Diversity and composition of culturable fungi in Horqin Sandy Land
ShaoKun Wang ,XueYong Zhao ,Hao Qu ,Jie Lian ,Fei Wang ,FengHua Ding
1.Urat Desert-grassland Research Station,Northwest Institute of Eco-Environment and Resources,Chinese Academy of Science,Lanzhou,Gansu 730000,China
2.Naiman Desertification Research Station,Northwest Institute of Eco-Environment and Resources,Chinese Academy of Science,Lanzhou,Gansu 730000,China
3.Key Laboratory of Stress Physiology and Ecology,Gansu Province,Lanzhou,Gansu 730000,China
4.Lishui University,Lishui,Zhejiang 323000,China
ABSTRACT Soil fungi play a key role in soil functional performance and ecological restoration.To understand the diversity and com‐position of culturable fungi in soils of Horqin Sandy Land,China,mobile dune,semi-fixed dune,fixed dune and sandy grassland were selected to investigate the soil fungal diversity using a traditional culture-dependent approach.ITS se‐quencing was applied to identify the fungal strains.The counts of culturable fungi increased significantly from mobile dune to sandy grassland along the gradient of sandy land restoration.The Shannon-Wiener,Simpson and Evenness indices of culturable fungi ranged from 1.26−1.71,0.22−0.37 and 0.83−0.87,respectively.A total of 27 fungal strains were isolat‐ed using dilution plate cultural technique.The 27 fungal isolates were clustered into three groups:Ascomycota,Basidio‐mycota and Mucoromycota at phylum level,indicating that Ascomycota was the dominant fungal phylum (88.9% of the total).The isolated fungi were grouped into 3 phyla,5 classes,6 orders,11 families and 13 genera.The results show that culturable fungi were diverse in sandy land soils and fungal isolates have potential function in lipid turnover,cellulose degradation and ethanol,glucose and fatty acid production.Future studies should be carried out to explore their ecological and biological function for degraded sandy land restoration.
Keywords:Culturable fungi;fungal diversity;ITS sequences;phylogenetic analysis;Horqin Sandy Land
1 Introduction
With the rapid development of molecular biologi‐cal techniques and the remarkable improvement of da‐ta analysis (Kochet al.,2021),the original method for culturable microorganism has long been neglect‐ed,and even been discriminated,especially in studies on soil microbial diversity analysis.It is a fact that the culture-dependent approach underestimates the micro‐bial population (Wardle and Lindahl,2014).However,research based on genomic DNA sequencing has the unavoidable fact that they destroy the organism and analyze the "dead body",thoroughly blind on the liv‐ing organism.Soil microorganisms play an active role in litter decomposition,nutrient cycling,soil structure formation and the interaction with plants and animals(Harris 2009;Van der Walet al.,2013;Jacobyet al.,2017;Jiaet al.,2020).The ecological functions are collaboratively performed by the living microorgan‐isms(Torsvik and Øvreås,2002).Although the cultur‐able microorganism only occupies less than 10% of the total in the soil,the ecological function of the cul‐turable microorganism should not be neglected(Hawksworth 2001;Wigginton and Amador,2020).
Soil fungi constitute more than 75% of the soil to‐tal microbial biomass,and play a key role in soil func‐tional performance and ecological balance mainte‐nance(Varelaet al.,2015).Fungal mycelium binds soil particles to reconstruct soil structure and systematically combined soil and plant through arbuscular mycorrhi‐za (Van der Heijdenet al.,1998;Peayet al.,2013;Gaoet al.,2019).Decomposers of organic matter are mainly fungi,consisting 78%−90% of the total de‐composers in grassland ecosystems (Zhanget al.,2013).Culturable fungal decomposers significantly accelerated straw decomposition rate in sandy crop‐lands (Wanget al.,2020).Changes in fungal diversity may alter soil fertility,stability and crop yield in agri‐culture ecosystems (Bardgettet al.,1993).The role of soil fungi is critical in arid and semi-arid environ‐ments by accelerating litter decomposition in sandy soil and improving nutrient uptake through mycorrhi‐za,and fungal diversity represented in soil activity of desert ecosystems (Martínez-García and Pugnaire,2011).However,most of the soil fungi are resistant to laboratory cultivation,and studies of their ecological function have long been hampered (Mueller and Schmit,2007;Gaoet al.,2019).
Horqin Sandy Land is located in the southeastern part of Inner Mongolia,and is one of the most vulner‐able areas in a typical semiarid area of northern Chi‐na.This area was once the most severely desertified region,and unsustainable land-use practices aggravat‐ed by climate change and population pressure shifted the original tree-scattered grasslands into degraded sandy grasslands and mobile dunes (Wang,2003;Zhaoet al.,2003).The Chinese government per‐formed the Fencing and Nongrazing Policy to restore the degraded sandy land since the 1970s.As a result,a large area of shifting dunes has gradually recovered into stabilized dunes and grasslands during the recent few decades (Zhaoet al.,2010;Zhaoet al.,2015).It is reported that the world is getting greener since 2000,and China is leading the"Greening Earth"most‐ly contributed from planting trees (42%) (Chenet al.,2019),and greening in Horqin Sandy Land is mostly due to degraded grassland restoration(Liet al.,2020).Vegetation composition and diversity shifted during the degraded sandy land restoration,accompanied by the increasing of soil nutrients,soil microbial and macrofaunal diversity (Zuoet al.,2012;Liet al.,2015;Liuet al.,2015;Wanget al.,2016).However,the study of culturable fungi and their diversity re‐main unexplored in this area.The purpose of this study is to isolate fungal strains in sandy soils,to ana‐lyze their diversity and composition,and to predict their potential function in Horqin Sandy Land.
2 Materials and methods
2.1 Study area
This study was conducted at Naiman Desertification Research Station (NDRS) of the Chinese Academy of Sciences,located in the southwestern part of Horqin Sandy Land in Naiman County,Inner Mongolia,China(120°19′40″E−121°35′40″E,42°14′40″N−43°32′20″N)(Figure 1).The climate is characterized as temperate,semi-arid continental monsoonal.Annual mean pre‐cipitation is approximately 366 mm,with 70%−80%from June to September.Annual mean potential evapotranspiration is 1935,five times more than annu‐al precipitation.Annual mean temperature is 6.4 °C,ranging from a monthly average minimum of −16.8 °C in winter to a maximum of 23.5 °C in summer.Annu‐al mean wind velocity ranges from 3.6−4.1 m/s.The zonal soil is classified as sandy chestnut,which is san‐dy in texture,loose in structure and light yellow in color,making the sandy soil vulnerable to wind ero‐sion (Liet al.,2003;Zhaoet al.,2003;Wanget al.,2016).The original landscape was dominated by san‐dy grassland with scattered trees (mostlyUlmusspp.).However,unsustainable land-use practices aggravated by climate change and population pressure shifted the tree-scattered grasslands into degraded sandy grass‐lands and mobile dunes.The mobile dunes have been gradually restored into semi-fixed and fixed dunes by the effort of the Chinese government and local people(Zhaoet al.,2015).

Figure 1 Study area of Horqin Sandy Land
2.2 Experimental design
2.2.1 Sites and soil collection
We selected mobile dune (MD),semi-fixed dune(SFD),fixed dune (FD) and sandy grassland (SG) as four representative habitats along a gradient of de‐graded sandy grassland restoration in Horqin Sandy Land.The vegetation cover is less than 10% in MD,and the dominant plant species isAgriophyllum squarrosum.SFD and FD were naturally restored from MD with vegetation coverage of 44%and 65%,respective‐ly.SG was enclosed from grazing with good grass quality,representing original natural vegetation in Horqin Sandy Land.Five replicates for each habitat were chosen as sampling sites.The vegetation and soil properties of the four habitats are listed in Table S1.At each site,we randomly established five 1m×1m quadrats in a relatively flat area.A pooled soil sample was derived from five soil cores to a depth of 0−20 cm in each quadrat using a 3 cm diameter soil auger.The soil samples were separated in sterilized ziplock bags after removing rocks and plant material by a 2 mm mesh.The samples were then transferred to the labora‐tory in a cooler and prepared for isolation of cultur‐able fungi.The auger and mesh were sterilized using 75% alcohol before sampling.Soil water content was measured at 105°C for 24 h in an oven.
2.2.2 Fungal cultivation and counts
Culturable fungi were determined as colony form‐ing unites (CFU) by the pour plate method,and fungi were cultured on the potato dextrose agar (PDA)medium within penicillin and chloromycetin to isolate diverse fungal morphotypes (Xu and Zheng,1986;Wanget al.,2011).10 g of fresh soil was suspended in 90 mL sterilized water and shaken to form 10%soil microorganism suspension.1 mL of the soil supernate was pipetted into 9 mL sterilized water and shaken to form 1% suspension,and then to 0.1% suspension.1 mL of the 1% and 0.1% dilutions were coated in PDA plates with three replicates for cloning fungi,re‐spectively.The plates were incubated at 28−30 °C for 4 −6 days under aerobic conditions.All plates were counted by CFU.The abundance of culturable fungi was expressed as per gram dry soil and calculated by the following formula:

whereArepresents fungal abundance,indicating total CFU in 1g dry soil.CFU is colony forming units in each plate.DR is dilution rate.SWC is soil water con‐tent of the soil sample.
Based on colors and morphological characteristics of the colonies,distinct fungal isolates were enumerat‐ed and selected for purification.Single strains were picked from the selected colonies and were incubated in PDA plates at 28 −30 °C for 4 −6 days to isolate pure fungal strains.
2.2.3 ITS identification
A small amount of fungal mycelium from the pu‐rified fungal strains were scraped by a lancet to ex‐tract genomic DNA using Omega Fungal DNA Maxi Kit (Omega Bio-Tek,USA) following standard pro‐tocol.Polymerase chain reaction (PCR) amplifica‐tion was conducted targeting fungal ITS rRNA gene using universal primers ITS1F (5'-TCCGTAGGT‐GAACCTGCGG-3') and ITS4 (5'-TCCTCCGCT‐TATTGATATGC-3') (Manter and Vivanco,2007).The PCR program was set with the following condi‐tions:initial denaturation for 5 min at 94 °C followed by 38 cycles of 40 at 94 °C,40 at 55 °C and 1 min at 72 °C,and a final extension at 72 °C for 10 min in a Biometra-Mastercycler (Eppendorf,Germany).The PCR products were sequenced in Sino Geno Max.The DNA sequences were aligned using basic local alignment search tool (BLAST) with GenBank data‐base of the National Center for Biotechnology Infor‐mation (NCBI) to determine the taxa of culturable fungi.MEGA 7 was used to build a phylogenetic Maximum Likelihood (ML) tree to compare closely related fungal species with the obtained sequences.
2.3 Statistical analysis
Shannon-Wiener (H),Simpson (D) and Evenness(E) indices were used to calculate culturable fungal diversity based on the abundance (A) of fungal iso‐lates (S) (Zhanget al.,2013;Venkatachalamet al.,2015).
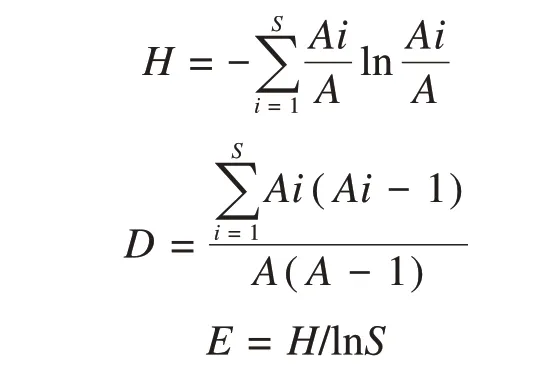
whereSis fungal species richness,Ais total abun‐dance of fungal isolates,andAiis the abundance of speciesi.
Diversity indices were expressed as mean±SE.Differences among the habitats were analyzed using one-way analysis of variance (ANOVA) and least-sig‐nificant-difference (LSD) tests.All significant differ‐ences are stated atp<0.05.The descriptive statistical parameters and significance tests were performed by Origin 8.0 and SPSS 17.0.
3 Results
3.1 Culturable fungal abundance
The culturable fungal abundance increased sig‐nificantly along the restoration gradient in Horqin Sandy Land (Figure 2).The number of representative CFU ranged from 1,342±275 per g dry soil in MD to 23,756±2,046 per g dry soil in SG.The culturable fun‐gal abundance in SG was 17.71,2.83 and 1.48 times more than that in MD,SFD and FD,respectively.It is clear that there are much less culturable fungi living in low-nutrient mobile dunes.However,the rare fun‐gal species living in mobile dunes may express a lownutrient tolerance function.
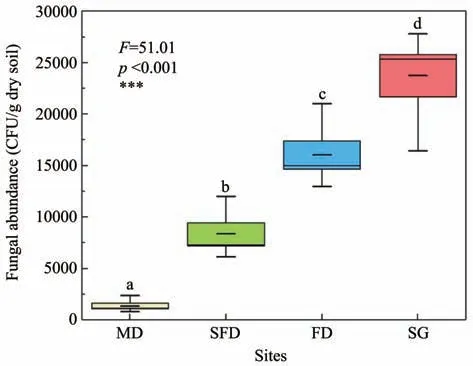
Figure 2 Culturable fungal abundance in the four habitats of Horqin Sandy Land
3.2 Diversity of culturable fungi
The Shannon-Wiener,Simpson and Evenness indi‐ces are presented in Figure 3.The Shannon-Wiener in‐dex increased significantly along the restoration gradi‐ent.The Shannon-Wiener index was 15.63%,35.75%and 32.20% higher in SFD,FD and SG than that in MD.The Simpson index decreased from MD to SFD,FD and SG.The Evenness index did not show any dif‐ference among the four habitats.
3.3 Phylogenetic analysis of culturable fungi
27 strains were isolated and purified from the sandy soil.DNA of the 27 isolated fungal species were extracted and sequenced to analyze their phylo‐genetic relationship.The obtained sequences were aligned using BLAST in NCBI.The sequences can be retrieved at GenBank submission in SUB9434782 from MW872373 to MW872399.The length of ex‐tracted DNA sequences of the 27 fungal isolates extended from 290 −558 bp,and the G+C content reached from 35.67%−60.05%.The result of BLAST indicated that most sequences were closely related to the fungal clones or isolates in GenBank (Table 1).S07 was 100% similar with the known fungal strain in GenBank (DQ339549.1),while S21 and S25 did not show strong similarity with the submitted strains in GenBank,presenting only 90.30% and 93.74%similarity with the known isolates MK299139.1 and JQ683240.1,respectively.

Table 1 Identification of DNA sequences and their taxonomic affiliations in NCBI using BLAST
The phylogenetic trees of the 27 fungal isolates and their most related strains are presented in Figure 4.It is clear that fungal isolates are clustered into three groups:Ascomycota,Basidiomycota and Mu‐coromycota at phylum level;and six groups:Pleospo‐rales,Eurotiales,Hypocreales,Sordariales,Filobasidi‐ales and Mucorales at order level.
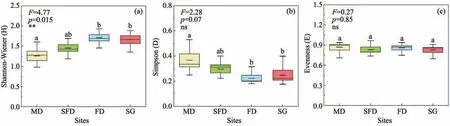
Figure 3 Culturable fungal diversity indices in the four habitats of Horqin Sandy Land
3.4 Composition of culturable fungi
Among the 27 isolated fungi,they were grouped in 3 phyla,5 classes,6 orders,11 families and 13 gen‐era (Figure 5).Ascomycota was the dominant fungal phylum,occupied 88.9% in the 27 isolates.At class level,Sordariomycetes and Eurotiomycetes dominat‐ed the culturable isolates,consisting 77.78% of the to‐tal.At order and family level,Hypocreales,Eurotiales and Nectriaceae,Aspergillaceae are dominate taxa.FusariumandPenicilliumoccupied over half of the isolated fungi at genus level.
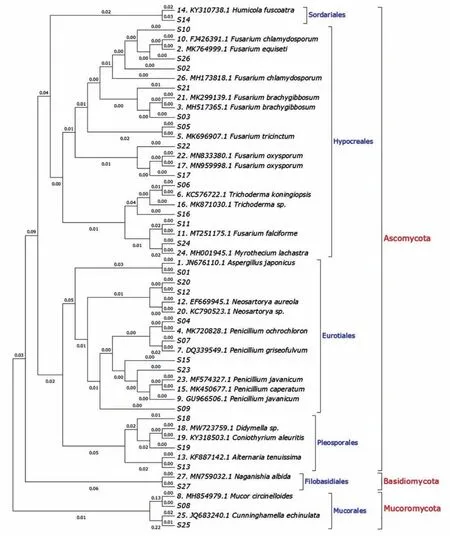
Figure 4 Phylogenetic Maximum Likelihood(ML)tree of the 27 fungal isolates and their most related strains in GenBank
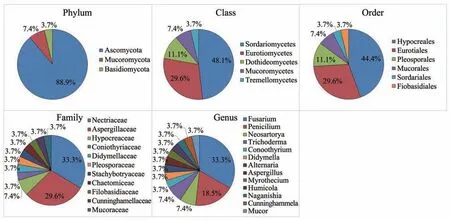
Figure 5 The proportion of different taxa at phylum,class,order,family and genus levels
4 Discussions
Plant species,cover and biomass increased along sandy land restoration (Zuoet al.,2012),and the plant species composition is closely related with mi‐crobial diversity and composition through litter input and root exudates.The different litter and root residu‐als are supposed to breed diverse fungal species(Shar‐maet al.,2015).Lower fungal abundance was detect‐ed in the mobile dunes due to its unstable cover and low nutrient content.However,the abundance of cul‐turable fungi presented 1,342±275 per g dry soil in the mobile dunes,and theses fungal species may have the ability to survive in the low-nutrient and severe environment.The culturable fungal abundance and di‐versity of Shannon-Winner index increased signifi‐cantly during the restoration process,which is similar with the former study using culture-independent ap‐proach (PCR-DGGE) (Wanget al.,2016).Our results show that Ascomycota (88.9%) is the dominant fun‐gal phylum using the culture-dependent approach,which is similar with that using PCR-DGGE.Howev‐er,the culture-dependent method detected a new phy‐lum (Mucoromycota) compared with the culture-inde‐pendent method.It is evidential that the culture-depen‐dent method should not be neglected in soil fungal di‐versity and composition studies.
Two of the 27 fungal isolates (S08 and S25) are clustered as Mucoromycota.S08 is annotated toMucor circinelloidesin the family of Mucoraceae.It is reported thatM.circinelloidescould produce extracellular endoglucanase to hydrolyze carboxy‐methyl cellulose and insoluble cellulose substrates to soluble cellodextrins (Saha,2004).A new study shows thatM.circinelloideswas promoted to be an engineered fungus.The recombinedM.circinelloidescould accelerate xylose uptake in corn straw hydrolysate by 70% and the increased utilization of xylose led to an increase in lipid production.There‐fore,the engineeredM.circinelloidesprovides a new way to produce lipids from lignocelluloses in industrial manufacture (Zhang and Song,2021).S25 is annotated toCunninghamella echinulatain the family of Cunninghamellaceae.C.echinulatais close‐ly related to lipid turnover (Papanikolaouet al.,2004;Fakaset al.,2007;Hoet al.,2010).It is also used as a model organism to hydroxylate 10-deoxoarte‐misinin(Fakaset al.,2007)and to biotransform betu‐linic acid into anti-inflammotory derivatives (Chenet al.,2021).We isolated an efficient cellulose de‐composing fungus,which is annotated toRhizomucor variabilisin the family of Mucoraceae (Mu‐coromycota) from the Horqin sandy cropland soil(Wanget al.,2020).TheR.variabilisstrain was applied to decompose maize straw and straw turn‐over significantly increased soil organic carbon and total nitrogen by 34.08% and 14.26%,indicating that the isolated fungus could potentially improve crop productivity and quality in sustainable agricul‐ture management.
S27 (Naganishia albida) was the only Basidiomy‐cota isolated from the sandy soil.It could ferment ag‐ricultural and industrial waste for lipid production(Sathiyamoorthiet al.,2020).
Ascomycota consists of approximately 6,600 gen‐era and is reported as the most abundant fungal phy‐lum in soil (Wijayawardeneet al.,2018),for in‐stance in forest (Huet al.,2014),grassland (Ochoa-Huesoet al.,2018),agriculture (Evanset al.,2020),desert (Sunet al.,2019) and even in alpine (Lenten‐duet al.,2011) and Arctic tundra ecosystems (Wal‐lensteinet al.,2007).The various species of Ascomy‐cota represent different functions in ecosystem ser‐vice and maintenance.Aspergillus japonicus(S01)could produce an endo-xylanase to immobilize ligno‐cellulose into small fractions of oligosaccharides(Mussattoet al.2009),and to degrade fruit peels within a wide pH stability (Da Silvaet al.,2019).Penicillium ochrochloron(S04) could produce plant cell wall degrading enzymes in ethanol production(Huanget al.,2014).Neosartorya sp.(S20)is report‐ed to promote cabbage growth and increase biomass(Hamayunet al.,2011).Humicola fuscoatra(S14)could produce eco-friendly cellulose nano fibers from agriculture wastes (Hasaninet al.,2018).Alternaria tenuissima(S13),derived fromArtemisia judaica,is an ideal candidate for potential industrial pro‐duction of antimicrobial agents with a variety of ap‐plications (Mousaet al.,2021).Coniothyrium aleuritis(S19) was isolated from a medicinal plant to pro‐tect the host plant against insects and pathogens by producing bioactive compounds (Manganyiet al.,2018).Trichoderma sp.(S16) was used to accelerate wheat straw methanolysis,and significantly enhance conversion,specificity and content of esters (Zhenget al.,2020).Trichoderma koningiopsis(S06) was used to control pine seedlings damping-off symptom and regulate active oxygen metabolism and rhizo‐spheric microbiome (Yu and Luo,2020).Fusariumis reported as a plant disease causing stack rot,plant wilt and dieback (Shanet al.,2016;Guoet al.,2020).However,some species present ecological and biolog‐ical functions.Fusarium equiseti(S02) was used as an antioxidant enzyme producer to peroxidate lipid in‐to glucose and maltose (Ayar Kayali and Tarhan,2004).Fusarium tricinctum(S05) is a metal-resis‐tant fungus isolated from a mine drainage environ‐ment (Bonillaet al.,2021).Fusarium falciforme(S11) is capable of biodegrading 4-t-octylphenol,which may cause adverse effects to the ecosystem and public health (Rajendranet al.,2020).Fusarium oxysporum(S17 and S22)is used to enhance bioetha‐nol production from rice-straw (Gupta and Chun‐dawat,2020).Members of the fungal genusMyrotheciumproduce structurally unique secondary metabo‐lites,which display promising biological and phar‐macological properties (Sunet al.,2020).However,the biological function ofMyrothecium lachastrae(S24)has not been reported.
According to the literature,fungal isolates in our study have potential ecological and biological func‐tion in lipid turnover,cellulose degradation and etha‐nol,glucose and fatty acid production in agricultural management and industrial production.These iso‐lates could be used in promoting plant growth,utiliz‐ing agricultural waste and producing antimicrobial components.
Shannon-Winner and Simpson indices are both widely used to represent community diversity.The Shannon-Winner diversity index emphasizes species richness,and higher species richness results in a high‐er Shannon-Winner index.The Simpson diversity in‐dex emphasizes species evenness,and higher Simp‐son index indicates that the abundance of each species in the community is even (Nagendra,2002).In our study,the fungal Shannon-Winner index increased,while fungal Simpson index decreased along sandy land restoration.The results indicate that fungal spe‐cies richness increased accompanied with increasing plant species richness along the restoration gradient.Also,heterogeneity is stronger in restored sandy land,resulting in lower Simpson index in fixed dunes and sandy grassland.
It is reported that fungal isolates are the same spe‐cies if the similarity of tested DNA sequence is ≥99%with the known fungal sequence in the Genbank data‐base;the isolates are in the same genera if the similari‐ty is 95%−99%,and they are in the same family if the similarity is 90%−95% (Landeweertet al.,2003).In our study,two isolates (S21,90.3% and S25,93.74%)did not show strong similarity with known isolates in the GenBank database,indicating that these isolates could be new species or clustered in new genera.Fur‐ther work should be done to explore their molecular classification and ecological function.
Theoretically,all living microbes could be cultured and isolated in the laboratory,given a suitable living en‐vironment.It is really difficult to mimic the exact soil environment for each living microorganism.However,this is not a reason to discriminate against traditional culture-dependent methods to study soil fungal com‐position,because of the accumulation of centuries of knowledge.We have long way to explore the un‐sealed knowledge and should focus on how to im‐prove and invent new cultural mediums and disclose the soil microenvironmental parameters in the labora‐tory,promoting our initiative to explore the vibrant soil microbiome.
5 Conclusions
Our study demonstrated that soil culturable fungal abundance and Shannon-Wiener diversity increased significantly,but Simpson diversity decreased from mobile dune to sandy grassland along the gradient of sandy land restoration.27 fungal strains were isolated using the culture-dependent approach.The fungal iso‐lates were clustered into three groups:Ascomycota,Basidiomycota and Mucoromycota at phylum level,and Ascomycota (88.9%) dominated the culturable fungal community.The isolated fungi were grouped into 3 phyla,5 classes,6 orders,11 families and 13 genera.The fungal isolates have a potential function in lipid turnover,cellulose degradation and ethanol,glucose and fatty acid production.Two isolates did not show strong similarity with known isolates in the GenBank database,indicating that these isolates could be new species or clustered in new genera.Further re‐search should be carried out to explore their ecologi‐cal and biological function for degraded sandy land restoration.
Acknowledgments:
This study was financially supported by the National Nature Science Foundation of China (41771117 and 41877540),the China National Key Research and Development Plan (2017FY100200),the Second Ti‐betan Plateau Scientific Expedition and Research program (2019QZKK0305) and the Key Research and Development project of Zhejiang Province(2018C02031).We thank all members of the Naiman Desertification Research Station and Urat Desertgrassland Research Station of CAS for their contribu‐tion in field and laboratory work.
 Sciences in Cold and Arid Regions2022年2期
Sciences in Cold and Arid Regions2022年2期
- Sciences in Cold and Arid Regions的其它文章
- Editor-in-Chief Yuanming Lai
- Estimating snow depth or snow water equivalent from space
- Influence of meteorological elements on chemical evolution of snow and ice of Urumqi Glacier No.1,eastern Tianshan Mountains
- Coarse fragment content influences estimates of soil C and N stocks of alpine grassland on the northeastern edge of Qinghai-Tibetan Plateau,China
- Effect of GGBS on performance deterioration of non-dispersible underwater concrete in saline soil
- Simulation assessment and prediction of future temperatures in Northwest China from BCC-CSM Model
