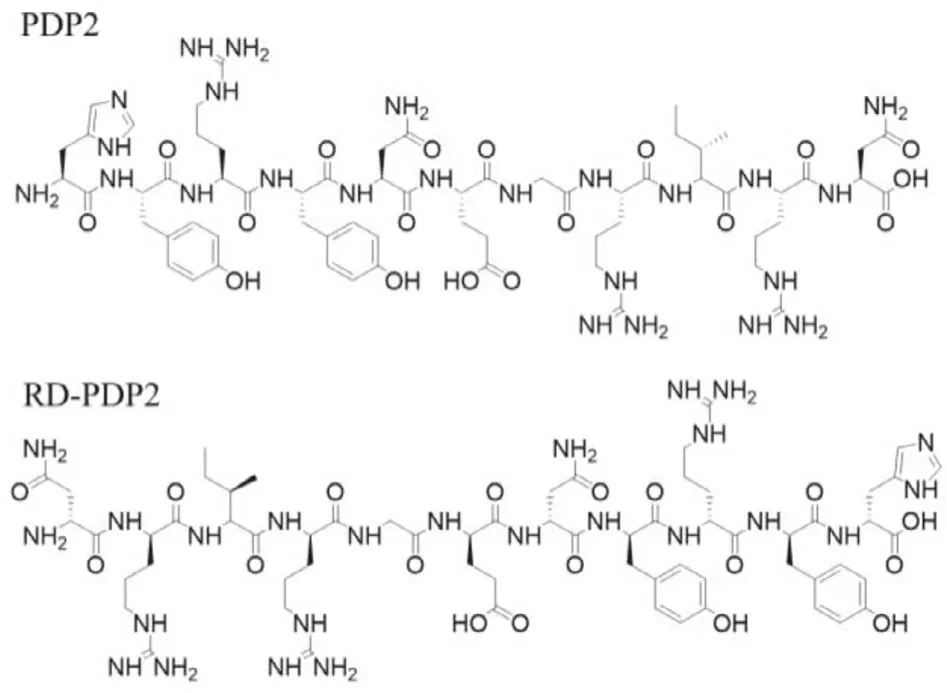
Fig.1.Chemical structures of HYRYNEGRIRN (PDP2) and D(NRIRGENYRYH) (RDPDP2).
Immunohistochemistry (IHC) is currently the commonly used method to determine the PD-L2 expression level of tumor tissues[21,22].However, similar to PD-L1, PD-L2 expression has also intratumoral heterogeneity and dynamic alteration along with cancer progression and treatment [22,23].Therefore, IHC has the limitation to detect the PD-L2 expression in clinical practice.In addition, since there are no strict criteria to define the PD-L2 level by IHC, the actual status of PD-L2 expression would be misinterpreted sometimes.Compared with IHC, the non-invasive specific nuclear imaging can provide the real-time, dynamic, and quantitative information of biomarkersin vivo[24].Until now, there is no report on PD-L2 targeting imaging probes.In this study, we screened a novel PD-L2-targeted peptide PDP2 by the one-bead one-compound (OBOC) library approach.Using the retro-inverso D-peptide of PDP2 (RD-PDP2) and PEGylation, we developed a Tc-99m-labeled SPECT probes (99mTc-PEG6-RD-PDP2) for the detection of PD-L2 expressionin vivo.The purpose of this study is to provide a non-invasive method for physicians to make an optimum selection of anti-PD-1 or anti-PD-L1 therapy.In addition,99mTc-PEG6-RD-PDP2 SPECT/CT imaging would be used to guiding the PD-L2-targeted therapy in the future.
Firstly, a peptide library with random amino acid residues at specific positions was constructed to create an 11-mer OBOC combinatorial library [25].Peptide beads with a high affinity toward PD-L2 could be covered with biotinylated human PD-L2 protein and recognized by streptavidin-coated magnetic beads in an affinity-dependent manner (Fig.S2 in Supporting information).A microarray chip was fabricated forin situtrapping and characterization of the positive peptides [26].After the Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDITOF-MS) spectra were characterized, we obtained 23 positive sequences and aligned them (Fig.S3 in Supporting information).The sequence PDP2 (sequence: HYRYNEGRIRN) was revealed to hit the highest probability analyzed by the software WebLogo.
The structure of peptide PDP2 is shown in Fig.1.To detect the affinity of peptide PDP2 toward human PD-L2 protein, we performed surface plasmon resonance (SPR) analysis on a Plexera PlexArray HT system (Plexera, USA).The dissociation constant (KD)value between the PDP2 and human PD-L2 protein was calculated as 57.1 nmol/L (Fig.S4A in Supporting information), which performed a good peptide-protein recognition behavior.We chose human serum albumin (HSA), the most abundant protein in plasma,to test the nonspecific absorption.The SPR curves portrayed as the nonbinding “square wave” (Fig.S4B in Supporting information),suggesting peptide PDP2 had no specific binding toward HSA.
PDP2 is a linear L-type peptide, which is sensitive to proteolysisin vivo.The radiotracer developed based on linear peptide often shows poor metabolic stability, resulting in insufficient accumulation and retention in tumors, which is an obstacle to its application[27].The presence of D-amino acid residues in peptide could improve their metabolic stability to proteolysis and alter the biological half-life [28].The retro-inverso D-peptide modification involves both inversion of amino acid stereochemistry and reversal of peptide sequence, which aims to create peptides that are stable toward proteolysis and display the same binding properties compared to the original L-peptides [29].Therefore, we first replaced the peptide sequence of L-type PDP2 with D-type amino acids to improve itsin vivostability, and then reversed the sequence of D-peptide D-PDP2 (D-amino acid sequence: HYRYNEGRIRN).The reversed Dtype peptide RD-PDP2 (D-amino acid sequence: NRIRGENYRYH, Fig.1) has a similar spatial configuration to L-type peptide PDP2, so as to maintain the PD-L2-targeting ability equivalent to L-type peptide PDP2.
The PD-L2-specific binding ability of peptide RD-PDP2 was evaluated in human NSCLC cell lines A549 (PD-L2-negative) and A549-PD-L2 (PD-L2-positive).Flow cytometry results confirmed that FITC-labeled RD-PPDP2 peptide (FITC-RD-PDP2) showed a significant difference in the binding of A549-PD-L2 and A549 cells (Fig.2A), which was consistent with the PD-L2 expression of cells measured by anti-PD-L2 antibody (Fig.S5 in Supporting information).Fluorescence staining results also showed that FITC-RD-PDP2 accumulated significantly on the cell membrane of A549-PD-L2 cells,while A549 cells showed almost no bound fluorescence signals(Fig.2B).In addition, the PD-L2-positive human NSCLC cell line H226 also had a significantly bound fluorescence signal of FITC-RDPDP2 (Fig.S6 in Supporting information).These results indicated that the binding of FITC-RD-PDP2 toward tumor cells was specifically mediated by PD-L2.
The99mTc/HYNIC system was used for the radiolabeling of peptides to fulfill the SPECT imaging [30].First, we synthesize the radiolabeled precursors using a standard procedure [31].Chelator HYNIC conjugate of RD-PDP2 (HYNIC-RD-PDP2) was synthesized as the route in Fig.S7A (Supporting information).After purified and lyophilized, we found that HYNIC-RD-PDP2 was insoluble in water, which limited it radiolabeling in aqueous solution.To improve its water solubility, we introduced PEG chain between the peptide and chelator.HYNIC-conjugated RD-PDP2 and PDP2 with PEG6linker (HYNIC-PEG6-RD-PDP2, HYNIC-PEG6-PDP2) were synthesized as the route in Fig.S7B (Supporting information).HPLC results showed that the purity of conjugates was all>95%.MALDITOF-MS results confirmed the molecular weight of the HYNIC conjugates (Fig.S8 in Supporting information).
99mTc-labeled HYNIC-PEG6-RD-PDP2 and HYNIC-PEG6-PDP2 tracer [99mTc(tricine)(TPPTS)-HYNIC-PEG6-peptide] (shortened as99mTc-PEG6-RD-PDP2 and99mTc-PEG6-PDP2, Fig.3A) were prepared in high yield (>95%) using the non-SnCl2formulation (Fig.3B).The molar activity of radiotracer was>35 MBq/nmol.99mTc-PEG6-RD-PDP2 and99mTc-PEG6-PDP2 were both stable in 25 nmol/L phosphate buffer (pH 7.4) containing cysteine (1.0 mg/mL) at 37°C for 4 h (Fig.S9 in Supporting information).However,in vivostability evaluation demonstrated the retroinverse D-peptide had favorable stability.99mTc-PEG6-RD-PDP2 was excreted by urine as the original drug at 6 h post-injection (p.i.),while99mTc-PEG6-PDP2 was completely degradedin vivoat 0.5 h p.i.(Fig.3B).
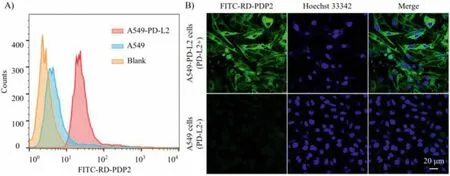
Fig.2.(A) Flow cytometry histograms of A549-PD-L2 (PD-L2-positive) and A549 cells (PD-L2-negative) with FITC-RD-PDP2.(B) Fluorescent staining of A549-PD-L2 and A549 cells with FITC-RD-PDP2 (green).
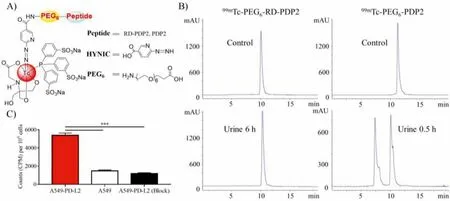
Fig.3.(A) Structure of 99mTc-radiolabeled complexes [99mTc(tricine)(TPPTS)-HYNIC-PEG6-peptide] (shortened as 99mTc-PEG6-RD-PDP2 and 99mTc-PEG6-PDP2).(B) Radio-HPLC chromatograms of 99mTc-PEG6-RD-PDP2 and 99mTc-PEG6-PDP2 in control and urine sample, respectively.(C) Binding of 99mTc-PEG6-RD-PDP2 to A549-PD-L2 (without or with unlabeled RD-PDP2 peptide blocking) and A549 cells (n=4; ***P <0.001).
The cell-binding specificity of99mTc-PEG6-RD-PDP2 was evaluated in both PD-L2-positive A549-PD-L2 and PD-L2-negative A549 cells (Fig.3C).The binding of99mTc-PEG6-RD-PDP2 to A549-PD-L2 cells was significantly higher than that to A549 cells (P <0.001).Excessive unlabeled RD-PDP2 can significantly inhibit the binding of99mTc-PEG6-RD-PDP2 to A549-PD-L2 cells (P <0.001).These results demonstrated that the binding of99mTc-PEG6-RD-PDP2 toward A549-PD-L2 cells was specifically mediated by PD-L2.
In vivotumor targeting capabilities of L-peptide and retroinverse D-peptide were determined in the A549-PD-L2 tumor model by nanoScanSPECT/CT (Fig.4).99mTc-PEG6-RD-PDP2 showed better tumor imaging intensity compared to99mTc-PEG6-PDP2 (Fig.4A).The SPECT-quantified tumor uptake of99mTc-PEG6-RD-PDP2 and99mTc-PEG6-PDP2 was 2.30%±0.11% ID/cm3and 0.73%±0.10%ID/cm3at 0.5 h p.i., respectively (P <0.001, Fig.4C).To assess the targeting specificity of D-peptide tracer, SPECT/CT with99mTc-PEG6-RD-PDP2 was also performed in the PD-L2-negative A549 tumors, and the tumor uptake of99mTc-PEG6-RD-PDP2 was significantly decreased (0.75%±0.07% ID/cm3) at 0.5 h p.i.(Figs.4B and C).The radioactive signals in A549-PD-L2 tumors were significantly blocked by co-injection of excess RD-PDP2 peptide (0.61%±0.06%ID/cm3) (Figs.4B and C).These results suggest that the uptake of99mTc-PEG6-RD-PDP2 in A549-PD-L2 tumor is PD-L2-specific.
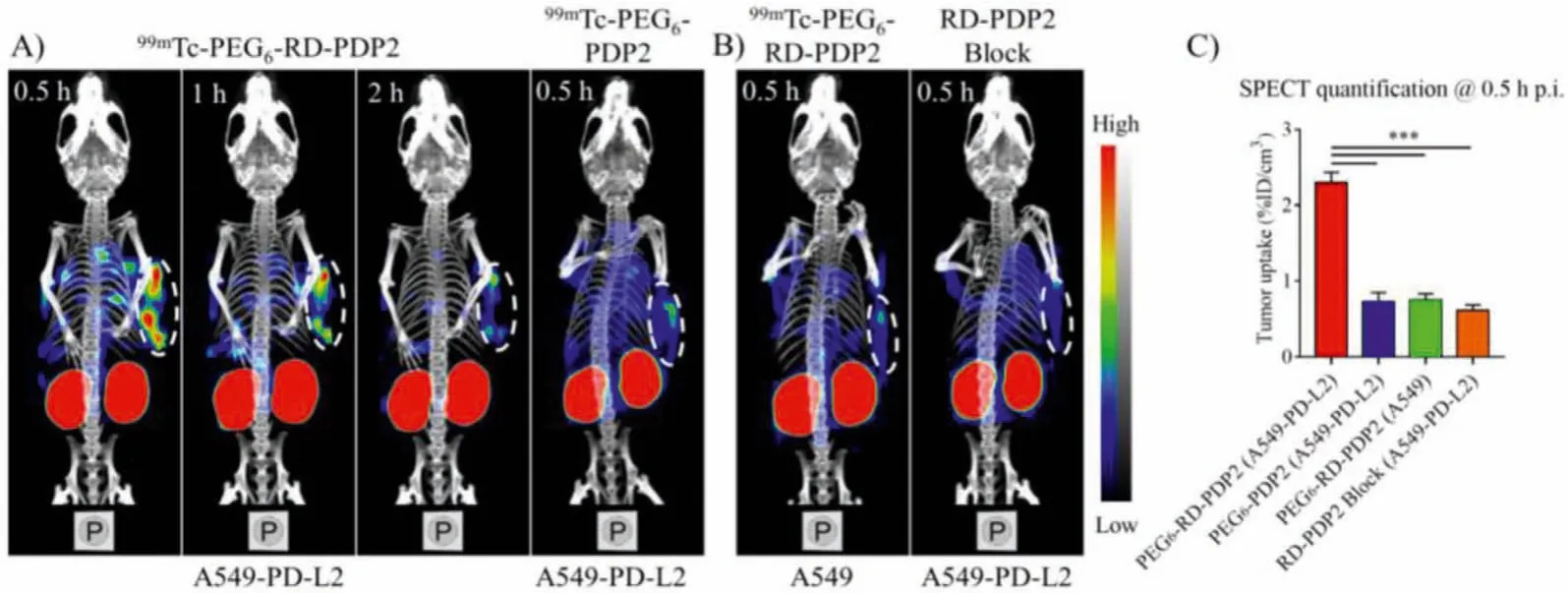
Fig.4.(A) Representative SPECT/CT images of 99mTc-PEG6-RD-PDP2 and 99mTc-PEG6-PDP2 in A549-PD-L2 tumor model.(B) Representative SPECT/CT images of 99mTc-PEG6-RD-PDP2 in A549 tumor model and in A549-PD-L2 tumor model with co-injection of excess cold RD-PDP2 peptide at 0.5 h p.i.(C) Quantification data of tumor uptake from imaging in panel A and B.The area shown in the white dashed circle indicate tumors (***P <0.001).
The biodistribution results were consistent with the SPECT imaging results (Fig.5).The uptake of99mTc-PEG6-RD-PDP2 in A549-PD-L2 tumors was 2.10%±0.15%, 1.10%±0.17%, and 0.51%±0.07% ID/g at 0.5, 1 and 2 h p.i., respectively (Fig.5A).The uptake of99mTc-PEG6-RD-PDP2 in A549-PD-L2 tumors was higher than that in normal organs such as the heart, lung, liver, and muscle at all tested time points.The uptake of99mTc-PEG6-RD-PDP2 in A549-PD-L2 tumors was significantly higher than that in A549 tumors (2.10%±0.15%vs.0.53%±0.06% ID/g,P <0.001, at 0.5 h p.i., Fig.5A), and the co-injection of excess RD-PDP2 and99mTc-PEG6-RD-PDP2 resulted in a significantly reduced tumor uptake(0.54%±0.08% ID/g,P <0.001, Fig.5B) at 0.5 h p.i., which further confirmed the specificity of the radiotracer to PD-L2.In addition,99mTc-PEG6-RD-PDP2 has a much higher tumor uptake than that of99mTc-PEG6-PDP2 (2.10%±0.15vs.0.64%±0.12% ID/g,P <0.001, Fig.5B) at 0.5 h p.i.Using reverse D-type of peptide sequence strategy,we have significantly improvedin vivostability of the radiotracer,thereby increasing its accumulation and retention in tumors.
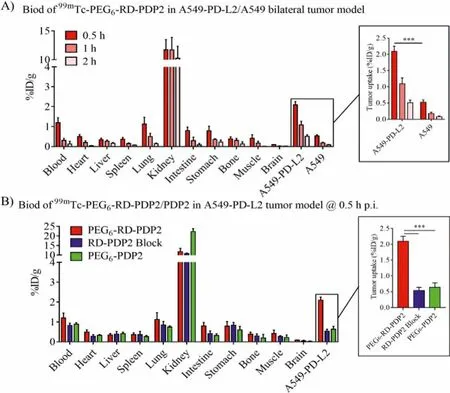
Fig.5.(A) Biodistribution of 99mTc-PEG6-RD-PDP2 in A549-PD-L2/A549 bilateral tumor model at 0.5, 1 and 2 h p.i.(B) Biodistribution of 99mTc-PEG6-PDP2 and 99mTc-PEG6-RD-PDP2 in A549-PD-L2 tumor model, as well as the blocking study by co-injection 99mTc-PEG6-RD-PDP2 and excess cold RD-PDP2 peptide in A549-PD-L2 tumor model at 0.5 h p.i.(***P <0.001).Inset: tumor uptake values from (A) or (B).
In summary, a novel PD-L2-targeted SPECT probe99mTc-PEG6-RD-PDP2 was designed and developed.This radiotracer can be easily prepared with high radiolabeling yield and possesses promising tumor targeting capability as well as favorable pharmacokinetics propertiesin vivo.99mTc-PEG6-RD-PDP2 SPECT/CT imaging accurately reflected the PD-L2 expression in tumors, demonstrating that the99mTc-PEG6-RD-PDP2 probe could be translated into the clinic to guide the immunotherapy of the PD-1/PD-L1 or PD-1/PDL2 axis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the National Natural Science Foundation of China(NSFC, Nos.92159201, 81630045 and 81927802 to F.Wang; No.81971676 to J.Shi; No.32027801 to Z.Hu), National Key R&D Program of China (No.2017YFA0205600 to F.Wang), Emergency Key Program of Guangzhou Laboratory, (No.EKPG21–16 to F.Wang),and Youth Innovation Promotion Association of Chinese Academy of Sciences (YIPACAS, No.2016090 to J.Shi) for financial support.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2022.02.068.
 Chinese Chemical Letters2022年7期
Chinese Chemical Letters2022年7期
- Chinese Chemical Letters的其它文章
- Professor Zhifang Chai: Scientific contributions and achievements
- Stable isotope labeling of nanomaterials for biosafety evaluation and drug development
- Emerging nanozymes for potentiating radiotherapy and radiation protection
- Recent progress of astatine-211 in endoradiotherapy: Great advances from fundamental properties to targeted radiopharmaceuticals
- Recent development in selective Tau tracers for PET imaging in the brain
- 64Cu radiolabeled nanomaterials for positron emission tomography(PET) imaging
