Optimizing Sample Size for Assessing the Genetic Diversity in Alfalfa (Medicago sativa) Based on Start Codon Targeted (SCoT) Markers
MAO Pei, SHEN Shu-heng, PU Jun, LOU Ke-ke, ZHOU Qiang, WANG Zeng-yu,SUN Juan*, LIU Zhi-peng*
(1. State Key Laboratory of Grassland Agro-ecosystems, College of Pastoral Agriculture Science and Technology,Lanzhou University, Lanzhou, Gansu Province 730020, China; 2. Grassland Agri-Husbandry Research Center, College of Grassland Science, Qingdao Agricultural University, Qingdao, Shandong Province 266109, China)
Abstract:Alfalfa (Medicago sativa L.) is one of the most economically valuable forage crops in the world. Due to its autotetraploidy and cross-pollination with a complex genome structure,genetic and breeding projects for alfalfa improvement have been significantly hindered. In the present study,six different germplasms of alfalfa including 600 individuals were used for sample size determination. The analysis of genetic diversity of alfalfa was performed with start codon targeted (SCoT) markers. From the PCR amplification,we selected 6 out of 56 primers that were used in previous studies. All analyzed markers were polymorphic,and 80 bands were identified,with 13.3 bands per locus detected. The percentage of polymorphic bands values were all 100%,which indicated that the markers were highly informative. The genetic diversity of alfalfa with 11 random sampling levels (10,15,20,30,40,50,60,70,80,90,and 100 genotypes) was compared in this study. When the sample size was increased to 40,there was a coverage of at least 95% genetic variation in the six germplasms. Additionally,based on the unweighted pair-group method with arithmetic averages (UPGMA) and STRUCTURE analysis,each cultivar in this study could be distinguished from others by utilizing a sample size of 40 individuals. The results of the present study showed that these SCoT markers would benefit the alfalfa research community for genetics and breeding. Furthermore,this study also established the foundation for future cultivar identification,resequencing and evolution in the alfalfa.
Key words:Medicago sativa;SCoT markers;Genetic diversity;Sample sizes
Alfalfa (Medicagosativa) belongs to the Fabaceae family;it is grown in 80 million ha worldwide and is one of the most economically valuable crops in the world[1]. Alfalfa has been an important component of sustainable agricultural systems for many years due to several key biological and agricultural features[2-3]. It is not only of huge value as a livestock feed,but also important for water and soil conservation,grass field rotation and soil improvement. Besides its applications in agriculture and animal husbandry,alfalfa also can be used for paper and biofuel production because of its high fiber content biomass,which makes alfalfa a potentially unique bioenergy crop[4-5]. Alfalfa was derived from a species complex (M.sativaspecies complex) that includes diploid and tetraploid interfertile subspecies[6]. Cultivated alfalfa is a perennial species with high genetic complexity at individual and population levels due to its autotetraploidy (2n=4x=32) and cross-pollinated (allogamous)[7]. Moreover,since the tetraploid structure and severe selfing depression of alfalfa,the study of population genetics is also complicated[8].
The genetic diversity ofM.sativahas been characterized by traditional morphological traits and molecular markers. Compared with conventional phenotyping assessment,molecular marker technology is an efficient supplement and alternative to morphological trait measurements,as the markers are easily detectable and stable in plant tissues regardless of environmental influences[9]. Various molecular assays have been successfully used to evaluate genetic diversity in alfalfa. These assays mainly focus on amplified fragment length polymorphism (AFLP)[10],random amplified polymorphic DNA (RAPD)[11],inter-retrotransposon amplified polymorphism (IRAP)[12],sequence-related amplified polymorphism (SRAP)[13],and simple sequence repeats (SSR)[14]. More recently,many new alternatives and promising molecular assays have emerged. Of these assays,start codon targeted (SCoT) markers are dominantly inherited,which has been developed based on the short conserved region flanking sequences of the adenine-thymine-guanine (ATG) start codon site[15]. SCoT markers display the following advantages:simple operation,low cost,high level of polymorphism,and high abundance in the genome[16]. SCoT markers open their way in various plant investigations:genetic diversity and structure analysis,identify cultivars,quantitative trait loci (QTL) mapping,and DNA fingerprinting,including wild rye (Elymussibiricus)[17-18],dandelion (Taraxacumpieninicum)[19],barb goatgrass (Aegilopstriuncialis)[20],cowpea (Vignaunguiculata)[21],potato (Solanumtuberosum)[22],maize (Zeamays)[23]and persian oak (Quercusbrantii)[16]. Nevertheless,as a remarkable tool in detecting genetic diversity and targeting the functional gene of various plant species,the application of the SCOT marker system in alfalfa is still limited.
The sample size of genotypes evaluated by each group plays an important role in the reliability of alfalfa population diversity research. The optimal sample size of each variety should be detected to achieve the optimum between reasonable labor and the reliability of the data set. While there are few reports on the variation of genetic diversity of different alfalfa varieties with the change of sample sizes. To establish an optimal sampling strategy,additional work regarding the optimal number of individual plants is needed. In the current study,the effectiveness of SCoT markers for the analysis of the genetic diversity of alfalfa and the genetic relationships among six alfalfa varieties were examined,with the goals of determining the appropriate number of individuals that should be sampled to reasonably represent the genetic diversity within a population. A focus on this point could be worthwhile to give a more complete judgment on the analysis by SCoT markers in alfalfa,and the detailed results provide an optimized sampling strategy,which is crucial for the genetic diversity investment of alfalfa for breeding purposes.
1 MATERIAL AND METHODS
1.1 Plant Materials and DNA Extraction
In the present study,six alfalfa varieties (‘Archer’,‘BOJA’,‘CUF101’,‘Maverick’,‘Ranger’,and ‘Zhongmu No.1’) were used in the experiment (Table 1). All varieties from a total of 600 individuals were used for the present study. For each of the six populations,the DNA of 100 individuals per population was extracted from young leaves and analyzed with six SCoT markers. A revised cetyltrime thylammonium ammonium bromide (CTAB) method was used for DNA extraction[24]. Then,the DNA quality and concentration were determined using a Nanodrop spectrophotometer (NanoDrop Products,Wilmington,DE,USA). Eventually,the final concentration of each DNA sample was diluted to 50 ng·μL-1and stored at —20℃ for further study.
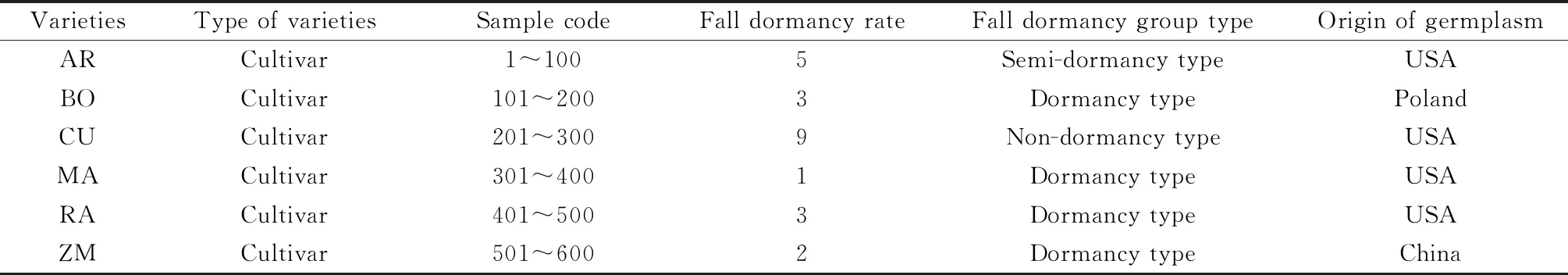
Table 1 Medicago sativa varieties used in the study
1.2 PCR Amplification
In this study,the PCR reactions were all carried out in volumes of 10 μL,containing 50 ng DNA,5.0 μL 2× Power Taq PCR Master Mix (Bioteke,Beijing,China),1.0 μL primer (10 μM) and 2.0 μL double-distilled water. The PCR programs consisted of an initial denaturation at 94℃ for 4 min,followed by 35 cycles of 1 min at 94℃,1 min at 50℃ and 2 min at 72℃,and the final extension at 72℃ for 7 min. After PCR amplification,fragments were separated in 1.4% agarose gel containing 0.14 μg·mL-1of Goldview through electrophoresis in 1× TBE buffer solution at 129 V for 2 h 15 min. DNA fragments were visualized under the ultraviolet (UV) light Gel Doc (TM) XR System (BioRad,Hercules,CA,USA),and photo documentation was obtained for each gel. Fifty-six SCoT primers from previous studies were selected[15,27],and the primers were tested and screened by agarose gel electrophoresis.
1.3 Data Analysis
First,the amplified bands of 6 SCoT markers were scored as present (1) or absent (0),and only reproducible bands were considered. Six hundred sampled individuals (one hundred genotypes from each variety) were used to analyze the genetic diversity of alfalfa using six SCoT markers. A series of random resampling of these data were made according to the 11 sample sizes (10,15,20,30,40,50,60,70,80,90,and 100 genotypes) chosen,with 20 replicates for each resampling. Then the genetic diversity parameters were calculated:Polymorphic information content (PIC) values were calculated according to the formula:PIC=1-p2-q2[28];wherepis the frequency of the present band andqis the frequency of the absent band. The Reserve Percentage (RP),which is an important indicator of the proportion of bands remaining in the sample population was calculated[29]. The resolving power of the primer (Rp) in each variety was measured by Rp=ΣIb[30]. In addition,the number of effective alleles (Ne),Shannon’s information index (I)[31]values,and the expected heterozygosity (He) were calculated with GenAlEx software 6.41[32].
To display variety relationships with different sample sizes,a dendrogram was constructed by Jaccard’s genetic similarity matrix using the unweighted pair group method with arithmetic mean (UPGMA),NTSYS version 2.11[33]. Furthermore,to visualize the population structure of the six alfalfa varieties,Bayesian cluster analysis using STRUCTURE v2.3.4 software[34],with the ‘admixture mode’,burn-in period of 10,000 iterations,and a run of 100,000 replications of Markov Chain Monte Carlo (MCMC) after burn-in,were carried out. For each run,20 independent runs of STRUCTURE were performed with the number of clusters (K) varying from 1 to 10. Maximum likelihood and delta K values were used to determine the optimum number of groups[34,35],and the most likely value of K was computed using STRUCTURE HARVESTER (http://taylor0.biology.ucla.edu/structureHarvester/)[36].
2 RESULTS
2.1 SCoT analysis
Examples of SCoT amplifications were shown in Fig.1,representing band patterns obtained with primers SCoT26. Primers producing banding patterns that were difficult to score and those with inconsistent amplification in all individuals were excluded. Only if the primers can produce clear and repeatable bands can they be used for further study (Fig.1). Finally,six of these primers were chosen for the analysis of genetic diversity parameters in six alfalfa varieties. The oligonucleotide sequences of the 6 SCoT primers were summarized in Table 2. As a result,a total of 80 bands were generated with the six SCoT primers ranging from 6 (SCoT10) to 19 (SCoT3 and SCoT6) with an average number of 13.3 bands per primer in the 600 genotypes of the six alfalfa varieties. The size of the amplified products ranged from 200 bp to 3 500 bp (Table 2). The percentage of polymorphic bands (PPB) of six SCoT primers were all 100%,indicating that the primers had high specificity in these populations. Meanwhile,the resolving power (Rp) of the 6 SCoT primers ranged from 1.89 (SCoT10) to 8.67 (SCoT3). The polymorphic information content (PIC) of six SCoT primers ranged from 0.30 to 0.49,with an average of 0.43. The primer SCoT6 had the lowest PIC (0.30),while the highest PIC was found in primer SCoT37 (0.49) (Table 2). The primers with higher PIC and Rp values have greater potential to investigate more individuals or sampling sites with a small number of primers.
The number of polymorphic bands,the percentage of polymorphic bands,the observed number of bands,the number of effective alleles,the expected heterozygosity,and Shannon’s Information index were calculated for each variety (Table 3). The number of polymorphic bands for varieties ranged from 52 (variety RA) to 70 (variety CU). The highest percentage of polymorphic bands was 100% for variety AR,while the lowest was 94.59% for variety CU. The observed number of bands ranged from 1.325 to 1.800. The number of effective alleles varied from 1.236 to 1.392. The highest Shannon’s Information index was recorded for variety CU (0.364) and the lowest for variety RA (0.232).
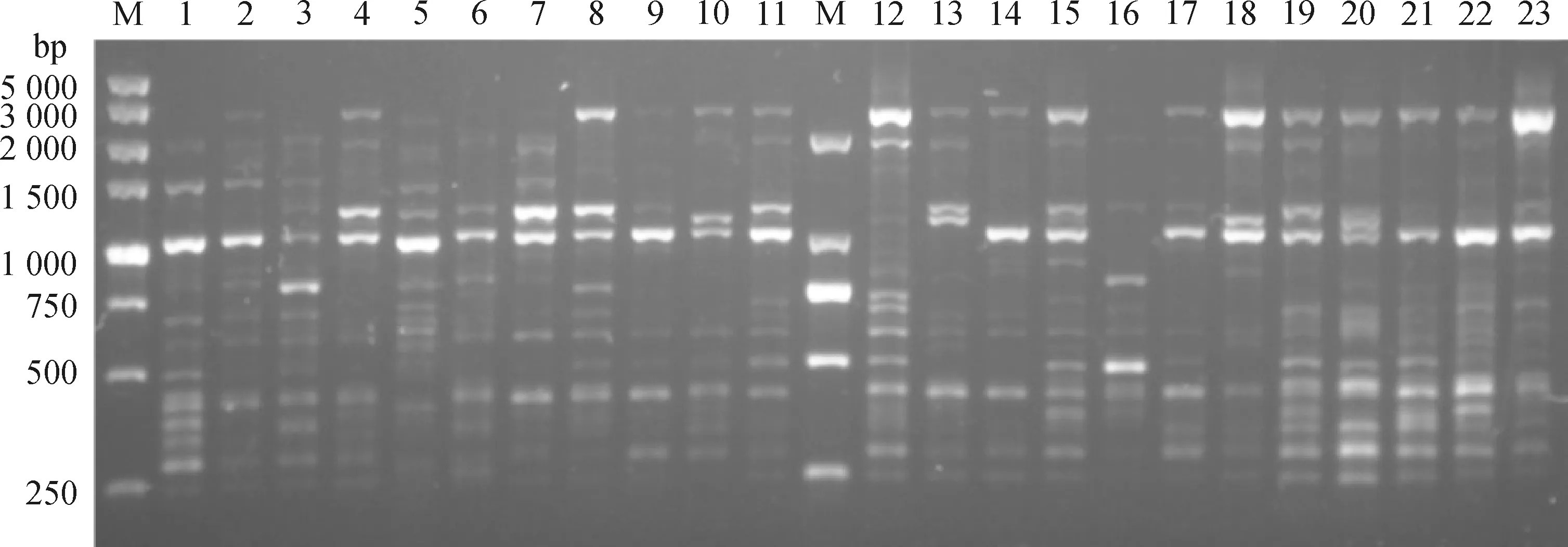
Fig.1 PCR-amplified band pattern generated in SCoT analysis by primers SCoT26 on DNA of 23 alfalfa individuals:molecular marker (100~5 000 bp) (M)

Table 2 Information for the six SCoT primers used in the genetic diversity analysis in six alfalfa varieties
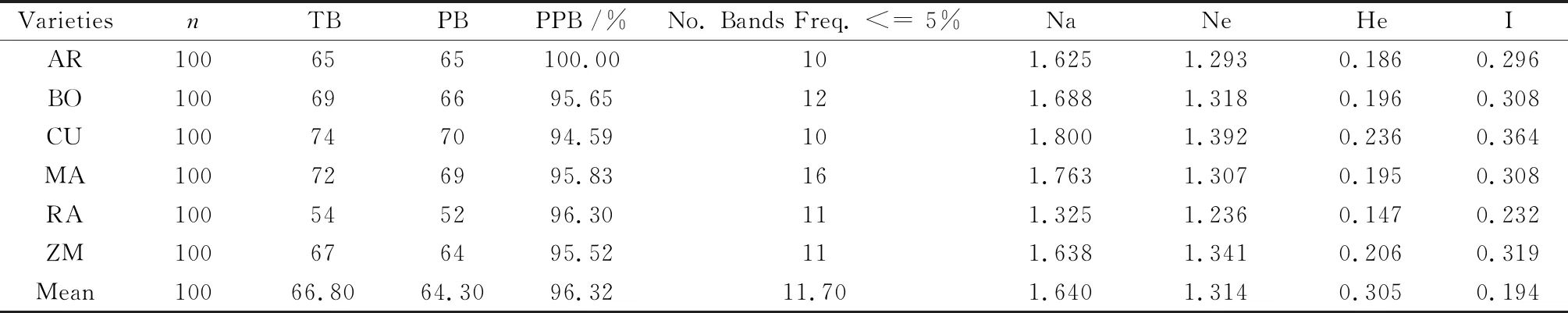
Table 3 Genetic variability within six alfalfa varieties detected by six SCoT markers
2.2 The Genetic Diversity Analysis of SCoT Markers at different sampling size
Understanding the genetic diversity and variation among and within varieties is of great significance for the selection of an effective strategy for conservation and sampling management[37]. As shown in Figure 2,the primers that we selected for this study had high PIC values in different sampling sizes under random strategy in each accession. Furthermore,the analysis results showed that with an increase of the sample size,the TB,Ne,He,and I of the six varieties increased gradually. However,when the sample size increased to a certain number,the value of the genetic diversity of the six germplasms did not increase (Tables 4~7). The Ne value of the six germplasms in different sampling sizes represented more than 95% of the genetic diversity in the total population when the sample size was 10 (Table 5). The I and He parameters increased with an increase in the sample size,and more than 95% of the genetic diversity per population was covered when the sample size exceeded 30 and 40 individuals (Tables 6 and 7,respectively). When the sample size increased up to 40 individuals,the RP value of the six germplasms represented more than 90% genetic diversity (Table 4).

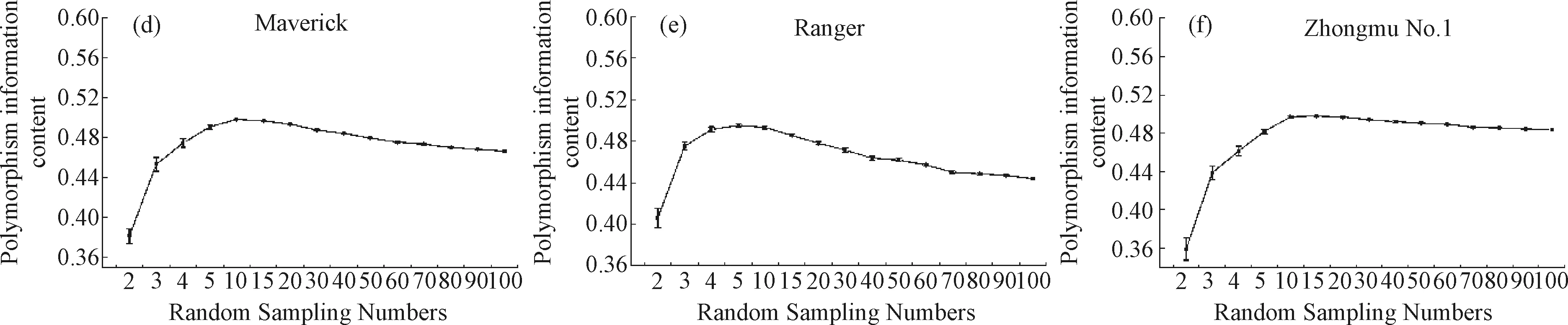
Fig.2 Polymorphism Information Content of the six accessions with different sampling sizes under random strategyNote:(a) ‘ARCHER’;(b) ‘BOJA’;(c) ‘CUF101’;(d) ‘Maverick’;(e) ‘Ranger’;(f) ‘Zhongmu No.1’
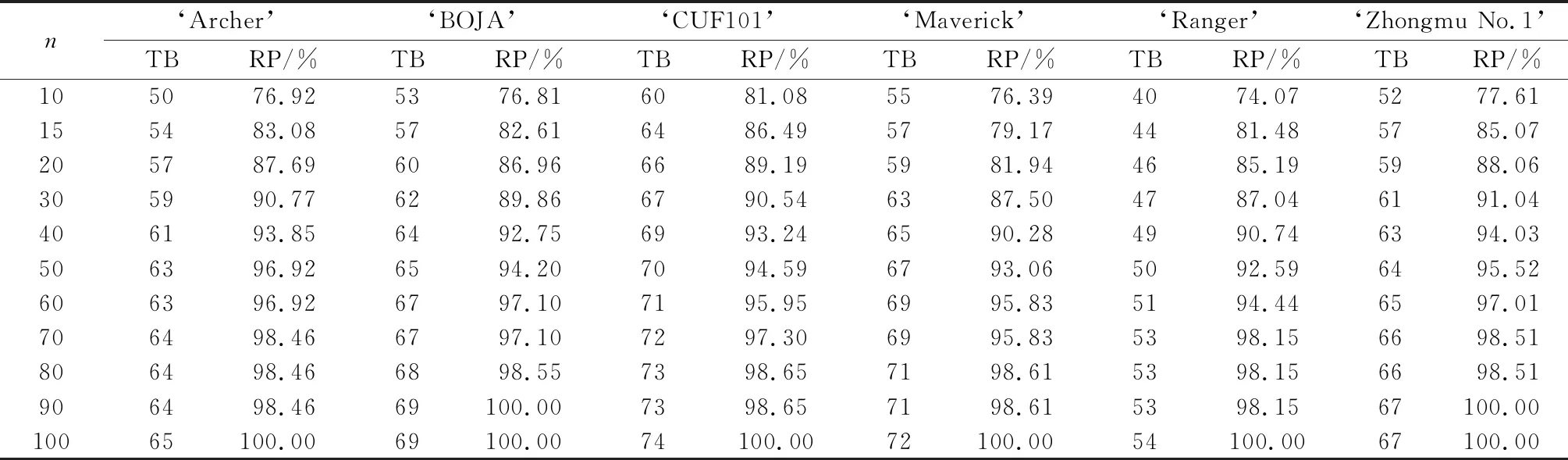
Table 4 The total number of bands (TB) and the Reserve Percentage (RP) with different sampling sizes (n) under random strategy in each variety

Table 5 The Number of Effective Alleles (Ne) and the Percentage of Genetic diversity capture (GC) with different sampling sizes (n) under random strategy in each accession

Table 6 The Shannon information index (I) and the Percentage of Genetic diversity capture (GC) with different sampling sizes (n) under random strategy in each variety
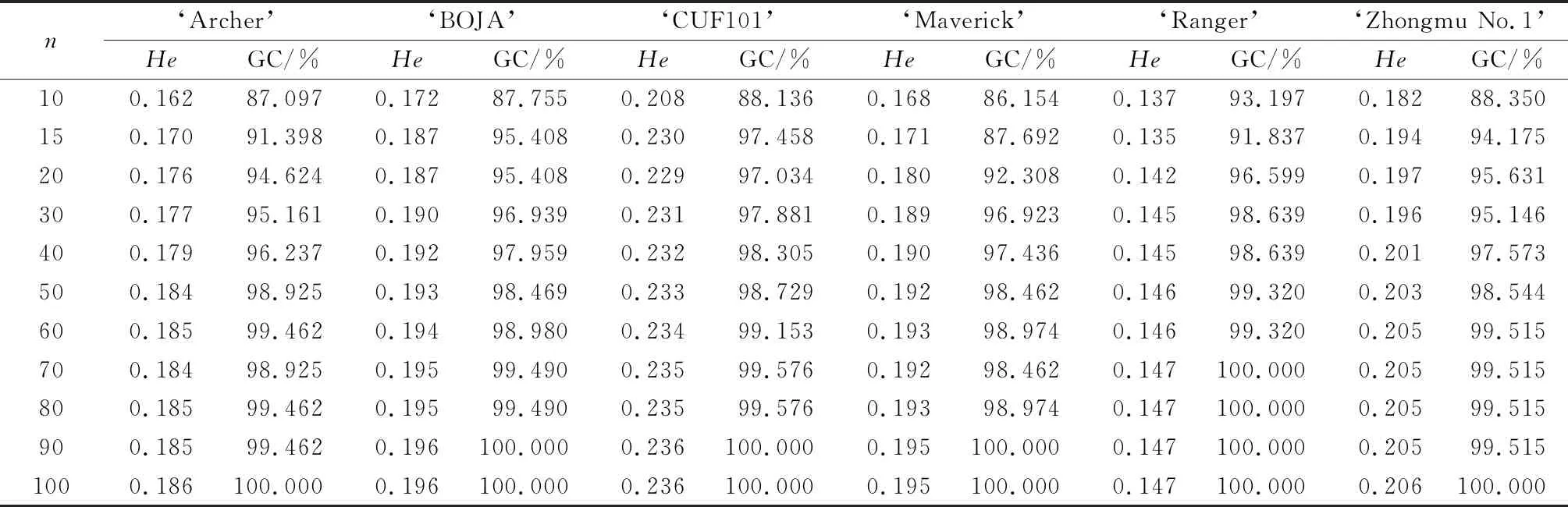
Table 7 The Expected Heterozygosity (He) and the Percentage of Genetic diversity capture (GC) with different sampling sizes (n) under random strategy in each variety
2.3 Cluster and Population Structure Analysis
To investigate the relationships between the different sampling levels,the software of Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and STRUCTURE 2.3.4 was used in this study. The cluster analysis carried out by the UPGMA method based on Jaccard’s genetic similarity,using arithmetic averages cluster analysis,showed that the sampling level of 40 individuals was consistent with that of 100 individuals,where 240 (Fig.3a) and 600 genotypes (Fig.3b) from six varieties were clustered into three major clusters (A-C). Cluster A was further divided into four subgroups. Among the six varieties of alfalfa,‘Archer’,‘BOJA’,‘Maverick’,and ‘Zhongmu No.1’ were assigned to Cluster A. Cluster B included one variety of Ranger,and the remaining variety,‘CUF101’,was assigned to cluster C (Fig.3).
Furthermore,to verify the feasibility of the proposed random sampling results,the genetic structure of the six populations was analyzed using STRUCTURE software based on maximum likelihood and delta K (DK) values for each k,calculated by STRUCTURE HARVESTER according to the DK method of Evanno et al. for 20 simulations[35-36]. The result showed that 240 samples could be divided into three main clusters,which indicated that all 6 varieties were divided into three subpopulations,corresponding to Cluster A,B and C with a few admixed lines,which was consistent with clustering based on genetic distance (Fig.4). Meanwhile,the sampling level of 40 individuals was consistent with that of 100 individuals (Fig.4).
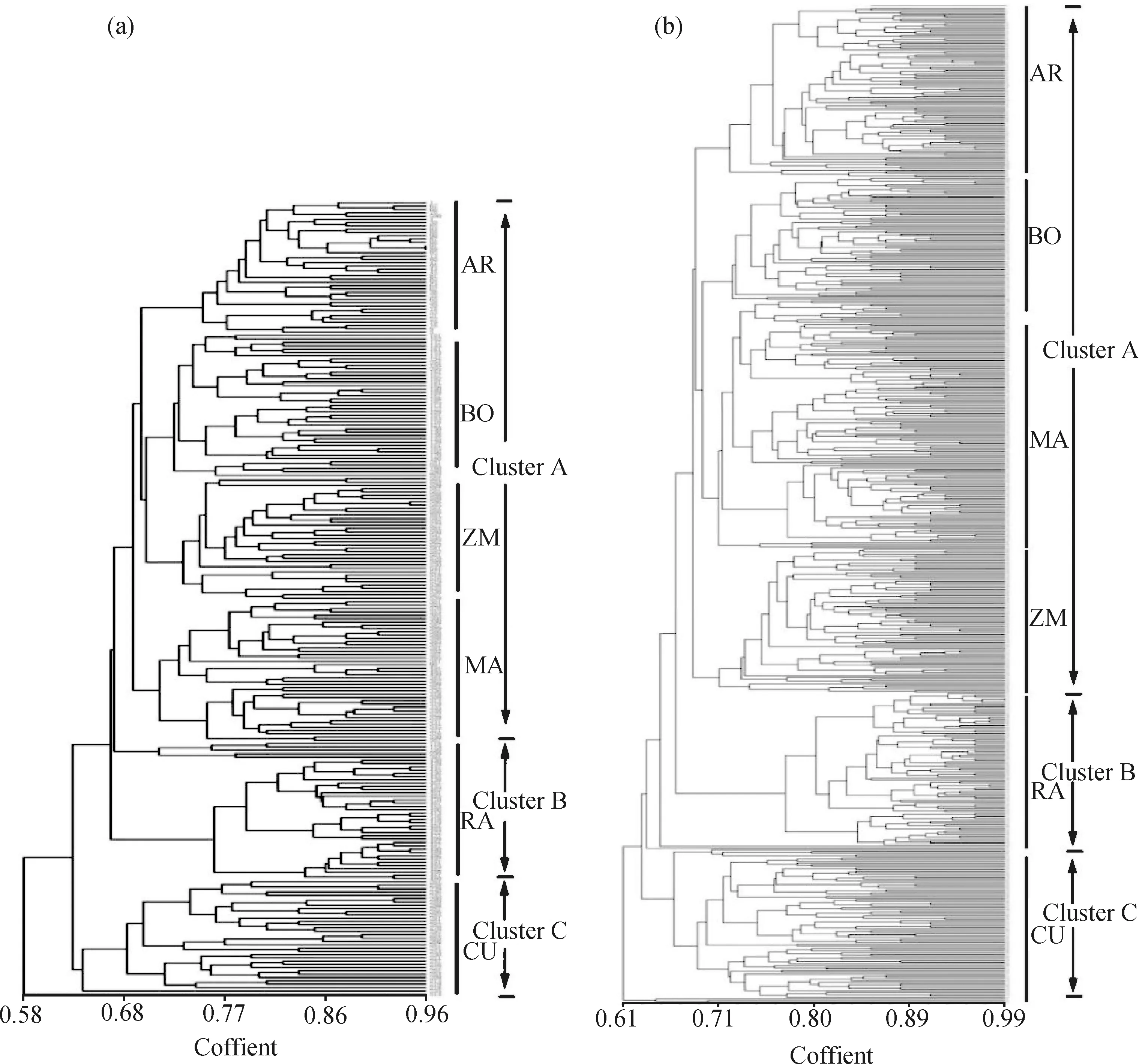
Fig.3 The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) dendrogram of alfalfa based on Jaccard’s genetic similarityNote:(a) 240 individuals from six germplasms;(b) 600 individuals from six germplasms
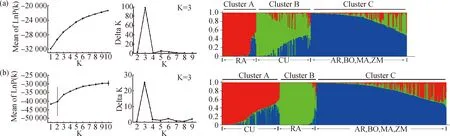
Fig.4 STRUCTURE analysis of the genetic structure of the six alfalfa varietiesNote:Each structure was described by using two graphics of the process to detect the optimum K value. One on the left side was the mean LnP(K) over 20 runs for each K value (1~10),another on the right side was the maximum delta K values used to determine the uppermost level of structure for K ranging from 2 to 9. (a) The genetic structure of 40 individuals for each of the six varieties as inferred by STRUCTURE with the SCoT markers data set;(b) The genetic structure of 100 individuals for each of the six varieties as inferred by STRUCTURE with the SCoT markers data set
3 DISCUSSION
In recent years,DNA-based molecular markers have developed rapidly,and have become a versatile tool in various fields like genetic engineering,plant breeding,and genetic diversity[38]. The SCoT marker system in the present investigation has also been used previously as an effective method of assessing genetic diversity in various plant species. ForE.sibiricusgenetic diversity analysis by using SCoT markers,10.8 bands per primer were generated,89.0% of which were polymorphic[18];Five SCoT markers generated 122 products,what in average gave 24.4 bands per primer. Moreover,the Percentage of Polymorphic Bands (PPB) of the primers were all 100%[27]. Similar observations have also been found in our study,six SCoT primers were used to generate 13.3 bands per primer,and a 100% PPB was obtained across 6 varieties of alfalfa (Table 2);The PPB values (100%) was higher than the previous studies of ISSR variation (PPB=78%)[39],RAPD variation (PPB=82.69%)[40],IRAP variation (PPB=79.73%)[12],and SRAP variation (PPB=49.22%)[41]. Additionally,SCoT markers were applied to alfalfa because of several advantages to other marker assays:lower cost than AFLP,easier development of species-specific primers than SSR,and higher reproducibility than RAPD[42-43]. The results of this study indicated that SCoT markers have enough power to detect the genetic structure and diversity of alfalfa. Meanwhile,the primers showed polymorphisms among six alfalfa varieties,which indicated that this type of molecular marker can distinguish six alfalfa varieties well.
The sample size is an important factor to consider for a reliable investigation of the diversity and differentiation of different plant species. Zhu et al. studied the population sampling strategy ofGlycinesoja,and found that the level of genetic diversity represented 95% of the population when the sample size reached 27 by using He and I like the response criteria. However,when TB was used as the evaluation indicator,the sample size needed to be increased to 52 to achieve 95% of the population’s genetic diversity[44]. In microsatellite studies that assess the genetic diversity ofMelospizamelodia,Pruett and Winker recommend that at samples of less than 40,the number of bands per locus is a poor measure of diversity[45]. These previous researches indicated that there are significant differences in genetic information among different species and the corresponding sample sizes. However,due to its cross-pollination and autotetraploid,the genome structure of alfalfa is very complicated[7,46]. In addition,alfalfa belongs to a species complex (M.sativaspecies complex) that includes diploid and tetraploid interfertile subspecies[6,47],and there is extensive gene flow between these subspecies[48]. Therefore,sampling strategies for self-pollinated and diploid plants,such asG.sojaandViciasativa,cannot provide a reference for alfalfa.
In the current study,we explored the sampling strategy of alfalfa using SCOT markers,and the relationship between sample size and genetic diversity was studied. To our knowledge,this is the first survey on the genetic variability of cultivated tetraploid alfalfa with different sample sizes based on SCoT analysis. Our results showed that the increase in genetic diversity was strongly correlated with an increase in the number of randomly collected samples when TB and the RP values were used as the criterion to estimate the genetic diversity of the six alfalfa populations. As indicated in Table 4,90% of the genetic variation of the germplasms can be achieved by sampling 40 individuals because the rare bands (gene frequencies lower than 5%) accounted for about 13.51%~23.22% of the total bands in each of the varieties. Furthermore,95% of genetic variation of germplasms could be attained if the number of samples increased to 70 individuals. The optimal number of samples in a population should cover more than 95% of the bands with a gene frequency greater than 5%[44]. Therefore,a sample size of 30~40 genotypes could represent 95% of the genetic diversity of all six alfalfa varieties according to the values of I and He.
Accurate genetic characteristics of alfalfa cultivars are necessary for plant breeders to distinguish between two cultivars,even closely related ones. Firstly,the results of UPGMA cluster analysis showed that the six alfalfa varieties could be clustered into three major groups. Cluster analysis with molecular data of 40 plant DNA samples divided the ecotypes into three groups that were consistent with 100 samples. Furthermore,this is also supported by the genetic clusters inferred from STRUCTURE analysis,wherein different varieties were grouped into three distinct clusters. The results showed that the sample size of 40 was sufficient to distinguish different alfalfa varieties.
The accuracy of genetic diversity analysis increases with the samples size taken from the population. However,the cost of genetic analysis increases with the increase in the number of samples. Large scale experiments,cultivar identification in outcrossing heterogeneous species is crucial if a lower labor cost is the target. Based on our results,we can conclude that different sample sizes represent different genetic diversity,40 individual DNA samples is an adequate number to represent most of the genetic diversity within a population,and to differentiate various alfalfa populations. Although the sampling strategy in the present study is based on alfalfa,these results can also be recommended for diversity studies of other tetraploid plant,such as potato,strawberry,and sugarcane.
4 CONCLUSIONS
In summary,SCoT markers in the present study proved useful in evaluating the relationship among different alfalfa cultivars,which showed a high level of polymorphism. At different sample sizes,the genetic variation and genetic relationships among six populations of alfalfa were efficiently determined using SCoT markers. A sample size of 40 individuals can cover more than 95% of the variation of a sample of 100 individuals based on the data provided. Besides,the among population distinctiveness is more evident for the two varieties with opposite fall dormancy (Ranger in cluster B and CUF 101 in cluster C) and less clear cut in cluster A grouping varieties with intermediate fall dormancy coming from different geographical origin.

