Brain delivering RNA-based therapeutic strategies by targeting mTOR pathway for axon regeneration after central nervous system injury
Ming-Xi Li, Jing-Wen Weng, Eric S. Ho, Shing Fung Chow, Chi Kwan Tsang
Abstract Injuries to the central nervous system (CNS) such as stroke, brain, and spinal cord trauma often result in permanent disabilities because adult CNS neurons only exhibit limited axon regeneration.The brain has a surprising intrinsic capability of recovering itself after injury. However, the hostile extrinsic microenvironment significantly hinders axon regeneration. Recent advances have indicated that the inactivation of intrinsic regenerative pathways plays a pivotal role in the failure of most adult CNS neuronal regeneration. Particularly, substantial evidence has convincingly demonstrated that the mechanistic target of rapamycin (mTOR) signaling is one of the most crucial intrinsic regenerative pathways that drive axonal regeneration and sprouting in various CNS injuries. In this review, we will discuss the recent findings and highlight the critical roles of mTOR pathway in axon regeneration in different types of CNS injury. Importantly, we will demonstrate that the reactivation of this regenerative pathway can be achieved by blocking the key mTOR signaling components such as phosphatase and tensin homolog (PTEN). Given that multiple mTOR signaling components are endogenous inhibitory factors of this pathway, we will discuss the promising potential of RNA-based therapeutics which are particularly suitable for this purpose, and the fact that they have attracted substantial attention recently after the success of coronavirus disease 2019 vaccination. To specifically tackle the blood-brain barrier issue, we will review the current technology to deliver these RNA therapeutics into the brain with a focus on nanoparticle technology. We will propose the clinical application of these RNA-mediated therapies in combination with the brain-targeted drug delivery approach against mTOR signaling components as an effective and feasible therapeutic strategy aiming to enhance axonal regeneration for functional recovery after CNS injury.
Key Words: axon sprouting; axon regeneration; brain targeted drug delivery; CNS injury; ischemic stroke; mTOR; nanoparticle; neural circuit reconstruction; PTEN; RNA-based therapeutics
Introduction
It is well known that the central nervous system (CNS) injury triggers spontaneous recovery. For instance, ischemic stroke alters the expression of genes that stimulate the growth of axons and dendrites, followed by circuit reconnection and synaptogenesis, pruning, and functional recovery (Cramer,2018). These molecular events often occur in the perilesional tissues, related ipsilateral areas and functionally related contralateral areas, and spinal cord (Carmichael et al., 2017). The nervous system has amazing plasticity,especially in the peripheral nervous system. Unfortunately, spontaneous recovery is often incomplete, leaving many patients with long-term disability.Axon regeneration after CNS injury is very limited because of the extrinsic and intrinsic factors. Extrinsic factors consist of the glial scar, myelin debris,and glia-secreted growth-suppressive molecules such as Nogo and myelinassociated inhibitors (He and Jin, 2016). Previous works have demonstrated that blocking these extrinsic inhibitory factors is insufficient to trigger axon regeneration after axotomy in the CNS. Intriguingly, recent findings have uncovered the pivotal role of intrinsic factors in stimulating axon regeneration in various CNS injury models (Fawcett, 2020). Among them, the mechanistic target of rapamycin (mTOR) pathway has been convincingly demonstrated to be one of the most important intrinsic regenerative pathways in almost all major CNS injuries such as stroke, brain, and spinal cord damages. In this review, we will discuss the recent findings of the mTOR pathway in association with axon regeneration and identify the promising targets after CNS injury. In addition, we will discuss the potential application of RNA-mediated strategy for these targets. Finally, we will review the recent advance concerning the novel brain delivery technology to facilitate the clinical application of these strategies.
Database Search Strategy
In this narrative review, we performed the NCBI PubMed database search for identifying English-language articles that reported the role of mTOR regenerative pathway in axon regeneration after CNS injury, RNA-based therapy, and the brain-targeted drug delivery from 2001-2021. Search terms included CNS injury, mTOR, axonal regeneration, axon sprouting, RNAmediated therapy, nanoparticles, brain-targeted delivery, ischemic stroke,spinal cord injury, retinal ganglion cell (RGC) repair, and traumatic brain injury. The authors also included primary research papers, case reports, and review articles. The titles and abstracts for inclusion were independently reviewed by the authors. Additional relevant articles were also identified from the referenced citations. For the DrugBank search, “siRNA” was the term used. For the search of clinical trials, we narrowed down to “Interventional studies” where “Intervention” is “siRNA”, and “Status” included completed and all kinds of recruiting modes. We filtered only “Interventional studies”where “Intervention” is “Antisense oligonucleotide”, and “Status” included completed and all kinds of recruiting modes.
Axonal Regeneration after Central Nervous System Injury
CNS injury after stroke induces a rapid loss of neurons and axons which accounts for the loss of nerve connections during the acute phase and subsequently induces various degrees of plasticity during the spontaneous recovery phase (Cramer, 2018). At the cellular level, stroke-mediated injury induces axon sprouting, dendritic branching, and synaptogenesis for remapping of neural circuits and re-construction of the lost connectivity(Carmichael et al., 2017). Axonal sprouting is fundamental to the spontaneous recovery after injury as the sprouting axons could establish new connections around the peri-infarct regions, or in areas in different lobes in the ipsilateral and contralateral hemispheres, depending on the size and location of the infarct. Axonal sprouting can also occur from the contralateral cortex to the de-afferent side of the cervical spinal cord (Carmichael et al., 2017). In contrast to stroke, most injured axons resulting from axotomy or trauma in the adult brain or spinal cord do not show significant spontaneous regeneration (He and Jin, 2016). Multiple challenges exist such as the formation of new growth cones at the severed axon stumps, polarization guided by the growth cones, and the requirement of axon extension over a long distance to reconnect with their targets. One of the most difficult hurdles is the synthesis of building materials such as cytoskeletons, neurofilaments,and plasma membrane in the regenerating axons (He and Jin, 2016). However,after the developmental stage, the activity of the intrinsic regenerative pathway is often diminished to avoid cellular overgrowth in the adult stage.Based on the reasoning above, the group of He examined the evolutionary conserved molecular pathways which may contribute to the diminished regenerative capacity in adult CNS neurons (Park et al., 2008). By using the genetic knockout approach in the adult retinal ganglion cells (RGCs) and optic nerve injury model, they examined the effect of knocking down the conserved growth control genes including Rb, P53, Smad4, Dicer, LKB1, and PTEN on axon regeneration. Surprisingly, they found that deletion of PTEN alone is sufficient to promote robust axon regeneration without manipulation of extrinsic factors in the lesion site (Park et al., 2008). They further confirmed that knockdown of TSC, another negative regulator of mTOR pathway, also produces a similar effect. Importantly, they showed that mTOR activity is suppressed in the axotomized RGCs of wild-type adult mice (Park et al., 2008).These results convincingly demonstrate that mTOR signaling is the intrinsic axon regenerative pathway and that reactivation of this pathway by targeting its negative signaling components can promote axon regeneration after adult CNS injury (Park et al., 2008). Since this pioneering work, substantial followup studies from different laboratories have confirmed the key role of mTOR pathway in axon regeneration in various CNS injury models (He and Jin, 2016;Al-Ali et al., 2017; Huang et al., 2019; Williams et al., 2020; Bhowmick and Abdul-Muneer, 2021; Ma et al., 2021).
In addition to axon regeneration, the administration of human recombinant insulin has been reported to fully restore mTOR activity in injured RGC which is accompanied by robust regeneration of dendrites and activation of synaptogenesis (Agostinone et al., 2018). By knocking down Raptor, the specific and essential mTORC1 component, the authors further demonstrated that insulin-dependent mTOR activation is required for restoration of dendritic branching, arbor complexity, and re-establishment of the field area. These findings indicate that reactivation of mTOR pathway can promote regrowth of dendrites and synaptogenesis, and restores circuit function after traumatic CNS injury as well as in neurodegenerative diseases such as glaucoma. After a stroke, the peri-infarct cortex responds by triggering axonal sprouting that leads to new functional connections (Carmichael et al., 2017). It has also been reported that the dendritic morphology changes in regions of axonal sprouting in the peri-infarct cortex (Agostinone et al., 2018). Since dendrites can be remodeled after the loss of afferent innervation, the remodeling dendrites and the spared or sprouting neurons could promote synaptogenesis. A series of seminal works performed by Carmichael’s group has shown that this limited capacity can be significantly increased by stimulating the intrinsic neuronal regrowth program in the motor, somatosensory, premotor cortex, and motor corticospinal projections in the cervical spinal cord (Carmichael et al., 2017).Not surprisingly, mTOR pathway is an ideal target for promoting stroke recovery as it has pleiotropic effects on multiple cellular processes involved in the neuronal repair. Physiologically, the peri-infarct region and white matter penumbra often experience partial ischemia which creates the inhibitory microenvironment for restricting axon regeneration. We have shown that ischemia induces mTOR inactivationin vitro(Tsang et al., 2018). It has also demonstrated that mTOR activity is significantly reduced in the peri-infarct tissue in a rat model of permanent distal middle cerebral artery occlusion(Xie et al., 2013). These results suggest that upregulation of mTOR pathway activity is a promising strategy for promoting axon regeneration and sprouting.Indeed, recent studies have provided compelling evidence supporting the role of mTOR pathway in axon sprouting after stroke (Li et al., 2015; Carmichael et al., 2017; Liu et al., 2017; Zareen et al., 2018; Chen et al., 2019). Liu et al.(2017) showed that post-lesional adeno-associated virus (AAV)-assisted coexpression of insulin-like growth factor (IGF1) and osteopontin in cortical neurons caused a robust regrowth of the corticospinal tract (CST) and the improved behavioral performance using a unilateral cortical stroke model. As IGF1 is a well-known activator of mTOR pathway, these results demonstrate that mTOR activation may promote corticospinal axon regrowth after ischemic stroke in adults for recovery. Given that the spontaneous CST axon sprouting after stroke injury is often insufficient to completely recover the lost function, the electrical stimulation at the motor cortex is known as a strategy to promote CST axon sprouting. It has been shown that chronic electric stimulation shifts the equilibrium of PTEN to its inactive phosphorylated form in the CST system and upregulates mTOR and Jak/Stat signaling activity which leads to enhanced CST sprouting and presynaptic connection (Zareen et al., 2018). Li et al. (2015) also reported that GDF10 expression is induced in the post-stroke peri-infarct brain tissue, where axonal sprouting occurs.GDF10 induces downregulation of PTEN and upregulates PI3K signaling that promotes specific axonal guidance (Li et al., 2015). The same group further used the isolated neurons that sprout a new connection in the cortex after stroke in aged animals and compared their transcription profile to that of the young stroke animals. This sprouting transcriptome revealed that IGF1 is uniquely induced in aged sprouting neurons, along with its downstream signaling molecules (Carmichael et al., 2017). These studies suggest that the axon-sprouting competent neurons require IGF1 and its downstream signaling such as mTOR pathway for the induced axon regeneration in aged individuals after stroke. Interestingly, activation of mTOR pathway by ginsenoside Rg1 can also promote angiogenesis in ischemic mice (Chen et al., 2019).
Possible Mechanisms of mTOR-Mediated Axon Regrowth
mTOR integrates diverse upstream signals including neurotrophic factors,neurotransmitters, growth hormones, nutrients and energy levels, and stress stimuli to regulate various aspects of growth, survival, metabolism, and cellular homeostasis (Tsang et al., 2007). mTOR forms two complexes, mTOR complex 1 (mTORC1) and mTORC2, with shared and distinct components.mTORC1 pathway is highly sensitive and inhibited by rapamycin and its derivatives, while mTORC2 is less sensitive to rapamycin treatment (Liu and Sabatini, 2020). Since most of the mTOR functions associated with CNS repair are found in mTORC1, we will focus on mTORC1 in this review.
mTORC1 promotes protein synthesis, cell growth, and proliferation by activating two well-characterized downstream substrates, S6K and 4E-BP,which are the key regulators of mRNA translation and protein synthesis (Liu and Sabatini, 2020) (Figure 1). As a well-studied mTOR target, S6K1 has been suggested as a positive effector of axon outgrowth. However, a recent study has challenged this idea by showing that PF-4708671, a selective S6K1 inhibitor, stimulates corticospinal tract regeneration and increases axon density beyond the injury site, and produces significant locomotory recovery(Al-Ali et al., 2017). Therefore, not all the positive effectors of mTOR could produce similar axon growth-promoting effects. We and others have recently found that mTORC1 regulates MAF1 and superoxide dismutase 1 (SOD1) as two immediate downstream effectors that mediate the key cellular processes including the biogenesis of ribosomes and translation machinery, metabolism,and redox homeostasis (Tsang et al., 2018; Willis, 2018; Wang et al., 2021).Although the precise mechanism by which mTOR pathway regulates axon regeneration remains unclear, a large body of evidence has suggested that mTOR pathway is intimately linked to axon regeneration after CNS injury through the following downstream regulations.
Synthesis of Building Blocks Essential for Axon Repair
In non-neuronal cells, mTOR has been implicated as a central regulator of anabolism such as protein, nucleotides, and lipid biosynthesis (Liu and Sabatini, 2020). It is worth noting that the ribosome is the ‘protein-production factory’ that translates mRNA codes into polypeptides, and thus ribosome availability determines the protein synthetic capacity of a cell (Tsang et al.,2007). We recently found that Sod1 is a conserved downstream effector of mTORC1 which regulates Sod1 activity in a nutrient-dependent manner(Tsang et al., 2018). A follow-up study further revealed that Sod1 regulates ribosome biogenesis and growth of Kirsten rat sarcoma viral oncogene homolog (KRAS)-driven lung cancer (Wang et al., 2021), suggesting that mTOR may fine-tune the rate of ribosome biogenesis via Sod1. As Sod1 catalyzes the conversion of superoxide to H2O2, it has been reported that Sod1 plays a role in the recovery of a mouse model of closed head injury (Beni et al.,2006). Interestingly, a recent study using unbiased mutagenesis screening inC. elegansdemonstrated that regulation of the biogenesis of ribosomes and translation machinery is coupled and crucial to hypoxic sensitivity (Itani et al., 2021). Although the detailed mechanism remains to be determined,neuronal mTOR is likely to play a critical role in ribosome biogenesis which is the determining factor for new protein synthesis and axon regrowth (Twiss and Fainzilber, 2009). Notably, a neuron should generate a large amount of ribosomes and other translation machinery to meet the demand for a high protein production rate for supporting the long-distance axon regeneration.We and others have identified that MAF1 is a conserved downstream effector of mTOR for regulation of RNA polymerase III-dependent transcription which controls the synthesis of 5S ribosomal RNA and transfer RNAs which are the essential components of ribosomes and translation machinery (Li et al., 2006;Wei et al., 2009; Michels et al., 2010; Willis, 2018) (Figure 1). We further found that MAF1 binds to the PTEN promoter to enhance PTEN promoter acetylation and increase PTEN expression (Li et al., 2016). Therefore,upregulation of mTOR pathway is likely to promote mTOR-mediated biogenesis of ribosomes and transcription machinery in support of axon regeneration.
Regulation of Local Protein Translation
mTOR has a well-known effect on cap-dependent protein translation which has been implicated in local protein synthesis in non-neuronal cells (Liu and Sabatini, 2020). Local biosynthesis is a plausible solution for the neurons to reduce energy-costly axonal transport from the cell bodies. Local protein synthesis mediated by mTOR activity in axons has been reported to affect pre-synaptic plasticity in the mature mammalian brain (Younts et al., 2016).A more recent study further demonstrated that mTOR activity is required for local translation, including its mRNA and the retrograde injury signaling molecules such as importin β1 and STAT3, in injured axons (Terenzio et al.,2018). Maintenance of the axonal pool of mTOR mRNA enables rapid and local upregulation of protein synthesis in injured axons. These results indicate that mTOR pathway plays a key role in the regulation of local translation and injured response of axon.
Growth cones form the axon projections that are required for establishing neural circuits during neurodevelopment and neural repair after injury. A recent study, using the growth cone sorting and subcellular RNA-proteomic mapping techniques, analyzed local transcriptomes and proteomesin vivoin the developing axon projections in the cerebral cortex (Poulopoulos et al., 2019). This study found that mTOR and the mRNAs that contain mTORdependent motifs for their translation are accumulated in these growth cones of developing projections, suggesting that mTOR-dependent local protein translation plays a crucial role in axon growth. Intriguingly, the authors identified that approximately 80% of all significantly growth cone-enriched transcripts contain the 5′ terminal oligopyrimidine (TOP) motif. The expression of TOP-transcripts, which include the ribosomal proteins and translation initiation factors, is tightly coupled to cell growth. Importantly, their expression is under direct control of mTOR (Tsang et al., 2007). The knockout approach was further used to demonstrate that mTOR signaling is necessary for trans-hemispheric axonal growthin vivo(Poulopoulos et al., 2019). These studies place mTOR and its pathway, and the supply of cellular translation machinery, at the leading edge of growing long-range axon projections,suggesting that mTOR plays an important role in neural repair after injury.
As mTOR is a key sensor for monitoring various pathophysiological conditions,the stress environment after stroke or traumatic CNS injury diminishes this important intrinsic regrowth pathway. The encouraging evidence discussed above raises the exciting prospect that reactivation of mTOR pathway can drive axonal regeneration following injury. As shown in Figure 1, the RNAmediated therapeutic strategies such as antisense oligonucleotide (ODN)by knocking down the negative regulators in mTOR pathway would be a promising approach. Since there are many negative inhibitory factors in the mTOR pathway, knocking down these factors can upregulate mTOR pathway activity, making the RNA-mediated knockdown approach particularly attractive. For this reason, we will focus on the use of RNA-based therapeutics and discuss the feasibility of targeting them as a promising therapeutic strategy for promoting neural repair after CNS injury.
RNA-Based Therapeutics
There are three classes of RNA-based therapeutics: antisense oligonucleotide(AS-ODN), RNA interference (RNAi-ODN), and aptamer. Here we will highlight the distinctive features of AS-ODN and RNAi-ODN. Readers who are interested in more information about them are recommended to refer to review articles devoted to each of these therapeutic platforms (Sherman and Contreras,2018; Setten et al., 2019; Roberts et al., 2020). The overarching goal of RNA-based therapeutics is to intervene disease pathways by modulating fundamental RNA-associated processes, including splicing, polyadenylation,protein translation, and mRNA degradation, through sequence-specific binding of oligonucleotides to targeted pre- or mature messenger RNAs (premRNAs or mRNAs).
Traditional small-molecule approaches achieve therapeutic outcomes by interfering the functions of disease-associated targets, usually proteins,using small organic drug compounds that interact with them. Such an approach requires the existence of viable drug binding pockets or receptors in the targets, which are hard to determine or even non-existent, leading to undruggable gene targets, e.g. oncogene KRAS (Kessler et al., 2019;Moore et al., 2020). RNA-based therapeutics, unlike small-molecule drugs that require structural information of the binding sites, derive targets from genomic sequence information, which is readily and cheaply available due to the advance of high throughput DNA sequencing with ever-decreasing cost.Choosing the unique signature sequence of a target’s mRNA requires much less effort than determining the optimal binding site of a target protein that fits the drug molecule. Sequence-based targeting also boosts RNA-based therapeutics the capability to differentiate wild-type and mutant targets.Recently, diseases have been reported to be associated with non-coding RNAs (Statello et al., 2021; Scicluna et al., 2020). In the absence of protein products, it further enhances the application of RNA-based therapeutics.Importantly, the manufacturing process and delivery platform of RNA-based therapeutics for various therapeutic areas, such as vaccines, cancer, etc., are chiefly identical, greatly streamlining effort in pharmacokinetics study. That said, the delivery of ODNs to a specific organ or tissue remains a challenging task (Didiot et al., 2016; Mignani et al., 2019; Tai, 2019). More details will be discussed below.
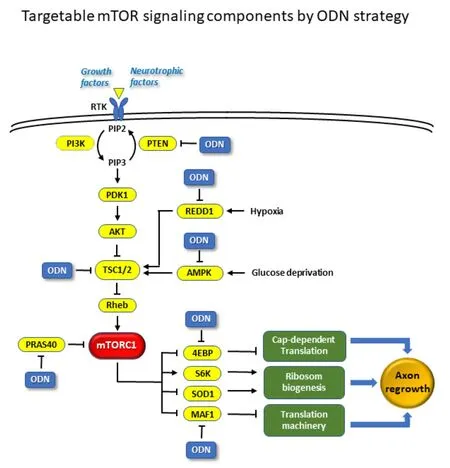
Figure 1 |A simplified diagram showing the mechanistic target of rapamycin(mTORC1) signaling pathway and the potential targets by RNA-mediated strategy such as the anti-sense oligonucleotide (ODN).Inhibition and activation are denoted by “⊥” and “↓”, respectively. 4EBP: Eukaryotic translation initiation factor 4E binding protein; AKT: a serine/threonine protein kinase encoded by the oncogene in the transforming retrovirus isolated from the thymoma cell line AKT-8, which is derived from the Stock A Strain k AKR mouse originally inbred in the laboratory of Dr. C. P. Rhoads by K. B. Rhoads at the Rockefeller Institute/protein kinase B; PDK1: protein 3-phosphoinositide-dependent protein kinase-1; PI3K: phosphoinositide 3-kinase; PIP2: phosphatidylinositol biphosphate; PIP3: phosphatidylinositol trisphosphate; PRAS40: proline-rich Akt substrate of 40 kDa; PTEN: phosphatase and tensin homolog; Rheb: ras homologenriched in brain; RTK: receptor tyrosine kinase;S6K: ribosomal protein S6 kinase; SOD1: superoxide dismutase 1; TSC: tuberous sclerosis complex.
Antisense Oligonucleotides
AS-ODN utilizes two independent mechanisms to achieve gene silencing:RNaseH-dependent pathway (Wu et al., 2004) and steric hindrance (Baker et al., 1997). AS-ODN is a single-strand nucleotide sequence of length 20-30 bases long where its sequence is antisense of the target mRNA. Details of binding site selection will be discussed in the next section. If RNaseH degradation mechanism is used, AS-ODNs are often, but not required,constituted of RNA and DNA nucleotides where two RNA segments are ligated at the two termini of a DNA segment, which is around ten bases long. The advantage of incorporating RNA nucleotides in the AS-ODN is to enhance binding affinity with the target mRNA (Monia et al., 1993), lowering the dosage requirement. To further enhance binding affinity and attenuate adverse immunological effects, modified RNA nucleotides are used. A ribonucleotide consists of three biochemical groups: a nucleobase, a ribose,and a phosphate group. There are chemical modifications on these groups that can boost stability by enhancing Watson-Crick base pairing (Geary et al.,2015). The modifications of the ribose include 2′-O-methyl, 2′-O-methoxyethyl,2′-fluoro, locked RNA (LNA), constrained ethyl bridged nucleic acid, and ethylene-bridged nucleic acid (Wan and Seth, 2016). For the nucleobase,the common modification is the 5-methyl pyrimidine. The modifications of the phosphate group that change the phosphodiester linkage between nucleotides are usually referred to as backbone modifications, which are achieved by phosphorothioate (Eckstein, 2014).
Once the target mRNA is bound by an AS-ODN, forming a double-stranded RNA-DNA hybrid that is recognizable by endogenous endonuclease RNaseH.Upon RNaseH binding, the RNA-DNA hybrid will be cleaved and, eventually,degraded. It is well established that RNaseH functions in the nucleus, but studies have shown that degradation can occur in the cytoplasm as well(Vickers and Crooke, 2014; Liang et al., 2017), easing delivery efficiency.
An alternative AS-ODN mechanism is achieved by imposing steric hindrance.Post-transcriptional RNA modifications that have been altered for therapeutic purposes are splicing (Lim et al., 2020), polyadenylation (Lee and Mendell,2020; Naveed et al., 2021), and 5′ UTR regulation (Sasaki et al., 2019). It is noteworthy that this approach is independent of RNaseH whereby the target mRNAs are not degraded, at least, not directly triggered by AS-ODN binding.At the time of writing, nine AS-ODN therapeutics have been approved by the U.S. Food and Drug Administration (FDA), and 19 are still under development,according to DrugBank (https://go.drugbank.com/) (Wishart et al., 2018)(Additional Table 1). Of the nine approved AS-ODNs, the only neurological drug is Intorsen (ATC N07XX15). Besides, ClinicalTrials.gov (ClinicalTrials.gov, 2021) shows 12 active interventional trials (Additional Table 2). We filtered only “Interventional studies” where “Intervention” is “Antisense oligonucleotide”, and “Status” included completed and all kinds of recruiting modes. The only neurological intervention trial undergoing is for Huntington’s disease. The scarcity of neurological AS-ODNs may reflect the challenge for ODN-based pharmaceutical products to cross the blood-brain barrier (BBB).
RNA Interference
Another class of RNA-based therapeutics is small interference RNA (siRNA)that harnesses the RNA interference (RNAi) cellular pathway. siRNA is a double-stranded RNA molecule. Each strand of the siRNA is 21 bps long, but the two strands only hybridize at positions 1 to 19, leaving two overhanging bases at the 3′ termini. One strand of a siRNA is named the guide strand or antisense strand, which is the strand that is utterly complementary to the target transcript. The other strand is called the passenger strand or sense strand in which no active role is played by it in the RNAi pathway (Elbashir et al., 2001 ).
siRNAs are first processed by Argonaute2 (ARGO2) in the cytoplasm. ARGO2 will remove one strand from the double-stranded siRNA molecule, forming the RNA-induced silencing complex (RISC). Whether the removed strand by ARGO2 is the guide strand or the passenger strand is arbitrary (Gregory et al., 2005). Further studies are needed to elucidate the discriminating factors underpinning the choice of strand made by ARGO2. From the therapeutic perspective, measures must be taken to safeguard no off-target effect can be induced by the passenger strand even though it does not interact with the target mRNA. RISC utilizes the sequence specificity of the guide strand to recognize the target transcript. Upon binding, enzymatic slicing activity of ARGO2 (Roberts, 2015) will be activated to cleave the target transcript at a position near the middle of the guide strand, i.e., between positions 10 and 11 (Schurmann et al., 2013), diminishing the biological functions of the transcript.
Similar to AS-ODNs, selected RNA nucleotides of the siRNA are substituted by modified RNAs such as 2′-O-methyl. Regarding the proportion and position of the modified RNAs that can achieve optimal results, it is beyond the scope of this review.
Five siRNA therapeutics are found in DrugBank, in which two have been approved by FDA (Additional Table 3). ClinicalTrials.gov indicates 20 active interventional trials (Additional Table 4). Thus far, siRNA therapeutics have yet to be developed in the neurological areas.
Workflow for Bioinformatic Identification of Target Sites
Figure 2 summarizes a multistep process for identifying putative target sites of a gene target. The process begins with the gene target postulated to achieve the therapeutic goal. The next step is to identify potential binding sites on the mature or pre-mRNA that are accessible to the ODNs. In this step, all cDNAs (isoforms) of the normal or mutated target gene will be retrieved from biological databases, such as NCBI RefSeq (O’Leary et al., 2016) and ENSEMBL(Howe et al., 2021). It is recommended to obtain isoforms from at least two databases for cross-validation purposes. More details will be discussed below.If the target is the mutant form of the transcript, the oligonucleotide target site must include the mutated sequence instead of the wild-type counterparts. If the type of mutation is insertion, the junctions of insertion become the signature sites of the target. For deletion, the flanking regions of the deleted segment are the only possible place to bind the ODN without interfering with the wild-type transcripts.
If the target is a specific unmutated isoform, all isoforms of the target gene must be aligned to figure out the isoform-specific regions. It is noteworthy that the presence of such region(s) is not guaranteed. Furthermore, the target regions should be free from genetic variations. Tools such as Ensembl Variant Effect Prediction (VEP) (McLaren et al., 2010) and BioMart (Smedley et al.,2015) are helpful to uncover such variations.
To streamline testing and avoid confounding factors, it is ideal to have a single set of ODNs to be tested in animal models, such as in mice, and in humans.Previous studies have shown many ultra-conserved regions were discovered among diverse species, e.g., humans, rats, mice, cows (Bejerano et al., 2004;Ho and Gunderson, 2011).
In the next step, the long target regions will be split into overlapping short sequences (potential target sites) with their length equal to the required length of the RNA therapeutic platform. The size of target sites ranges from 9 to 20 bases with siRNA near to the low-end and AS-ODN near to the high-end.After that, we will examine the biochemical propensities of each potential target site, such as GC%, melting temperature, and self-annealing (or selfdimerization) potential.
Those potential target sites that have fulfilled the above criteria will be checked further for sequence specificity. Word-based alignment tools such as BLASTN (Sayers et al., 2021) can be used to search against the genome and transcripts. It is noteworthy that as target sites are usually short, word-based alignment algorithms tend to bias perfect matches in the middle of a target site sequence while leaving mismatches and/or gaps at the two termini.Besides searching for homologous sites in transcripts, it is advisable to expand the radar to the whole genome as homologous sites present, for example, in introns and splice site junctions may attenuate unintended normal splicing process.
Once the specificity of candidate target sites has been warranted, their accessibility to ODNs will be assessed. Here we will focus on RNA secondary structures and RNA binding proteins (RBPs). A valuable data source for RNA secondary structures is RNA Atlas of Structure Probing or RASP (Li et al.,2021), which has compiled a large number of relevant experimental data generated by different techniques on various cell lines and species, including humans, mice, plants, bacteria, fungi, and viruses. Besides data browsing, the RASP portal also caters to large volume data download via their programmatic interface. RASP’s data is formatted in the popular BED format. Further options are provided for users to download data from a specific species, experimental technique, cell line, data type (which means scoring method), etc. With the RNA secondary structure data, candidate target sites can be assessed if significant secondary structures can hinder binding in the proximity.
Moreover, the binding of RBPs may obstruct ODNs in accessing the target sites. ENCODE3 project (www.encodeproject.org) (Davis et al., 2018) is a valuable source of RBP binding sites (Yee et al., 2019). At the time of writing,the ENCODE3 project has archived results from 737 experiments on 258 RNA binding proteins produced by seven assays on five cell lines. (ENCODE Project Consortium, 2012) Similar to secondary structure data, RBP binding data is formatted in BED and available for download.
At the end of the pipeline, the candidate target sites that pass the above multilevel of vetting will be handed over for experimental validation.
Antisense Oligonucleotide Targets of Phosphatase and Tensin Homolog
As previously discussed, numerous reports have demonstrated that PTEN deletion could promote robust and long-distance axon regeneration in adult CNS neurons in various injury models. Hence, antagonizing the expression of the wild-type gene is a viable therapeutic goal. Here, we will use PTEN as an example to illustrate how the pipeline produces candidate target sites for the AS-ODN platform.
The first step is to collect all wild-type isoforms of PTEN. We identified human PTEN isoforms from NCBI RefSeq and ENSEMBL databases. RefSeq database suggests three PTEN transcripts (NM_000314, NM_001304717,and NM_001304718). It is noteworthy that even NM_000314 is longer than NM_001304717 by only one base, the nature of variations (a singlebase indel and a substitution) lead to substantial differences between the two proteins. (NM_000314 and NM_001304717 encode PTEN (NP_000305)and PTEN-L (NP_001291646), respectively). However, the ENSEMBL database portrays a different picture. As shown in Figure 3A, PTEN has six isoforms ENST00000371953.8, ENST00000462694.1, ENST00000472832.2,ENST00000487939.1, ENST00000498703.1, and ENST00000618586.1 but only NM_000314 and ENST00000371953.8 share a high degree of similarity.To resolve the discrepancy above, we used expression data to ascertain if isoforms are genuinely transcripts or artifacts. cDNAs were searched against a normal tissue expression dataset using the BLASTN suite-SRA. For instance, to verify ENST00000487939.1 or PTEN-204, its sequence was searched against the SRA experiment SRX4794864, which contains short reads from normal human brain tissue. A list of expression datasets originating from normal human tissues can be found in Suntsova et al. (2019).
For illustration, we will focus on the three isoforms documented in NCBI RefSeq. The exon within the blue box (Figure 3A) is unique to NM_001304718,whereas the exons inside the red box represent a region that can be used to differentiate NM_001304718 from the other two isoforms. However, no distinct regions exist that can isolate NM_000314 or NM_001304717 alone.If the goal is to mitigate the expression of all isoforms, all regions outside the red and blue boxes are suitable places to look for target sites. To target either NM_000314 or NM_001304717, the region with the single-base indel or the single-base substitution mentioned above remains the only option.
In summary, this example illustrates the importance of using multiple data sources to collect the most extensive set of transcripts. Even though database providers uphold high annotation standards, variations in supporting experimental data, relevant literature, and reviewing procedures may inevitably contribute to disparate results. Moreover, the PTEN example has revealed the challenge of isoform-specific targeting through a single ODN target site (Figure 3A). Two options can resolve the issue, either eliminating isoforms that are probably the results of splicing artifacts with the support of evidence or designing the minimum numbers of ODNs that target all isoforms but on separate sites.
In the next step (Figure 2), we will use the antisense strand of the target region(s) to churn out a list of overlapping fragments of size L, where L is the required length of an ODN binding site. In other words, two immediate adjacent fragments share L-1 bases in common except the 5′- and 3′-most bases. Suppose the length of binding site L is 20. Our goal is to identify shared ODN binding sites among all three isoforms. As the first and second exons of PTEN are shared among the three transcripts, we will design target sites residing in them as an example to demonstrate the bioinformatic pipeline
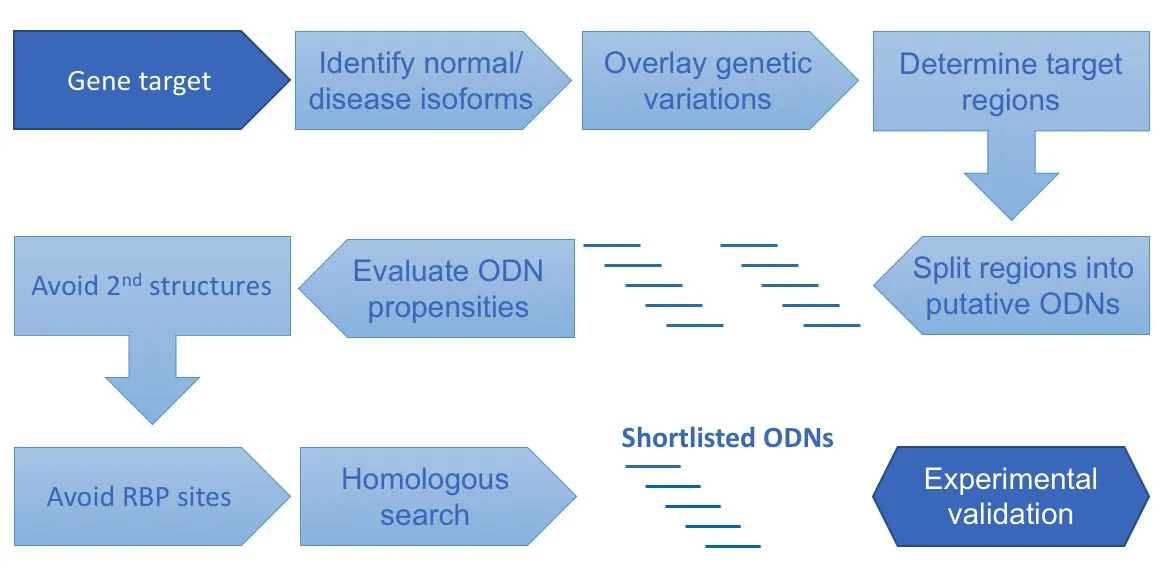
Figure 2 |Workflow for bioinformatic identification of RNA-based therapeutic target sites.The pipeline is designed to address three major concerns: accessibility, specificity, and binding affinity. Avoiding RNA secondary structures and sites being blocked by RNAbinding proteins (RBPs) are vital to ensure that the oligonucleotide (ODNs) can hybridize with the target site in the absence of interference. To eliminate the off-target effect,ODNs must specifically bind to the target site. As such, isoforms, genetic variations of the target gene must be incorporated for sequence comparison, followed by sequence homology searching. The last factor is the sequence propensities of the binding sites such as GC-content, CpG induced immunity, and self-annealing potential. In this multistep process, a large number of ODN binding sites are eliminated, alleviating effort for experimental validation.
● Lower the “Expected threshold” to 0.01, demanding statistically significant hits, and it will expedite searching as well.
● Choose the smallest “Word size”.
● Uncheck all the filters and masks so that the entire genome and transcriptome are searched.
The number of putative ODNs is usually large, manual screening is prohibitive for a sizable report as such. Therefore, it is recommended to download the hit-table in a readily computer-readable format, e.g., .csv, so that hit information can be extracted by a simple Python script, for example.
Note that the BLAST algorithm pivots on word seeding and a pre-built genomic index. The genomic index registers genomic positions of words(k bases long sequences or seeds where k is the “Word size” parameter mentioned above) in the genome. A query sequence is translated into a series of overlapping words according to the exact way discussed above for generating putative AS-ODNs. The algorithm retrieves the genomic positions of these words from the genomic index if found. These positions will become the initial places (seeds) of alignments where the algorithm will extend the alignment at the two ends of the seed in both directions. The key advantages of this algorithm are the runtime speed, parallelizability, and reduced memory requirement of long queries compared with Smith-Waterman local alignment.However, BLAST is biased in weighing perfect matches in the middle of a short query sequence (the seed), penalizing highly homologous hits with mismatches or gaps lying evenly in the middle of the ODNs.
RNA-Based Therapeutics Approved or Under Clinical Trials
depicted in Figure 2. The total length of the two exons is 1009 bps, resulting in 990 putative antisense ODNs. Figure 3B displays the first five putative antisense ODNs that target the 5′-end of exon 1.
These putative ODNs will be checked if their sequence propensities fulfill criteria such as the minimum GC content and melting temperature (Tm),which are essential in securing binding. To avoid self-dimerization, ODNs that share a high degree of self-complementary must be eliminated. For example,the PTEN fragment GCAGCCGCCGCGGCCGCCGC is 80% complementary to itself. ODNs forming into secondary structures is another property that should be avoided. There are open-source tools that calculate the minimum free energy of an RNA sequence, such as ViennaRNA and RNAlib (Lorenz et al.,2011). ODNs that share high homology to non-target genes or their regulatory regions may lead to the off-target effect, so the pipeline must identify and discard them. For the PTEN example, putative ODNs were aligned to the human genome and the human transcriptome using NCBI BLASTN (Camacho et al., 2009). The former reveals potential bindings to the regulatory, exonic,and intronic regions of genes, while the latter provides additional exonexon splicing junction information. Since these ODNs are short, BLASTN will automatically readjust or optimize search parameters that facilitate their alignments against large sequence databases. Despite that, additional adjustments are recommended as shown below:
● Choose the “Somewhat similar sequences” program (BLASTN).
● Reduce the “Max target sequences” to 10 from the default 100 as it will speed up the searching process.
There are twenty-eight antisense oligonucleotide (AS-ODN) drugs registered in DrugBank, where nine of them are approved, as shown in Additional Table 1. The majority of them belong to cancer treatment. Whereas siRNA drugs,only five are found, out of the five, two have been approved. Since 2018,twelve active AS-ODN clinical trials have been conducted, spanning phases I to III (Additional Table 2). The conditions cover from rare diseases to cancer to common diseases.
Twenty-one active clinical trials have been found for siRNA since 2012, more than AS-ODN. Similar to AS-ODN, trials span all three phases.
Challenges of Delivering RNA Therapeutics to the Brain
Despite RNA therapy having emerged as an attractive strategy for the treatment of various diseases, their clinical application is hindered by several hurdles from the viewpoints of drug delivery. They are negatively charged and hydrophilic macromolecules which are difficult to be taken by cells. Also, they are biologically unstable with a short half-life due to rapid degradation by nucleases, and subsequent renal clearance. Moreover, naked RNA therapeutics can induce undesired off-target side effects and immune responses by binding to Toll-like receptors (Bishop et al., 2015). In particular,the instability of RNA in the host would limit the effectiveness of the neural repair. While there is no clinical study currently available to evaluate this issue, several lines of evidence from preclinical studies have convincinglysuggested that RNA-mediated knockdown of target genes could promote significant neural repair and regeneration. For example, it has been reported that shRNA-based knockdown of CCR5 through intracranial injection induces robust axonal sprouting and regeneration in the mouse cerebral cortex after stroke and induces motor recovery and improves cognitive decline after TBI(Joy et al., 2019). Axon regeneration after optic nerve injury in mice was also reported by intravitreally delivery of shRNA against PTEN (Duan et al., 2015).Additionally, the antisense oligonucleotide delivery by intracerebroventricular injection to mice has demonstrated effectiveness for the treatment of spinocerebellar ataxia type 2 (Scoles et al., 2017). To overcome the RNA instability issue, several approaches can be used, such as chemical modifications of RNAs as mentioned above to make them more stable in the peripheral blood or cerebrospinal fluid. Moreover, the drug administration frequency, dosage, and carriers can be adjusted to meet the therapeutic level.Currently, there are several clinical trials using the antisense oligonucleotides and administered intrathecally in patients for treating Huntington’s disease.These results suggest that the RNA-based knockdown approach is promising to induce neural repair.
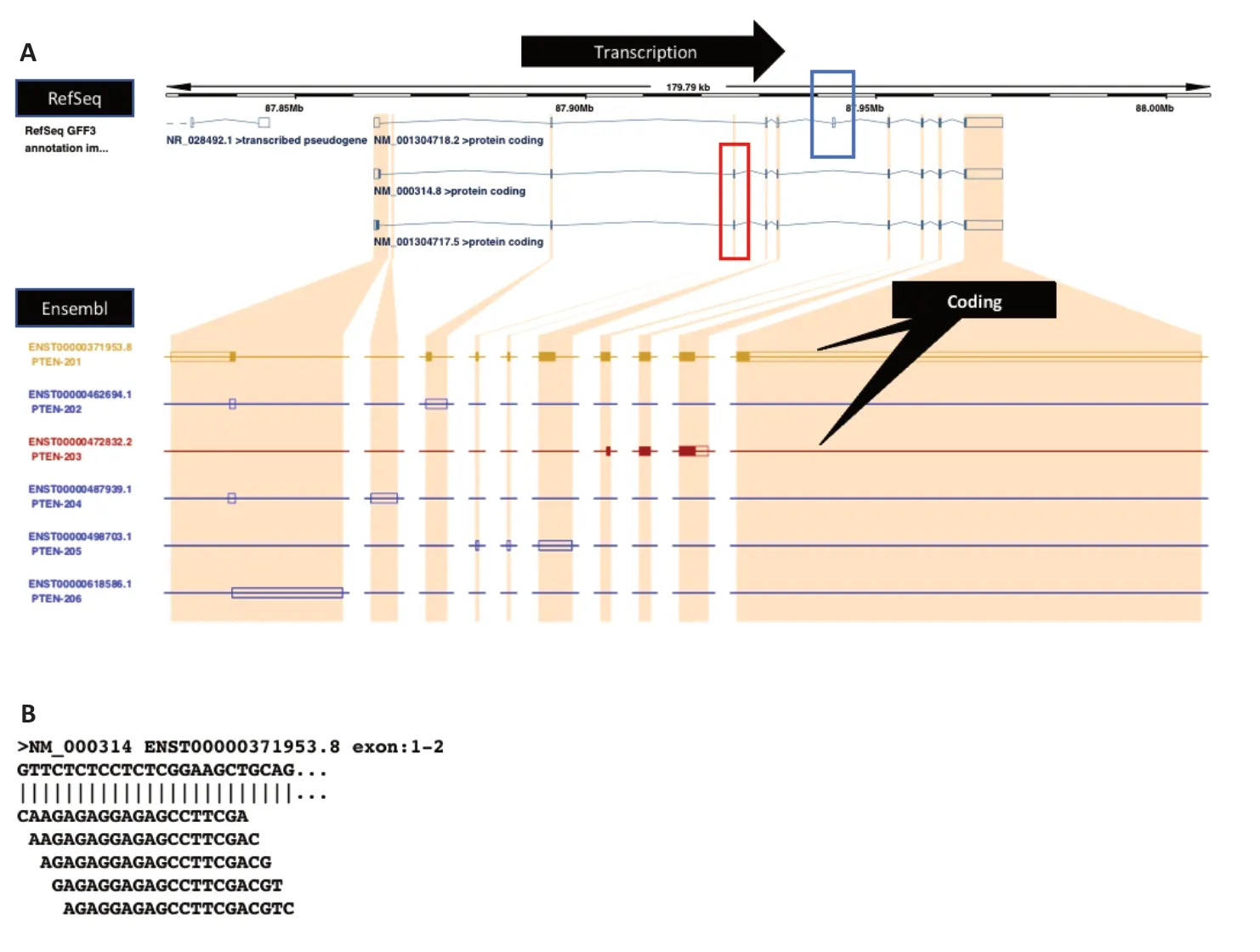
Figure 3 |Gene structure of the human phosphatase and tensin homolog (PTEN)transcripts.(A) Transcripts found in the National Center for Biotechnology Information (NCBI) RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/) can be drastically different from ENSEMBL for the same PTEN gene. The blue box highlights the exon that is unique to isoform NM028492. Whereas, the red box indicates the two exons present only in the other two isoforms. (B) Five putative oligonucleotides (ODNs) which are antisense to the beginning of exon1 of NM_000314.
The BBB represents another major obstacle to successful RNA delivery to the brain through the systemic circulation. It is comprised of brain capillary endothelial cells, astrocytes, pericytes, microglia, neurons, and mast cells,synergistically acting as a dynamic interface to control the exchange of substances between the blood and the brain. The tight junctions formed between the adjacent endothelial cells restrict the entry of most of the therapeutic agents, only allowing passive diffusion of small water-soluble molecules but to a little extent. Moreover, lipid-soluble molecules with molecular weights of less than 400 Da are favorable to cross the BBB through transmembrane diffusion (Crawford et al., 2016). Even though the efflux transporter expressed on the surface of endothelial cells, such as P-glycoprotein and multidrug resistance-associated proteins, can extrude these foreign molecules passing the BBB. Carrier-mediated transport,transporter-mediated transcytosis, and adsorptive-mediated transcytosis are known mechanisms to facilitate the BBB penetration, but are selective to a few essential compounds like glucose, insulin, and albumin. Cell-mediated transcytosis is another possible route for immune cells, and pathogens across the BBB. Recent evidence also confirms the BBB dysfunction in central nervous system (CNS) disorders including Alzheimer’s disease (AD),Parkinson’s disease (PD), stroke, etc. (Sweeney et al., 2018). Although the disease-induced BBB disruption is commonly assumed to promote the entry of therapeutic agents into the CNS, it complicates the process by altering the expression of tight junctions and transporters on the BBB.
The research of Stoicea et al. (2016) suggests that the BBB allows the exchange of miRNA between the cerebrospinal fluid and the blood, making it a promising biomarker for the diagnosis of CNS diseases. However, miRNA possesses a more diluted concentration in the blood in health conditions and is prone to leak from the brain in pathological conditions. On the other hand,siRNA that has a similar molecular weight of miRNA (~13 kDa) cannot cross the BBB (Mathupala et al., 2006).
Current Strategies for RNA Therapeutics Overcoming the Blood-Brain Barrier
One of the obvious strategies is to bypass the BBB, which can be achieved through the alteration of the administration route. The intracerebral administration is the most direct approach to delivering therapeutics into the brain, which can be further assisted with stereotaxic surgery for more precise delivery, implantation of drug reservoirs such as Gliadel® wafers for less frequent dosing, and convection-enhanced delivery for creating a positive hydrostatic pressure to drive the agents to the targeted site (Oh et al., 2020; Saraiva et al., 2016; Yu et al., 2017). The intrathecal injection is also practical to deliver the drug into the CNS to circumvent the BBB.However, these methods are all invasive, requiring specific techniques but also bringing a painful experience to the patients, resulting in an increased burden of healthcare professionals and patient non-compliance. Safety issues related to the risk of infection and traumatic injuries are also considerations.More recently, the intranasal administration has appeared to be a promising alternative to circumvent the BBB. It is a non-invasive route that allows the drug to target the brain directly through olfactory and trigeminal pathways.Although this neuronal connection is feasible to deliver a wide variety of substances including the nucleic acids to the brain, it also possesses some limitations like small nasal volume, enzymatic degradation, and mucociliary clearance. Advanced drug delivery systems such as nanoparticles are commonly employed to overcome these problems (Rodriguez et al., 2017;Hao et al., 2020).
BBB disruption is another strategy to improve drug delivery to the brain.Both chemical and physical means are available to enhance the permeability of the BBB. Mannitol is the most studied agent that can osmotically disrupt the BBB by shrinking the endothelial cells, opening the tight junctions, and subsequently promoting the passive diffusion of large molecules, such as siRNA, across the BBB (Park et al., 2015). Vasoactive agents like bradykinin,cereport and histamine were also studied for BBB disruption (Borlongan and Emerich, 2003; Liu et al., 2010). While they could effectively permeabilize the brain tumor capillaries, the poor effect was shown on brain endothelial cells. Focused ultrasound is another approach to transiently opening the tight junctions (Downs et al., 2015). Nowadays, it is facilitated by microbubbles with diameters of 1-10 μm so that less acoustic energy is required. Nevertheless,the disruption of BBB can not only allow for the entry of therapeutics but also increase the risk of the entry of undesired neurotoxins and micro-embolism.It may also cause the inflammation of the BBB and the disturbance of glucose uptake, leading to dysfunction of the brain.
Even if the RNA therapeutics can reach the CNS through the methods mentioned above, only restricted amounts of them may enter the targeted cells to exhibit their therapeutic effects. They are easily degraded by extracellular nucleases, and hard to penetrate the lipid cell membrane, owing to their hydrophilic and negatively charged nature. After intracellular uptake,they can still be degraded by endolysosomal enzymes (Bishop et al., 2015).To resolve these problems, various chemical modifications are introduced.For example, terminal modification (3′ and 5′) using polyethylene glycol (PEG)and sugar molecules are able to improve their stability and cellular uptake,while conjugation of the targeting ligands like mipomersen and tri-antennary N-acetyl galactosamine can promote their cellular uptake. In addition, partial replacement of the nucleosides with pseudouridine is effective to reduce the immunogenicity of the RNA therapeutics (Miele et al., 2012). However,chemical modification is a critical technique as gene modulation activity of the nucleic acids should be preserved, and the improvement of the stability and cellular uptake is limited.
Viral vectors represent the most efficient gene transfer vehicles since they naturally evolve to infect cells. Commonly used viral vectors are developed from adenovirus, adeno-associated virus, and retrovirus, particularly lentivirus. Notwithstanding the promising transfer capacity of the viral vectors,they also possess significant disadvantages like limited loading capacity and strong immunogenicity. Their native tropism often does not match the therapeutic need, i.e., they are required to be engineered to target cells that they do not infect naturally. Additionally, the production of viral vectors is of large difficulty and high cost to be scaled up (Waehler et al., 2007).
Therefore, nanoparticle-based vehicles have progressed substantially in recent years, aiming to transport RNA therapeutics to the brain. They are colloidal carriers with sizes ranging from 1 to 1000 nm, and of a synthetic or natural origin, which is much more biocompatible than viral vectors. They can be easily engineered to achieve the requirements for better delivery of nucleic acids, including stability, high encapsulation efficiency and loading capacity controlled and sustained cargo release, as well as specific cell targeting. Also,with the assistance of the other strategies mentioned above, the delivery efficiency of nanoparticles can be further improved. Building on these merits,many research groups are currently working on the development of various nanoparticles for RNA therapeutics delivery to the brain (Additional Table 5).
Nanoparticles for RNA Therapeutics Delivery to the Brain
Polymeric nanoparticles
Polyethylenimines (PEIs) are the most widely used polymer for RNA therapeutics transportation. It is a polycationic polymer rich in amine groups,and available in both linear and branched forms with different molecular weights. As a positively charged polymer, PEI can complex with nucleic acids electrostatically and efficiently enter the cells through endocytosis.Subsequently, PEI can retain a weak-base buffering property to protect nucleic acids from endolysosomal degradation, and induce the ‘proton sponge effect’,resulting in the burst of endosome and release of cargos to the cytoplasm(Vermeulen et al., 2018). PEI nanoparticles were found to efficiently deliver siRNAs to different cells in the brain after intranasal administration (Rodriguez et al., 2017). However, the low biocompatibility and non-biodegradability have limited its clinical application. The cytotoxicity of PEI is generally considered to be increased with the increase of molecular weight and positive charge density, but the transfection efficiency is diminished with low molecular weight. Hence, measures are taken to modify the PEI. After conjugation with negatively charged deoxycholic acid, the cytotoxicity of PEI (12 kDa)nanoparticles was significantly decreased while its high transfection efficiency was preserved (Oh et al., 2020). For the PEI with a low molecular weight (800 Da), cell-penetrating peptide like iNGR was proposed for surface modification to achieve efficient targeting delivery (An et al., 2015).
As an FDA-approved biomaterial, poly(lactide-co-glycolide) (PLGA) is one of the most investigated polymers for clinical usage. Despite its virtue of better biocompatibility and biodegradability than PEI, PLGA is difficult to encapsulate nucleic acids due to its negatively charged nature. Therefore, cationic compounds such as protamine sulfate were employed to coat the surface of PLGA nanoparticles to improve the loading of miRNA (Saraiva et al., 2016).
Polyethylene glycol (PEG), a Generally-Regarded-as-Safe (GRAS) hydrophilic polymer, has been tremendously used for conjugation (known as ‘PEGylation’)to tackle problems with drug delivery. With the shield of PEG, nanoparticles can be protected from aggregation, opsonization, and phagocytosis. On the other hand, it can also minimize the toxicity of polymers like PEI. In light of these merits, PEG was conjugated to PEI to enhance the biocompatibility of nanoparticles. Park et al. (2015) further formulated a rabies virus glycoprotein(RVG) peptide, a neuron-targeting peptide, and mannitol, an osmotic agent into the PEG-PEI nanoparticles. As a result, this delivery system significantly improved the siRNA delivery targeting the brain (Park et al., 2015). RVG29 modified PEG-PLGA nanoparticles synthesized by Hao et al. (2020) also showed significant symptom alleviation for ischemic brain injury via nasal route. Our recent study further demonstrated that the fatty acid-conjugated PEG nanoparticles could improve drug delivery to the brain (Li et al., 2018). It is noteworthy that long-term knockdown of PTEN in the non-neuronal brain cells such as astrocytes could induce gliomas (Wu et al., 2020). To avoid this potential negative effect, these nanoparticles conjugated with the neuron targeting peptides would be desirable to promote neural regeneration with reduced risk of oncogenesis.
While PEI, PLGA, and PEG are all synthetic polymers, chitosan is a natural polymer commonly used for gene delivery. Chitosan is a polysaccharide derived from chitin with a highly biocompatible, biodegradable, and nonimmunogenic nature. Like PEI, it can form a complex with RNA therapeutics through the electrostatic interaction between its amine groups and the anionic nucleic acids. This strong interaction, on the other hand, is one of the drawbacks of chitosan, making it hard to release the loaded cargo into the cytoplasm. Chitosan also allows for surface modification by varied materials like transferrin antibodies and bradykinin B2 antibodies for brain targeting (Gu et al., 2017).
Lipid-based nanoparticles
Liposomes are small vesicles composed of single or multiple phospholipid bilayers with an aqueous core where allows for RNA therapeutics encapsulation (Wei et al., 2016). It is widely used for gene delivery and has already been available on the market for mRNA vaccines. Also, solid lipid nanoparticles with a solid hydrophobic core coated with a phospholipid monolayer are capable of transporting nucleic acids, where siRNA were electrostatically bound to the outer surface (Erel-Akbaba et al., 2019). The lipid-based nanoparticles generally have a very stable structure to protect nucleic acids from degradation, but their half-lifein vivois restricted by their fast clearance in the liver and spleen. To overcome this limitation, PEGylation acts as a golden standard to prolong their circulation time. In addition, they can be functionalized with various targeting molecules such as T7 peptide and iRGD (a tumor-penetrating peptide) for higher transfer efficiency to the brain and the tumor (Erel-Akbaba et al., 2019; Wei et al., 2016).
Recently, a new class of lipopolymeric nanoparticles has been investigated for RNA therapeutics delivery, thus combining the advantages of both polymers and lipids. A lipopolymeric nanoparticle was successfully formulated by Yu et al. (2017) using epoxide-terminated lipids and low-molecular-weight polyamines, exhibiting therapeutic benefits to the attenuation of brain tumor growth via the CED method.
Inorganic nanoparticles
Inorganic materials have been utilized for nanomedicine development,including iron oxide, gold, silica, etc. They are biodegradable, biocompatible,and easy to control their sizes and morphology. Among them, iron oxidebased nanoparticles have been proposed for magnetic purposes and received the most attention. With the combination of cationic polymers like PEI, high encapsulation efficiency of RNA therapeutics could be achieved.Increased brain tumor cell apoptosis was observed with PEI-coated ZnFe2O4nanoparticles carrying lethal-7a miRNA (Yin et al., 2014).
Peptide-based nanoparticles
Peptides were commonly used to directly conjugate with RNA therapeutics or nanoparticles as a functional moiety. However, recent advances in nanotechnology have employed peptides as the backbone of the nanoparticles, forming a complex with nucleic acids by non-covalent interaction. Kwon et al. (2016) engineered a series of peptide-based nanoparticles with RVG as the neuron targeting peptide and transported it as the intracellular trafficking peptide. After intravenous injection of the optimized nanoparticles, a significant accumulation of siRNA was observed in the injury site of the brain.
Nucleic acid-based nanoparticles
As nanotechnology evolves, RNA has been engineered as a distinctive nanomaterial. The RNA nanoparticles can be programmed with desired characteristics including size and structure, for specific applications. They are thermodynamically stable and water-soluble, so PEGylation is not required.Although original RNA is prone to degradation by nuclease, 2′-Fluoro (2′-F)modifications of U and C nucleotides are commonly used to minimize the degradation, while retaining the authentic functions of both the scaffold and the incorporated molecules (Liu et al., 2011). The packaging RNA (pRNA), an element of the bacteriophage non-coding RNA, is a versatile nanomaterial with a three-way junction (3WJ) motif that can allow the assembly of three RNA modules to form multivalent pRNA-3WJ nanoparticles (Shu et al., 2011).Lee et al. (2015) have generated a folic acid (FA)-conjugated pRNA-3WJ nanoparticle based on phi29 DNA packaging motor, which achieved targeted delivery of siRNA to the brain tumor cells through systemic administration.
Spherical nucleic acids are an emerging nanotechnology platform for the delivery of biologicals. They consist of an inorganic nanoparticle, especially gold nanoparticle, as the core, as well as a highly oriented, dense layer of oligonucleotides such as DNA, siRNA, and mRNA, as the shell. They are frequently shielded by PEG to extend the half-life in the systemic circulation(Kouri et al., 2015). Unlike the linear nucleic acids, spherical nucleic acids are less susceptible to nuclease degradation and ready to enter > 50 types of cells via caveolae-mediated endocytosis. However, after cell internalization, they tend to accumulate in the endosomes. In a recent study, Melamed et al. (2018)have wrapped the spherical nucleic acids with PEI to promote endosomal escape, thereby enhancing the transfection efficiency. Interestingly, they also found that the cytotoxicity of PEI was dramatically diminished after its conjugation to spherical nucleic acids.
Exosomes
Exosomes are nanosized lipid vesicles naturally secreted from almost all the cell types, carrying non-coding RNAs for intercellular communication.Nowadays, they have been extensively studied as a unique vehicle for various therapeutics. Thanks to their endogenous origin, exosomes elicit a minimal immunogenic response and are stable in the systemic circulation. They are also believed to possess specific cell selectivity. Exosomes isolated from the bEnd.3 cell medium successfully facilitated the transportation of siRNA to the brain of zebrafish (Yang et al., 2017). Researchers also modified the exosomes with RVG to achieve brain targeting effect (Cooper et al., 2014; Liu et al.,2015). However, critical issues associated with the choice of exosome origin and the procedure of cargo loading do exist as a barrier for exosomes to reach maximum potential in clinical application.
Conclusion and Future Perspectives
Compelling evidence has demonstrated that reactivation of the regenerative mTOR pathway can substantially promote axon regeneration, neural repair,and functional recovery after CNS injury. We specifically discussed the potential of PTEN as a promising target using the antisense oligonucleotide approach. We have discussed the characteristics of RNA-based therapeutics and a bioinformatics pipeline to predict a manageable number of viable ODN binding sites, saving the effort of experimental validation. Identifying putative ODN binding sites requires significantly less effort than identifying binding pockets used by small molecule drugs. As datasets from large-scale RNAprotein interactions and RNA folding studies are available, and more are in the making, an accurate genome-wide landscape of ODN binding sites can be obtained, accelerating the development of RNA therapy. While this review focuses on the RNA knockdown approach, the mRNA transient expression is an alternative approach to promote neural regeneration. For example, transient transgene expression of AKT or other positive upstream regulators of mTOR pathway could be considered. However, the RNA-based transgene expression,such as the expression of viral spike protein for the COVID-19 vaccine, can only be expressed at a very low level for a short time to avoid potential cytotoxicity and cytokine storm. In addition, there are other challenges and obstacles associated with mRNA therapy, including mRNA’s molecular size, charge,intrinsic instability, and targeted delivery issue. Importantly, with the success of COVID-19 mRNA-vaccines, it is evident that it is viable and safe to deliver mRNA molecules into the human body via the appropriate vehicle, making this technology a promising therapeutic tool for a comprehensive application.Pharmaceutical companies such as Moderna and Translate Bio are developing an mRNA-based therapy for treating cystic fibrosis. That has demonstrated the enthusiasm of this strategy. Considering the scant examples of nonvaccine mRNA-based transient transgene expression, the antisense and miRNA approaches focused in this review are currently a more feasible approach.On the other hand, it is noteworthy that although promising, targeting mTOR pathway (e.g. by PTEN deletion) is not without potential limitations.Concerning the effect on brain development, neuron-specific PTEN mutant mice cause seizures, ataxia, and brain enlargement with increased soma size and dysplasia of neural cell populations by 9 weeks and such abnormality is resembling Lhermitte-Duclos disease (Backman et al., 2001). Hyperactivation of mTOR signaling by selective deletion of PTEN in the granule cells was reported to form aberrant neuronal connections and disrupt the normal suppressive function of the dentate gyrus, resulting in epileptogenesis (Pun et al., 2012). Regarding the studies in conditional deletion of PTEN in adult mice, it has been observed that the corticospinal tract axons emanating from PTEN-deleted cortical motoneurons in adult mice have thicker axons without triggering compensatory increases in myelination. Also, unilateral deletion of the same PTEN knockout mice results in impaired motor coordination (Gallent and Steward, 2018). Sinton and colleagues reported that the conditional inactivation of PTEN in adult mice can induce symptoms reminiscent of human autism spectrum disorder such as seizures, macrocephaly, anxiety, and social interaction deficits (Kwon et al., 2006). A follow-up study further indicated that the maintenance of circadian rhythms may be impaired by PTEN mutation(Ogawa et al., 2007). Thus, the transient PTEN knockdown seems to be critical to control the optimal effect on the neural repair without causing undesirable effects. Compared with virus-mediated knockdown of PTEN, our ODNmediated approach would be clinically easier to maintain the transient and dosage of PTEN downregulation. While RNA therapy appears to be effective for CNS diseases, delivery of RNA oligonucleotides to the brain remains a huge challenge. Judicious design in formulation and particles using nanotechnology could be a promising direction for developing the next generation of RNA therapy for the treatment of CNS injury.
Author contributions:Review conception and design by CKT, SFC, ESH. All authors contributed towards literature retrieval, editing and critically revising the manuscript for important intellectual content. All authors approved the final version of this manuscript for publication.
Conflicts of interest:The authors declare that there are no conflicts of interest associated with this manuscript.
Availability of data and materials:All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Table 1:RNA-based therapeutics from DrugBank (https://go.drugbank.com/).
Additional Table 2:RNA-based therapeutics from ClinicalTrials.gov.
Additional Table 3:siRNA therapeutics from DrugBank (https://go.drugbank.com/).
Additional Table 4:siRNA therapeutics from ClinicalTrials.gov.
Additional Table 5:Recent studies on the nanoparticles for RNA therapeutics delivery to the brain.
- 中国神经再生研究(英文版)的其它文章
- Functional in vivo assessment of stem cell-secreted prooligodendroglial factors
- iGluR expression in the hippocampal formation, entorhinal cortex,and superior temporal gyrus in Alzheimer’s disease
- Exploiting Caenorhabditis elegans to discover human gut microbiotamediated intervention strategies in protein conformational diseases
- N-methyl-D-aspartate receptor functions altered by neuronal PTP1B activation in Alzheimer’s disease and schizophrenia models
- Aminopeptidase A and dipeptidyl peptidase 4: a pathogenic duo in Alzheimer’s disease?
- Ubiquitin homeostasis disruption,a common cause of proteostasis collapse in amyotrophic lateral sclerosis?