Inhibition of LncRNA Vof-16 expression promotes nerve regeneration and functional recovery after spinal cord injury
Xiao-Min Zhang , Li-Ni Zeng , Wan-Yong Yang, Lu Ding , Kang-Zhen ChenWen-Jin Fu, Si-Quan Zeng, Yin-Ru Liang Gan-Hai Chen , Hong-Fu Wu
Abstract Our previous RNA sequencing study showed that the long non-coding RNA ischemia-related factor Vof-16 (lncRNA Vof-16) was upregulated after spinal cord injury, but its precise role in spinal cord injury remains unclear. Bioinformatics predictions have indicated that lncRNA Vof-16 may participate in the pathophysiological processes of inflammation and apoptosis. PC12 cells were transfected with a pHBLV-U6-MCS-CMV-ZsGreen-PGK-PURO vector to express an lncRNA Vof-16 knockdown lentivirus and a pHLV-CMVIE-ZsGree-Puro vector to express an lncRNA Vof-16 overexpression lentivirus. The overexpression of lncRNA Vof-16 inhibited PC12 cell survival, proliferation, migration, and neurite extension, whereas lncRNA Vof-16 knockdown lentiviral vector resulted in the opposite effects in PC12 cells. Western blot assay results showed that the overexpression of lncRNA Vof-16 increased the protein expression levels of interleukin 6, tumor necrosis factor-α,and Caspase-3 and decreased Bcl-2 expression levels in PC12 cells. Furthermore, we established rat models of spinal cord injury using the complete transection at T10. Spinal cord injury model rats were injected with the lncRNA Vof-16 knockdown or overexpression lentiviral vectors immediately after injury. At 7 days after spinal cord injury, rats treated with lncRNA Vof-16 knockdown displayed increased neuronal survival and enhanced axonal extension. At 8 weeks after spinal cord injury, rats treated with the lncRNA Vof-16 knockdown lentiviral vector displayed improved neurological function in the hind limb. Notably, lncRNA Vof-16 knockdown injection increased Bcl-2 expression and decreased tumor necrosis factor-α and Caspase-3 expression in treated animals. Rats treated with the lncRNA Vof-16 overexpression lentiviral vector displayed opposite trends. These findings suggested that lncRNA Vof-16 is associated with the regulation of inflammation and apoptosis. The inhibition of lncRNA Vof-16 may be useful for promoting nerve regeneration and functional recovery after spinal cord injury.The experiments were approved by the Institutional Animal Care and Use Committee of Guangdong Medical University, China.
Key Words: apoptosis; functional recovery; inflammation; long non-coding RNA ischemia related factor Vof-16; nerve regeneration; nerve repair; neurite extension; neuronal survival; proliferation; spinal cord injury
Introduction
Spinal cord injury (SCI) is a severe central nervous system injury with a poor prognosis that typically results in irreversible sensory and motor dysfunction below the damaged area,resulting in heavy burdens on patients and society due to the lack of effective treatment methods (Sweis and Biller,2017). Surgical interventions, such as surgical decompression and spinal stability reconstruction, and pharmacological interventions, such as methylprednisolone and neurotrophic factors, are commonly used for SCI treatment (Zhu et al.,2014; Assunção-Silva et al., 2015). Cell replacement therapies and tissue engineering technologies have also received much attention in recent years (Kabu et al., 2015; Zhou et al.,2018). However, current treatment options remain far from satisfactory, and none of them effectively address the complex and progressive molecular alterations that are involved in the pathological processes following SCI. Therefore, studies focused on understanding the complex pathophysiological mechanisms underlying SCI could facilitate the discovery of novel targets for the treatment of SCI and nerve repair after injury (Venkatesh et al., 2019).
Recently, an increasing number of researchers have begun to pay attention to the roles played by long non-coding RNAs (lncRNAs) in SCI (Ding et al., 2016; Gu et al., 2017).Emerging results from RNA sequencing and microarray studies have suggested that lncRNA expression continuously changes in the spinal cord before and after SCI, and these differentially expressed lncRNAs have been associated with critical pathways and molecular networks involved in chronic and subchronic stages of SCI (Shi et al., 2018; Li et al., 2019;Wang et al., 2019). Studies have indicated that changes in the expression of lncRNAs might participate in SCI pathogenesis and influence key pathophysiological processes (Wang et al., 2015; Duran et al., 2017; Shi et al., 2019). Moreover,our previous results revealed significant changes in the expression levels of 16 lncRNAs (eight upregulated and eight downregulated) following SCI compared with those in the normal group. Among these 16 identified lncRNAs, ischemiarelated factor Vof-16 (lncRNA Vof-16) attracted our attention(Ding et al., 2020). LncRNA Vof-16 is a 2017-bp lncRNA located on chromosome 8 in Rattus norvegicus and was first recorded in the Rat Genome Database in 2003 (RGD: 628754). Tohda and Watanabe (Tohda and Watanabe, 2004) reported that lncRNA Vof-16 was primarily expressed in the hippocampus,the tenia tecta, the piriform cortex, and the area around the aorta. Additionally, lncRNA Vof-16 has been shown to play roles in neurodegenerative disease, vasculitis of the central nervous system, and other neuropathic diseases associated with neuronal inflammation (Tohda and Watanabe, 2004;Schiavone et al., 2017). However, the significance of the observed change in lncRNA Vof-16 expression following SCI has not been further investigated. Therefore, we explored the mechanism underlying the upregulation of lncRNA Vof-16 expression following SCI, which is involved in the regulation of nerve regeneration and functional recovery.
We investigated changes in lncRNA Vof-16 expression at the lesion site through the construction of knockdown and overexpression lentiviral vectors, which were used to transfect neuron-like PC12 cells and transplanted into SCI model rats with complete transection at T10. We observed the effects of changing lncRNA Vof-16 expression levels on neuron survival and proliferation, neurite extension, and functional recovery to explore the mechanisms underlying the functional effects of lncRNA Vof-16 upregulation after SCI.
Materials and Methods
Bioinformatics analysis
A previous study from our laboratory demonstrated that the expression profiles of lncRNAs and mRNAs in the rat spinal cord of a complete transection model through RNA sequencing analysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses suggested that lncRNAs play an important role in the early immuno-inflammatory response after SCI (Ding et al.,2020). To explore the role played by lncRNA Vof-16 in the pathogenesis of SCI, bioinformatics analysis was performed to predict the molecular mechanisms and pathways of lncRNA Vof-16 after SCI. MicroRNA (miRNA) families that potentially interact with lncRNA Vof-16 were predicted using both RNAInter (http://www.rna-society.org/rnainter/) and miRDB bioinformatics software (MicroRNA Target Prediction Database, http://mirdb.org/). Proteins that may be potentially targeted by the identified lncRNA Vof-16-interacting miRNAs were identified through the online prediction algorithm TargetScan, version 7.2 (http://www.targetscan.org/vert_72/),and the miRDB database (MicroRNA Target Prediction Database, http://mirdb.org/) and presented in a Venn diagram. The intersection between the two predicted groups of proteins was identified as most likely to represent the lncRNA Vof-16 target gene. Subsequently, KEGG pathway and GO analyses of the intersecting target genes were performed using the STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string-db.org/) and the Metascape database (http://metascape.org). Based on the order ofP-values, a visual network diagram was constructed using a network, a histogram, and bubble diagrams, and the potential mechanism for lncRNA Vof-16 in the treatment of SCI was determined.
Construction of lentiviral vectors
To evaluate the role played by lncRNA Vof-16 on SCI, we constructed lncRNA Vof-16 lentiviral vectors forin vitrotransfection into cells or injection into rats for changing the expression of lncRNA Vof-16. All lncRNA Vof-16 lentiviral vectors were constructed and synthesized by Hanbio Biotechnology Co., Ltd. (Shanghai, China), including an lncRNA Vof-16 knockdown lentiviral vector using the pHBLV-U6-MCSCMV-ZsGreen-PGK-PURO vector (5′-CCG ACA AAG TTA TTG GTC AT AGC AA-3′) and an lncRNA Vof-16 overexpression lentiviral vector using the pHBLV-CMVIE-ZsGree-Puro vector.In addition, a green fluorescent protein-tagged lentiviral vector was constructed to verify the efficiency of transfection,knockdown, or overexpression. Empty vectors were used as negative controls. The original lentiviral vector titers contained 2.0 × 108TU/mL.
Cell culture
PC12 cells were originally obtained from the Chinese Academy of Sciences (Shanghai, China). PC12 cells are a commonly used cell model forin vitroneuronal research (Genchi et al., 2015; Hoop et al., 2017; Gao et al., 2018), including the fields of neurophysiology and pathology, and have been used to explore various nervous system diseases and treatment strategies. Briefly, cells were maintained in RPMI-1640 medium(Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (100 U/mL,Sigma-Aldrich, St. Louis, MO, USA) at 37°C with 5% CO2.
Cell grouping and transfection
PC12 cells were randomly divided into the following groups:Control group (no treatment); NC-Knockdown group(transfected with knockdown negative control vector);Knockdown group (transfected with lncRNA Vof-16 knockdown lentivirus); NC-Overexpression group (transfected with overexpression negative control vector); and Overexpression group (transfected with lncRNA Vof-16 overexpression lentivirus). Cell transfection was conducted in 6-well plates (1× 105cells/well) using a serum-free medium (Gibco), according to the manufacturer’s instructions. The concentration of lncRNA Vof-16 lentivirus or negative control vector was 2 ×108TU/mL (multiplicity of infection = 20). PC12 cells in each group were cultured in a mixture containing the desired lentiviral vector and serum-free RPMI-1640 medium (Gibco).After 6 hours of transfection, the medium containing 2%fetal bovine serum (Gibco) equal to half of the total culture volume was added. The medium was replaced with fresh medium containing 10% fetal bovine serum after 24 hours of transfection and cultured for 48–72 hours after transfection.Quantitative real-time polymerase chain reaction (qRT-PCR)was used to detect the transfection efficiency of lncRNA Vof-16 lentiviral vectors in PC12 cells. The resulting expression levels induced by the lncRNA Vof-16 lentiviral vectors in PC12 cells can be found inAdditional file 1.
Cell viability assay
PC12 cell viability was measured 1, 3, and 5 days after transfection using a Live/Dead Cell Vitality Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, PC12 cells were plated on the surface of a 22-mm square coverslip and allowed to adhere for 24 hours. Cells were then rinsed with phosphate-buffered saline (PBS) and incubated with 150 µL of the live/dead assay combined solution for 30 minutes at room temperature. Afterward, the coverslips on which the PC12 cells were grown were mounted onto slides with Fluorescence Mounting Medium (DAKO, Copenhagen,Denmark) and observed under a fluorescence microscope(DMi8, Leica, Wetzlar, Germany). To perform the analysis of each experiment, images were captured from a minimum of 6 random fields/group, and the numbers of living cells (red,excitation and emission wavelengths of 488 nm and 660 nm,respectively) and dead cells (green, excitation and emission wavelengths of 480 nm and 530 nm, respectively) were counted.
Cell proliferation assay
The cell proliferation analysis was conducted using Ki67 (a cell proliferation marker; Miller et al., 2018) staining 1, 3, and 5 days after transfection. In brief, PC12 cells were fixed with 4% paraformaldehyde for 10 minutes and permeabilized with 0.3 % TritonTMX-100 for 15 minutes. Cells were stained with 10 µg/mL anti-mouse/rat Ki67 fluorescein isothiocyanate(Cat# 11-5698-82, monoclonal antibody; 1:100; eBioscience,Waltham, MA, USA) and simultaneously counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 1:5000; Cell Signaling Technology, Danvers, MA, USA) for 2 hours at room temperature. The coverslips on which the PC12 cells were grown were carefully mounted onto slides using a fluorescence mounting medium (DAKO). Under a fluorescence microscope, the nuclei containing green fluorescence dots were categorized as proliferating and dividing cells, and the proportion of proliferating cells relative to total nuclei, which appeared blue, was calculated as the cell proliferation rate.For analysis, images were captured from a minimum of 6 random fields in each group, and the proliferation index was defined as the percentage of Ki67 and DAPI double-positive cells in each group.
Cell migration assay
Cell migration abilities were evaluated using a wound-healing assay (Grada et al., 2017). The cell wound-healing assay was performed on confluent PC12 cell monolayers cultured in serum-free RPMI 1640. A P10 pipette tip was used to score four equally sized vertical scratches in each well. After scratching, the cells were rinsed with PBS three times and cultured in serum-free medium. The healed state of the scratches was observed using an inverted microscope (ECLIPSE TS100-F, Nikon, Tokyo, Japan), and photographs were taken every 12 hours during the 24-hour incubation. For analysis,images of at least six random fields/group were used. The relative percentage of each wound area occupied by migrating cells was calculated relative to the original wound area using ImageJ software, version 1.8.0 (National Institutes of Health,Bethesda, MD, USA).
Immunocytochemical staining
At 1, 3, and 5 days after transfection, PC12 cells were washed with PBS for 5 minutes and fixed with 4% paraformaldehyde for 15 minutes. Cells were permeabilized with 0.3% Triton X-100 for 15 minutes and incubated with PBS containing 5%bovine serum albumin for 2 hours at room temperature. The fixed cells were washed 3 times with PBS containing 0.1%Triton X-100 and incubated overnight with anti-microtubuleassociated protein 2 (MAP-2; Cat# AB5622, polyclonal antibody, rabbit anti-rat, 1:1000; Millipore, Bedford, MA,USA) at 4°C. The cells were incubated with an Alexa Fluor 488-conjugated donkey anti-rabbit antibody (Cat# A21206,1:500, Thermo Fisher Scientific, Waltham, MA, USA; excitation and emission wavelengths of 488 and 525 nm, respectively) at room temperature for 2 hours. Nuclei were stained with DAPI(1:5000, Cell Signaling Technology; excitation and emission wavelengths of 340 and 488 nm, respectively) for 15 minutes.At least 6 microscopic fields from each sample were observed under a fluorescence microscope, and images were obtained at 40× (Leica LAS X software, Wetzlar, Germany). Five PC12 cells with the longest protrusion lengths were selected from at least 100 cells, the mean fluorescence density was calculated for each visual field, and the average protrusion length was measured by ImageJ software 1.8.0 version.
SCI rat model and grouping
A total of 102 adult, female, specific-pathogen-free,Sprague-Dawley rats (aged 8–10 weeks; weighing 180–200 g) were purchased from the Experimental Animal Center of Southern Medical University (Guangzhou, China) and housed at 23–25°C under a 12-hour light/dark cycle. The rats were randomly divided into 6 groups (n= 17/group),as follows: Sham operation group (Sham group); PBS group(PBS group); knockdown negative control lentivirus group(NC-Knockdown group); lncRNA Vof-16 knockdown lentivirus group (Knockdown group); overexpression negative control lentivirus group (NC-Overexpression group); lncRNA Vof-16 overexpression lentivirus group (Overexpression group).Before injection, lentiviral vectors (2 × 108TU/mL) were mixed with 0.01 M PBS (Servicebio, Wuhan, China) on ice to obtain a final lentiviral vector concentration of 2 × 107TU/mL. In the Sham group, the rats were only subjected to laminectomy. In the PBS group, the rats were subjected to SCI, followed by the injection of 10 µL PBS at the complete transected SCI site after hemostasis. For the lentivirus groups, the corresponding 10µL lentiviral vector was injected into the SCI lesion gap after hemostasis.
The surgical procedure used for the T10 complete transection SCI was performed in the rats as described in our previous report (Wu et al., 2013). All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (Publication No. 85-23, revised 1996)and were approved by the Institutional Animal Care and Use Committee of Guangdong Medical University, China. Briefly,rats were anesthetized by intraperitoneal injection with 1%sodium pentobarbital (40 mg/kg), and an approximately 2-cm midline incision was made at the level of T10 under aseptic conditions. The T10 vertebral body of each rat was located by counting the ribs. A laminectomy was performed at the T9–10 level to expose the dorsal aspect of the spinal column. The spinal cord was completely traversed with a surgical blade,and a 2-mm block of the spinal cord tissue was removed.After hemostasis, the rats received 10 µL of PBS or lentivirus at the lesion gap. Finally, the muscle tissue and skin were closed in separate layers with sutures, and the animals were returned to their cages to recover. Except for the Sham group,all rats were completely paralyzed immediately post-surgery,demonstrating the successful generation of an SCI model. For the first 3 days after surgery, all rats received intraperitoneal injections of penicillin-streptomycin (10,000 U/mL; Sigma-Aldrich) daily to avoid infection. The bladders of all rats that underwent SCI had to be emptied manually using a defecation promoting method three times daily until normal micturition reflexes developed.
Tissue preparation
Eleven rats in each group were randomly sacrificed 7 days after SCI. Four rats from each group were used for immunofluorescence staining and fluorescentin situhybridization (FISH); four rats from each group were used for qRT-PCR; the remaining three rats were used for western blot assay. All rats were evaluated for hind limb motor function by assessing Basso, Beattie, and Bresnahan (BBB) scores at 1 and 7 days. The remaining rats were used to evaluate hind limb motor function at 2, 3, 4, 5, 6, 7, and 8 weeks after SCI.
At 7 days after spinal cord injury, each rat was anesthetized with an overdose of 1% sodium pentobarbital (10 mg/kg), the diaphragm was cut, and the pericardium was opened. The right atrial appendage of each rat was cut and perfused with 150 mL of sterile 0.9% normal saline at the same time. After the eラuent liquid became clear, the tissue near the injury site at the T10 level (5 mm above and below the injury epicenter,resulting in a 10 mm length of the spinal cord) was quickly placed in an Eppendorf tube containing protein lysis buffer(n= 3/group) or RNA lysis buffer (n= 4/group) and stored at–80°C. In addition, the rats used for immunofluorescence staining and FISH (n= 4/group) were perfused with 300 mL of 4% paraformaldehyde (Solarbio, Beijing, China) until the tissues hardened. The tissue near the injury site at the T10 level (5 mm above and below the injury epicenter) was then removed and placed in paraformaldehyde overnight. Thetissue was dehydrated in 30% sucrose solution, embedded in optimum cutting temperature compound, cut into continuous 14-µm frozen sections with a microtome (CM1850, Leica), and processed for FISH and immunofluorescence staining.
qRT-PCR
LncRNA Vof-16 expression was determined by qRT-PCR at 7 days post-SCI (n= 4/group). Total RNA was extracted from normal rat (before SCI) or SCI rat tissue at the T9–11 level using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For RNA analysis, 500 ng total RNA was reverse transcribed into complementary DNA (cDNA) with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) after the determination of RNA concentration (500–800 ng/µL) and purity (A260/A280= 2.0;A260/A230= 2.0). The reverse transcription procedure included 15 minutes of reverse transcription at 37°C and 5 seconds of reverse transcriptase inactivation at 85°C. Subsequently, real-time PCR was performed for cDNA amplification with FastStart Universal SYBR Green Master(Roche, Basel, Switzerland). The following primer sequences were used: Rat lncRNA Vof-16, forward 5′-TGT CCG AGT GAT GGG AAT AC-3′ and reverse 5′-ATG CTA ACA TCC TGT GCC TAC-3′; rat glyceraldehyde 3-phosphate dehydrogenase (Gapdh),forward 5′-CTG GAG AAA CCT GCC AAG TAT G-3′ and reverse 5′-GGT GGA AGA ATG GGA GTT GCT-3′. Gapdh served as an endogenous control. The levels of lncRNA Vof-16 expression were determined using the 2–ΔΔCtcalculation method, and the results are presented as fold changes relative to the levels in the control group.
Fluorescence in situ hybridization
Spinal cord segments 10 mm in length near the injury site at the T10 level were dissected from rats (n= 4/group) and fixed with 4% paraformaldehyde for 10 minutes. Sagittal sections 7-µm-thick were heated in a microwave oven at 300 watts for 7 minutes in citrate buffer solution, pH 3.5, cooled naturally,and then incubated with proteinase K (1:2000; Sigma-Aldrich)at 37°C for 8 minutes. Each section was incubated in 50 µL hybridization buffer (Cat# G3016-3, Servicebio, Wuhan, China)at 37°C for 1 hour, and treated with hybridization buffer containing oligonucleotide probes (8 ng/µL; Servicebio) at 37°C for 12 hours overnight. The hybridization buffer was blotted off, and the sections were washed three times with 2×,1×, and 0.5× saline sodium citrate, respectively. Each section was then incubated in 30 µL biotinylated anti-digoxin (1:50) at 4°C overnight, washed three times with 0.1 M PBS, incubated with a streptavidin-biotin-peroxidase complex at 37°C for 30 minutes, and rinsed four times in 0.1 M PBS. The secondary antibody (goat anti-rat conjugated to Alexa Fluor 568; Cat#A10042; 1:500; Thermo Fisher Scientific) was incubated at room temperature for 50 minutes in the dark. DAPI (1:5000;Cell Signaling Technology; excitation and emission wavelengths of 340 and 488 nm, respectively) was used to stain the nuclei before capturing images. We randomly chose eight visual fields from each section, and 6 sections near the central region of the spinal cord were investigated in each rat. Fluorescence was observed using a fluorescence microscope. LncRNA Vof-16 probes were designed as follows: 5′-CY3-TTG AAG CGT CTT CTA CCA AGT GGT GCC AAG TA-CY3-3′ (CY 3: excitation and emission wavelength 560 nm and 590 nm, respectively).
Western blot assay
Proteins were extracted from PC12 cells (harvested 72 hours after transfection) and T9–11 spinal cord tissues from rats(harvested day 7 after SCI) using radioimmunoprecipitation assay lysis buffer (10×; Millipore), supplemented with phenylmethylsulfonyl fluoride (PMSF, 1:100; Millipore) and mammalian protease inhibitor mixture (1:100, Biocolor Bioscience, Shanghai, China). Extracted proteins were analyzed by western blot analysis (n= 3/group). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membrane (0.2 µm; Millipore). Polyvinylidene fluoride membranes were incubated with the following primary antibodies for 12 hours at 4°C: MAP-2 (Cat# AB5622;polyclonal antibody; rabbit anti-rat; 1:1000; Millipore),neuronal nuclei (NeuN; Cat# MAB377; monoclonal antibody;mouse anti-rat; 1:1000; Millipore), interleukin-6 (IL-6; Cat#sc-57315, monoclonal antibody, mouse anti-rat, 1:1000;Santa Cruz Biotechnology, Hercules, CA, USA), tumor necrosis factor-α (TNF-α; Cat# bs-0078R; polyclonal antibody; rabbit anti-rat; 1:400; Bioss, Beijing, China), Caspase-3 (Cat# 9662;polyclonal antibody, rabbit anti-rat; 1:1000; Cell Signaling Technology), Bcl-2 (Cat# bs-0032R; polyclonal antibody,rabbit anti-rat; 1:400; Bioss), Bax (Cat# CPA3797; polyclonal antibody; rabbit anti-rat; 1:1000; Bioworld, St. Louis Park, MN,USA), and GAPDH (Cat# MA5-15738; monoclonal antibody;mouse anti-rat; 1:1000; Thermo Fisher Scientific), followed by incubation with corresponding secondary antibodies horseradish peroxidase goat anti-mouse/rabbit IgG (Cat#AS003; 1:5000; ABclonal, Cambridge, MA, USA) for 2 hours at room temperature. GAPDH was utilized as a reference protein.The protein blots were visualized using chemiluminescence reagent (Millipore), and protein levels were normalizing against those for GAPDH.
Immunofluorescence staining
Rat spinal cord tissues from the lesion epicenter were collected 7 days post-SCI (n= 4/group). The frozen sections were thawed at room temperature for 30 minutes. The sections were washed three times (for 10 minutes each) with 0.1 mM PBS containing 0.1% Triton X-100, incubated in proteinase K(1:2000, Sigma-Aldrich) at 37°C for 10 minutes, and blocked with 5% bovine serum albumin, 5% newborn donkey serum,and 0.02% Triton X-100 in PBS at room temperature for 2 hours. The tissue sections were then incubated with primary antibodies specific to neurofilament protein-200 (NF-200; Cat#N4142; monoclonal antibody; rabbit anti-rat; 1:400; Sigma),CD31 (Cat# AF3628; polyclonal antibody; goat anti-rat; 1:200;R&D Systems, Minneapolis, MN, USA), or NeuN (monoclonal antibody; mouse anti-rat; 1:200; Millipore) overnight at 4°C.The tissue sections were then incubated with secondary antibody (donkey anti-rabbit conjugated to Alexa Fluor 488,Cat# A21206; donkey anti-goat conjugated to Alexa Fluor 568,Cat# A11057; or donkey anti-mouse conjugated to Alexa Fluor 568, Cat# A10037; 1:500; Invitrogen) at room temperature for 2 hours in the dark. DAPI (1:5000; Cell Signaling Technology)was used to stain the nuclei before capturing images. The number of immunostained cells was counted in each of six random fields per section, and all images were acquired using a fluorescence microscope.
Functional assessment
We used the BBB scale to evaluate functional motor recovery at 1, 2, 3, 4, 5, 6, 7, and 8 weeks after surgery. Behavioral testing was performed on all rats at 1 week and then on only six randomly selected rats from each group at other time points. To ensure the objectivity of behavioral scoring,two independent investigators who were blinded to the experimental conditions observed the movement and scored the motor function of the rats according to the BBB scoring standard (Song et al., 2016). During the assessment, the rats were allowed to move and explore freely in an open field. The final score for each rat was obtained by averaging the values from both investigators.
Statistical analysis
Numerical data from at least three individual experiments are presented as the mean ± standard deviation (SD). The results were analyzed with GraphPad Prism 5.0 (GraphPad Software,Inc., La Jolla, CA, USA) and SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA). Differences between the experimental and control group were tested by Student’st-tests. Multiple group comparisons were analyzed by one-way analysis of variance, followed by Tukey’spost hoctests. Significant differences in functional assessments were assessed using repeated-measures two-way analysis of variance, followed by Tukey’spost hoctest. Significance was set toP< 0.05.
Results
The biological function of lncRNA Vof-16 correlates with inflammatory and apoptotic pathways
Using RNAInter and miRDB bioinformatics software, 42 miRNA families were predicted to bind to lncRNA Vof-16, of which 3 intersection miRNAs were identified by both programs,including rno-miR-346, rno-miR-205, and rno-miR-874-3p. A total of 877 proteins were predicted to interact with lncRNA Vof-16, and 148 proteins were identified by both the TargetScan and miRDB online databases as potentially interacting with lncRNA Vof-16 (Figure 1A). The STRING and Metascape websites were used to analyze the functional enrichment of the 148 identified target genes (Figure 1B–D).Figure 1Bshows a network diagram of the potential KEGG signaling pathways associated with lncRNA Vof-16.The size of the pathway was determined by the number of molecular targets involved. Excluding ossification, the negative regulation of catabolic processes, and the lipid biosynthetic process, which signaling pathways with little relevance for SCI, a total of 13 core potential signaling pathways were identified (Figure 1B). The GO enrichment analysis results demonstrated that the likely target genes for lncRNA Vof-16 are involved in the regulation of multiple biological processes, such as macromolecule modifications,cell communication, and protein modification processes(Figure 1C). As shown inFigure 1D, signaling pathways such as the phosphoinositide 3-kinase (PI3K)–protein kinase B (Akt) signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, inflammatory mediator regulation of transient receptor potential (TRP) channels, and the advanced glycation endproducts signaling pathway were identified, which are primarily associated with inflammation and apoptosis (Nakajima et al., 2015; Zhang et al., 2015; Fu et al., 2018; Shimizu et al., 2018), may be the primary signaling pathways through which lncRNA Vof-16 exerts effects on the pathophysiology of SCI. These findings provide a novel perspective on the underlying mechanisms of lncRNA Vof-16,but further validation with experimental evidence remains necessary to explore the functions of lncRNA Vof-16 in SCI.
LncRNA Vof-16 overexpression decreases neuronal viability and proliferation in vitro
A series of analyses were performed to determine the cellular function of lncRNA Vof-16 using rat PC12 cells transfected with lncRNA Vof-16 overexpression or knockdown lentivirus.As shown inFigure 2AandB, cells were evaluated for live/dead staining and Ki67 staining, 1, 3, or 5 days after 72 hours of transfection. The live/dead staining showed an increase in the cell death rate for the lncRNA Vof-16 overexpression group compared with the control group (P< 0.001;Figure 2C). In contrast, we observed that cell viability was elevated in the lncRNAs Vof-16 knockdown group compared with that in the control group (P< 0.01;Figure 2C). Except for the overexpression group, Ki67 staining indicated that PC12 cell proliferation decreased in a time-dependent manner (P<0.01;Figure 2D). The reduction in cell proliferation plateaued 5 days after transfection in the lncRNA Vof-16 knockdown group compared with the control group (P> 0.05;Figure 2D).These results suggested lncRNA Vof-16 knockdown increased the survival and proliferation of PC12 cells, whereas lncRNA Vof-16 overexpression was not conducive to cell survival and proliferation.
LncRNA Vof-16 knockdown ameliorates cell migration and neurite extension in PC12 cells
A wound-healing assay was used to detect the migration abilities of PC12 cells after changing lncRNA Vof-16 expression levels, in which the cell fusion area was used to represent the migration ability (Figure 3A). Images were acquired at 0,12, and 24 hours. As demonstrated inFigure 3B, the woundhealing assay revealed a reduction in the migration ability of PC12 cells transfected with the lncRNA Vof-16 overexpression lentivirus compared with the control group, accounting for 57.21 ± 3.02% of the scratch area relative to the 0-hour point(P< 0.05 for the overexpression group). Moreover, the lncRNA Vof-16 knockdown group filled 55.5 ± 2.5% (P< 0.001) and 95± 6.25% (P< 0.01) of the wounded area after 12 and 24 hours relative to 0 hour, respectively, which was a significant increase compared with the control group (Figure 3B). These results demonstrated that lncRNA Vof-16 knockdown promoted cell migration, whereas the overexpression of lncRNA Vof-16 had the opposite effect.
We next examined the effects of lncRNA Vof-16 on neurite extension using staining for MAP-2, a neuronal dendritic marker (Shafit-Zagardo and Kalcheva, 1998) that is expressed in neuronal cell bodies and dendrites and plays an important role in nerve regeneration and network reconstruction (Figure 3C). The results showed that neurite extension in the lncRNA Vof-16 knockdown group increased significantly, by almost 2-fold (205.4 ± 22.57 µm) compared with the control group(Figure 3D), whereas treatment with the lentiviral negative controls had no significant effect on neurite outgrowth.However, the neurites in the lncRNA Vof-16 overexpression group were shorter (46.20 ± 5.79 µm) than those in the control group (Figure 3D). The results of western blot analysis were consistent with the immunocytochemical staining results(Figure 3E), demonstrating that the expression levels of NeuN,an excellent neuronal marker (Duan et al., 2016), and MAP-2 were significantly increased in the lncRNA Vof-16 knockdown group than in the control group (Figure 3F), indicating an increase in neurites. These data suggested that inhibiting the expression of lncRNA Vof-16 in PC12 cells was beneficial for neurite outgrowth.
LncRNA Vof-16 contributes to an inhibitory pathologicalmicroenvironment through the inflammation and apoptosis pathways
To assess the results of the bioinformatics analysis (Figure 1), the mechanisms through which lncRNA Vof-16 exerted effects on the inflammatory and apoptosis pathways during SCI were further dissected by using western blot to detect the expression levels of the inflammation-related proteins TNF-α and IL-6, the pro-apoptosis factors Bax and Caspase-3, and the anti-apoptosis protein Bcl-2 in PC12 cells. As shown inFigure 4, compared with the control group, the overexpression of lncRNA Vof-16 increased the levels of the inflammatory factors TNF-α (P< 0.01) and IL-6 (P> 0.05), enhanced the expression levels of the pro-apoptosis factor Caspase-3 (P< 0.05), and inhibited the expression of the anti-apoptotic factor Bcl-2 (P<0.01), which indicated that the overexpression of lncRNA Vof-16 in PC12 cells enhanced inflammation and apoptosis. The knockdown of lncRNA Vof-16 showed the opposite trends (P< 0.05). Collectively, these results suggested that lncRNA Vof-16 influences neuroinflammation and apoptosis, contributing to a disadvantageous environment that aggravates the pathophysiology of SCI.
LncRNA Vof-16 is differentially upregulated and involved in neuronal apoptosis and inflammatory responses after SCI
The expression of lncRNA Vof-16 in rats before SCI (Normal group) and after SCI (SCI group) was examined by FISH and RNA analysis (Figure 5). As shown inFigure 5A, lncRNA Vof-16 was differentially upregulated during the course of SCI.Compared with the Normal group, FISH quantification showed that the lncRNA Vof-16 expression level increased dramatically in the SCI group (Figure 5B). Similar results were observed with the qRT-PCR assay, in which high levels of lncRNA Vof-16 were observed in injured tissue derived from rats after 7 days post-SCI (P< 0.01vs. the SCI group;Figure 5C).
To determine whether lncRNA Vof-16 affects biological behaviors through the regulation of inflammation and apoptosis pathwaysin vivo, western blot analysis was performed 7 days post-SCI in rats from different treatment groups. As shown inFigure 5D, the expression level of the pro-inflammatory cytokine TNF-α was reduced in the lncRNA Vof-16 Knockdown group, whereas the expression levels of TNF-α and Caspase-3 were upregulated and the level of Bcl-2 was downregulated in the lncRNA Vof-16 Overexpression group compared with the Sham group (Figure 5E). These results suggest that the upregulation of lncRNA Vof-16 after SCI plays a negative regulatory role by aggravating the local inflammatory microenvironment and inhibiting the survival of spinal cord neurons, whereas inhibiting the expression of lncRNA Vof-16 may improve the inflammatory microenvironment, promoting nerve regeneration and repair after SCI.
Intrathecal injection of lncRNA Vof-16 knockdown lentivirus facilitates neuron survival and neurogenesis post-SCI
The loss and death of large numbers of neurons after SCI remains a major obstacle to functional recovery. To further investigate the effects of lncRNA Vof-16 lentivirus on spinal cord neuronsin vivo, we used the neuron-specific antibody NeuN (Duan et al., 2016) to perform the immunofluorescence staining of spinal cord neurons (Figure 6A). As shown inFigure 6B, the number of neurons in the PBS group was significantly lower than that in the Sham group (25 ± 3 neurons/visual field,vs. 39 ± 3 neurons/visual field,P< 0.01), suggesting that a large number of neurons died and were lost after the spinal cord transection injury. In addition, compared with the Sham group, the number of neurons in the lncRNA Vof-16 Knockdown group increased significantly (35 ± 6.25 neurons/visual field,P< 0.05), whereas the number of neurons in the lncRNA Vof-16 Overexpression group decreased significantly(15.67 ± 1.53 neurons/visual field,P< 0.01). The western blot results also indicated that the downregulation of lncRNA Vof-16 expression in rats was associated with the promotion of spinal cord neuron survival, whereas the overexpression of lncRNA Vof-16 was associated with increased spinal cord neuron loss (Figure 6C).
The nerve fiber regeneration in each group was investigated 8 weeks after SCI by immunofluorescence staining against the nerve fiber marker NF-200 and the vascular marker CD31(Mohamadi et al., 2018) (Figure 7A). The results showed an increase in the linear distribution of cord-like nerve fibers in the spinal cord tissue of the Sham and lncRNA Vof-16 Knockdown groups, whereas the other groups presented fewer nerve fibers with scattered distributions (Figure 7BandC). These results suggested that thein vivotreatment with the lncRNA Vof-16 knockdown lentivirus was able to promote the regeneration of nerve fibers after SCI. In addition, the morphological analysis of CD31 immunofluorescence staining(vascular marker) preliminary showed that the blood vessels in the Sham group were primarily microvessels, based on vessel diameter, with a few vasodilating phenomena observed near the injury site after SCI (Figure 7D).
Knockdown of lncRNA Vof-16 promotes functional recovery in the rat SCI model
The effects of lncRNA Vof-16 on locomotor recovery after SCI were evaluated weekly using the BBB scale (Figure 8A).We found that all rats subjected to spinal cord contusions were paralyzed in both hind limbs starting from the secondday post-injury (Figure 8B). As shown in Figure 8B, hind limb locomotor activity improved gradually during the experimental period, as demonstrated by the increase in BBB scores. No significant differences in BBB scores were observed across the various SCI groups one week after SCI (Figure 8B). The rats in all SCI groups showed significant functional recovery at 4–8 weeks. However, the functional recovery in the lncRNA Vof-16 Overexpression group was remarkably slower than that for the other SCI groups (P< 0.01;Figure 8B), whereas the functional recovery in the lncRNA Vof-16 Knockdown group appeared to be faster than that of other groups starting 5 weeks post-surgery and became significantly improved starting at week 7 compared with the other SCI groups (P<0.01;Figure 8B). By week 8, significant differences in the BBB scores were observed between the lncRNA Vof-16 Knockdown and Overexpression groups (8.8 ± 0.98vs. 4.3 ± 8.2,P< 0.01).To further assess the effects of lncRNA Vof-16 on neurological recovery, the BBB scores were calculated for rats in each group 8 weeks after surgery. As shown inFigure 8C, the proportion of rats with a score less than or equal to 5 in the lncRNA Vof-16 Overexpression group reached 66.7%, but no animals in the groups without overexpression achieved this score. In contrast, the BBB scores in the lncRNA Vof-16 Knockdown group were all greater than 8, whereas only 16.7% of all animals in all groups except the knockdown group reached this level. These data indicated that the SCI model was established successfully and that the inhibition of lncRNA Vof-16 in SCI rats could improve the recovery of motor function.

Figure 1|Bioinformatics analysis identifies lncRNA Vof-16 enrichment in inflammation- and apoptosis-related pathways.

Figure 2|Changes in lncRNA Vof-16 expression affect neuronal survival and proliferation in vitro.

Figure 3|LncRNA Vof-16 affects cell migration and neurite outgrowth in PC12 cells.
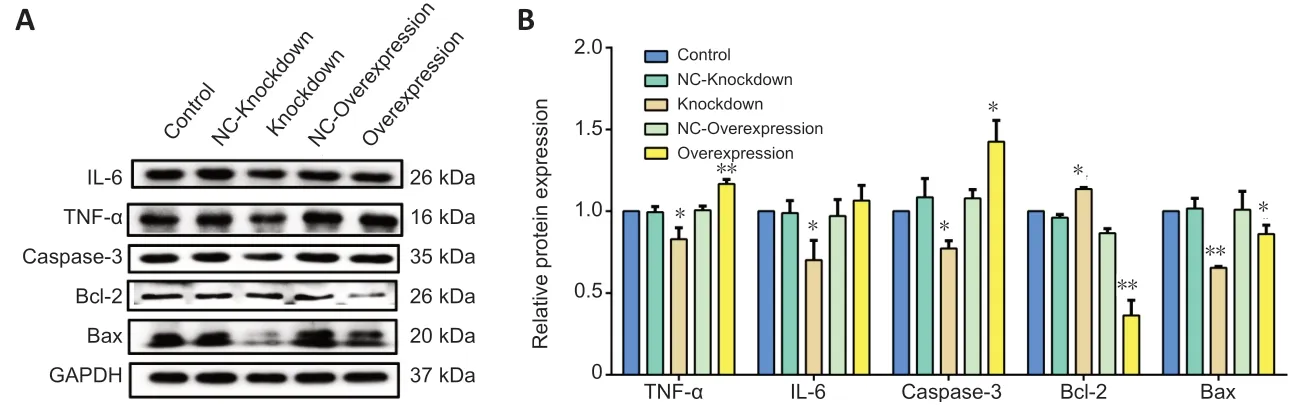
Figure 4|LncRNA Vof-16 is associated with inflammation and apoptosis pathways in vitro.
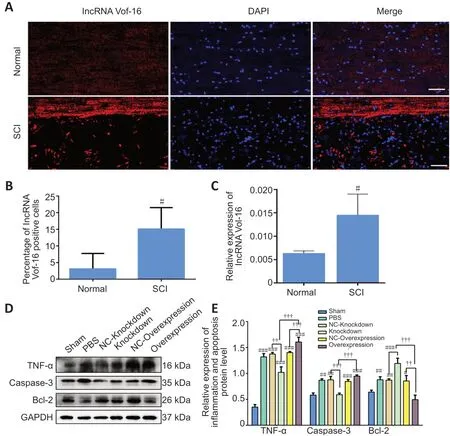
Figure 5|LncRNA Vof-16 is significantly upregulated and aggravated neuroinflammation and apoptosis after SCI.

Figure 6|Local delivery of the lncRNA Vof-16 knockdown lentivirus is beneficial for neuronal survival following SCI.

Figure 7|Transplantation of lncRNA Vof-16 knockdown lentivirus promotes the outgrowth of regenerative fibers in rats with SCI.

Figure 8|LncRNA Vof-16 knockdown improves functional recovery after SCI.
Discussion
SCI often results in physical impairments, including temporary or permanent paralysis, the loss of sensory functions, urinary incontinence, and neurological defects (Zhang et al., 2017).The atrophy and death of motor neurons after injury represent notable pathological mechanisms resulting in the loss of motor function in the inner muscles (Wu et al., 2012). Strong inflammatory responses and increased immune stress prevent further tissue damage but can negatively impact the survival of damaged motor neurons after SCI. Therefore, improving the local microenvironment and protecting injured neurons is crucial to SCI repair. Our data indicated that the inhibition of lncRNA Vof-16 could effectively improve the survival and proliferation of PC12 cells and significantly increased the number of spinal cord neurons in the injured area of SCI rats.
Moreover, the regulatory roles played by lncRNAs in SCI have been widely reported (Wang et al., 2019). For instance, the knockdown of the lncRNA brain-derived neurotrophic factorantisense (BDNF-AS) has been shown to induce cell apoptosis in acute SCI (Zhang et al., 2018). The lncRNA zinc finger protein 667-antisense RNA 1 (ZNF667-AS1) has also been shown to inhibit the inflammatory response and promote SCI recovery (Li et al., 2018b). We previously demonstrated that lncRNA Vof-16 was upregulated after SCI (Ding et al.,2020). In this study, we found that the increased expression of lncRNA Vof-16 reduced cell viability, proliferation, migration,and neurite extension. Combined with the results of the bioinformatics analysis, we speculate that these effects may be mediated by the induction of inflammation and apoptosis. The western blot analysis revealed the increased expression of Bcl-2 and the reduced expression levels of TNF-α and Caspase-3 in spinal cord lesions from the lncRNA Vof-16 Knockdown group, whereas the lncRNA Vof-16 Overexpression group showed the opposite trend. Many proapoptotic pathways are mediated by signaling molecules that are produced during neuroinflammation (Shabab et al., 2017).Thus, we speculate that lncRNA Vof-16 may participate in promoting an inhibitory and pathological microenvironment by releasing pro-inflammatory/pro-apoptotic cytokines that are not conducive to neuron survival, proliferation, migration,or neurite extension. Some researchers have studied the regulatory mechanisms through which TNF-α is involved in apoptosis after SCI and found that the expression of TNF-α was significantly upregulated 24 hours after electrical stimulation was applied to the tails of SCI rats, which was accompanied by neuronal and microglia apoptosis, as assessed by histochemical analysis (Garraway et al., 2014). These findings suggested that the increased expression of TNF-α does not favor neuronal survival, which is consistent with our research results. Ourin vitroandin vivowestern blotting analyses showed that the overexpression of lncRNA Vof-16 increased the release of the local inflammatory factor TNF-α in the lesion area of SCI rats. Moreover, the relative expression level of TNF-α protein in the lncRNA Vof-16 Overexpression group was significantly higher than that in the lncRNA Vof-16 Knockdown group, and the immunofluorescence staining results showed that the upregulation of TNF-α was not conducive to neuron survival. These findings suggested that lncRNA Vof-16 exacerbates SCI by promoting the inflammatory and apoptotic responses of nerve cells. However, the underlying signaling pathways remain to be further investigated.
Based on the above research, our data suggested that lncRNA Vof-16 functions in an inflammatory role in SCI, closely related to the inflammatory response. The inhibition of the pro-inflammatory environment in the injured spinal cord has become a key therapeutic target for the promotion of cell survival and the reduction of neuronal degeneration.Microglia/macrophage-associated inflammation appears to be a vital mechanism related to neuronal degeneration and regeneration (Zhou et al., 2014). After SCI, microglia activate the intracellular inflammatory cascade by engulfing necrotic cell debris and releasing large numbers of pro-inflammatory factors, including IL-6 and TNF-α, which intensify the inflammatory response in the tissue microenvironment,damaging the surrounding normal spinal cord tissue (Kong and Gao, 2017). Microglia transcribe IL-1β and TNF-α mRNA within 1 to 12 hours after SCI, which induces the cytokine cascade (Schnell et al., 1999). Therefore, lncRNA Vof-16 may also be associated with microglia/macrophages, although the correlation between lncRNA Vof-16 and immune cells remains unclear. Further clarification of the interactions between lncRNA Vof-16, inflammatory cell types, and inflammatory factors may contribute to the understanding of SCI pathogenesis and further clarify the sources of lncRNA Vof-16 after SCI. Understanding the relationship between lncRNA Vof-16 and dynamic changes in the immune system may help identify methods to reduce immune-mediated apoptosis,improve the survival rate of nervous tissue, and promote repair after SCI.
The most intuitive phenomenon in the repair of SCI is the restoration of motor function. To further determine the harmful role played by lncRNA Vof-16 in SCI, we examined the BBB scores of SCI rats and found that rats with overexpressing lncRNA Vof-16 presented weaker motor functional recovery after SCI. Meanwhile, rats with lncRNA Vof-16 knockdown presented remarkable motor functional recovery. Axon regeneration is an essential condition for the recovery of motor function after SCI, but the development of syringomyelia commonly occurs after SCI, preventing the spontaneous regeneration of axons from the corticospinal tract through the injury site, disrupting neural circuit reconstruction (Li et al., 2018a). Hence, one of the main challenges following SCI is the reconstruction of structural and functional connections between nerve cells. Researchers have attempted various therapeutic strategies, but only a few have successfully achieved significant nerve fiber regeneration, both inside and outside of the SCI epicenter,and the recovery of motor function has not been satisfactory(Zhou et al., 2019). We found that lncRNA Vof-16 knockdown promoted the extension of neurite outgrowth in PC12 cells.In line with thesein vitroresults, we observed that lncRNA Vof-16 knockdown promoted the regeneration of nerve fibers after SCI in a rat model. These findings suggested that the inhibition of lncRNA Vof-16 is beneficial for the extension of neuronal axons. Uncontrolled neurite growth or random neurite sprouting can result in unintended consequences.For example, the sprouting of nociceptive afferents and propriospinal ‘relay’ neurons below the injury site are major contributors to the development of autonomic dysreflexia,a leading health risk associated with SCI (Eldahan et al.,2020). Patients with life-threatening autonomic dysreflexia frequently complain of a sudden onset, severe, throbbing headache with cerebrovascular signs, such as hemorrhage and seizure. Moreover, immunofluorescence staining showed that the predicted increase in angiogenesis was not observed in the lncRNA Vof-16 Knockdown group after SCI. In contrast,the increased number and extension of regenerated nerve axons were not associated with the formation of peripheral blood vessels, suggesting that the inhibition of lncRNA Vof-16 does not promote nerve regeneration by increasing microvascular formation. Interestingly, we observed that a small number of blood vessels were dilated near the injury site after SCI, consistent with the newly reported observation of microvasodilation in the SCI region, which might promote the phagocytosis of microvascular endothelial cells and the degradation of myelin fragments through the autophagosome pathway, promoting inflammation and abnormal angiogenesis(Zhou et al., 2019).
This study investigated the function and mechanism of lncRNA Vof-16 for the first time in SCI. We constructed an SCI rat model and showed that rats injected with an lncRNA Vof-16 knockdown lentivirus presented significantly better recovery after SCI, which suggested that the early inhibition of lncRNA Vof-16 promoted nerve repair after SCI. Targeting lncRNA Vof-16 might benefit nerve repair and functional recovery to some degree. Although this experiment has deepened the understanding of the pathological mechanisms associated with SCI to a certain extent, this study has some limitations.First, we have not determined the precise mechanism through which lncRNA Vof-16 regulates the inflammation and apoptosis signaling pathways. In addition, SCI is an extremely complicated pathophysiological process, and the repair of SCIrelated damage is influenced by many factors. Thus far, no effective treatments for SCI have been discovered. In-depth research exploring the pathological mechanism of SCI remains necessary to identify potential key regulatory factors, which are necessary to identify effective targets for the treatment of SCI.
Author contributions:Study design and funding support: HFW; in vivo experiment implementation: LNZ, XMZ, WYY, LD; in vitro experiment implementation: KZC, WJF, SQZ, YRL; manuscript writing: XMZ; manuscript revising: HFW, GHC. All authors approved the final version of the paper.
Conflicts of interest:The authors declare that they have no competing interests.
Financial support:This work was financially supported by the National Natural Science Foundation of China, No. 82071374 (to HFW); Characteristic Innovation Project of Colleges and Universities in Guangdong Province of China, No. 2018KTSCX075 (to HFW);the Key Project of Social Development of Dongguan of China, No.20185071521640 (to HFW); College Students Science and Technology Innovation Cultivation Project in Guangdong of China, Nos.pdjh2020b0257 (to HFW), pdjh2020b0263 (to HFW); College Students Innovative Experimental Project in Guangdong Medical University,China, Nos. ZZDS006 (to HFW), ZYDS005 (to HFW), ZYDB004 (to HFW),FYDY003 (to HFW); College Students’ Science and Technology Innovation Training Project, Nos. 202010571027 (to HFW), 202010571054 (to HFW),202010571055 (to HFW), 202010571084 (to HFW), 202010571099(to HFW), GDMU2019054 (to HFW), GDMU2019055 (to HFW),GDMU2019099, GDMU2019123 (to HFW), GDMU2019027 (to HFW),GDMU2019084 (to HFW); and the Scientific and Technological Projects of Dongguan City, No. 202050715023190 (to WJF). The funding sources had no role in study conception and design, data analysis or interpretation,paper writing or deciding to submit this paper for publication.
Institutional review board statement:The animal experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of Guangdong Medical University, China.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix,tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewers:Florence E. Perrin, Université de Montpellier, France;Timothy Kopper, University of Kentucky, USA.
Additional files:
Additional file 1: The successful construction of lncRNA Vof-16 overexpression and knockdown lentivirus in vitro and in vivo.
Additional file 2: Open peer review report 1 and 2.
- 中国神经再生研究(英文版)的其它文章
- Inhibition of CXCR4/CXCL12 signaling: a translational perspective for Alzheimer’s disease treatment
- The new wave of p75 neurotrophin receptor targeted therapies
- The anatomical, electrophysiological and histological observations of muscle contraction units in rabbits:a new perspective on nerve injury and regeneration
- Intranasal insulin ameliorates neurological impairment after intracerebral hemorrhage in mice
- Lycium barbarum extract promotes M2 polarization and reduces oligomeric amyloid-β-induced inflammatory reactions in microglial cells
- Exosomes derived from bone marrow mesenchymal stem cells protect the injured spinal cord by inhibiting pericyte pyroptosis

