基于全基因组重测序的山羊产羔数性状关键调控基因的筛选
李恒,字向东,王会,熊燕,吕明杰,刘宇,蒋旭东
基于全基因组重测序的山羊产羔数性状关键调控基因的筛选
李恒1,字向东1,王会2,熊燕1,吕明杰1,刘宇1,蒋旭东1
1西南民族大学动物科学国家民委重点实验室,成都 610041;2西南民族大学青藏高原动物遗传资源保护与利用教育部重点实验室,成都 610041
【目的】对产羔数不同的山羊进行全基因组重测序分析,挖掘参与调控川中黑山羊产羔数性状关键调控基因,为解析山羊产羔数性状遗传机制及分子遗传改良提供理论依据。【方法】选择6只产4—6羔的川中黑山羊为高繁组(high fecundity, HF)和6只产单羔的川中黑山羊为低繁组(low fecundity, LF),采集颈静脉血液样本提取基因组DNA,构建350 bp双末端测序文库,利用Illumina HiSeq PE150平台对12个文库进行全基因组重测序。测序产出的净数据经BWA软件比对至山羊参考基因组ARS1,所获得的高质量SNPs通过两种全基因组扫描分析方法(、)的综合分析确定候选区域,候选区域的注释基因分别利用g:Profiler和KOBAS在线数据库进行GO分析与KEGG通路分析,以筛选调节川中黑山羊产羔数性状候选基因。为了进一步鉴定调节山羊产羔数目的关键遗传标记,根据基因组重测序变异分析,对繁殖候选基因的同义与非同义SNPs进行定位筛选,后续将12个山羊样本的扩增产物进行Sanger测序以验证重测序结果。【结果】12只山羊共获得431.50 Gb 净数据,经变异检测与注释发现,HF组山羊共发现7 771 417个单核苷酸多态性(single nucleotide polymorphism, SNPs),LF组山羊检测到8 935 907个SNPs,且LF组各类SNPs 均多于HF组。设置同时达到top 5%最大值和top 5%最小值的窗口为候选区域,在低杂合性、高遗传分化的区域共注释130个强选择信号,其中HF组、LF组以及共享窗口的注释基因分别为84、59和13个,经GO富集与KEGG通路分析发现,19个候选基因参与川中黑山羊的繁殖、繁殖过程和胚胎发育等调控,包括11个HF组特异性候选基因(和),5个LF组特异性候选基因(、、、和)和3个HF组与LF组共享窗口基因(、和)。同时,大多数GO分析,如G蛋白偶联受体活性、激素反应和神经肽信号通路等,都包含这19个候选基因。此外,14个HF候选基因有9个显著富集在代谢途径、神经活性配体-受体相互作用、糖胺聚糖-硫酸乙酰肝素/肝素的生物合成、钙离子信号通路、cAMP信号通路和叶酸生物合成等KEGG通路中(<0.05)。19个繁殖候选基因中共有2个同义突变(,10 A4662G)与2个非同义突变(G529A,),且仅定位于HF候选基因中。Sanger测序发现,、和突变位点均可检测到多态性,与基因组重测序结果一致,其中G529A多态性导致第177位丙氨酸突变为苏氨酸,多态性导致翻译提前终止。【结论】本研究共发现11个HF组特异性候选基因,推测是川中黑山羊多羔性状的关键调控基因,外显子G529A与外显子A281T突变可能是调控山羊多羔性状的关键遗传标记,在改良山羊繁殖性能方面具有较大的应用价值。
川中黑山羊;基因组重测序;多羔性状;候选基因
0 引言
【研究意义】产羔性状是山羊的重要经济性状,提高窝产羔数不仅可以提高出栏率,提高经济效益,而且还可以提高选择强度,加快山羊育种进程。产羔数是一个低遗传力(0.08—0.18)的限性性状,难以用常规育种方法进行快速改良,但适合用标记辅助选择(marker-assisted selection, MAS)等分子育种技术来改良[1]。实现MAS的先决条件是找到与数量性状基因座相连锁的分子遗传标记,而要找到辅助选择的分子标记,则需要解析山羊高繁殖力的分子遗传基础和形成原因。【前人研究进展】在过去20多年里,在一些绵羊品种的突变体中已成功分离鉴定出控制卵泡发育和排卵数的多胎基因,主要包括骨形态发生蛋白受体-1B(BMP receptor-1B,)基因的1个突变(FecB)[2]、骨形态发生蛋白-15(bone morphogenetic protein-15,)基因的6个突变(FecX、FecX、FecX、FecX、FecX和FecX)[3-4]和生长分化因子-9(growth and differentiation factor-9,)基因的2个突变(FecG和)[5-6],这些突变与绵羊的多羔性状密切相关。但是,迄今为止,在已开展过检测的印度[7]、伊朗[8]、泰国[9]和我国[10-11]的30多个山羊品种(类群)中均未检测到这些突变,排除了这些突变作为山羊品种多羔性状主基因的可能性,山羊多羔性状的遗传机制有待研究。山羊全基因组测序技术的发展与完善,为发掘参与调控川中黑山羊产羔数性状关键调控基因提供了全新的视角[12]。对崂山奶山羊高、低繁殖力的两个极端种群进行基因组重测序,发现细胞周期蛋白B2(cyclin B2,2)雄激素受体(androgen receptor,)腺苷酸环化酶1(adenylate cyclase 1,)SMAD家庭成员2(SMAD family member 2,)等基因在高繁殖力群体中被特异性选择[13]。丝氨酸/苏氨酸激酶3(serine/threonine kinase 3,)蛋白磷酸酶3催化亚基α (protein phosphatase 3 catalytic subunit alpha,)等96个候选基因与大足黑山羊产羔数性状显著相关[14],其中也与阿尔巴斯绒山羊产羔数性状相关[15]。进一步分析67个单核苷酸多态性(SNP)与大足黑山羊产羔数的关联,发现半胱氨酰转运RNA合成酶2(cysteinyl-tRNA synthetase 2,)Ⅰ型血小板结合蛋白基序的解聚蛋白样金属蛋白酶(ADAM metallopeptidase with thrombospondin type 1 motif 14,)和甲基转移酶 25(methyltransferase like 25,)基因编码区SNP与头胎产羔数目显著相关[16]。对头胎产单羔、双羔、三羔的三组济宁青山羊进行全基因组扫描,结果在双羔和三羔群体中发现酪氨酸激酶受体(KIT proto-oncogene, receptor tyrosine kinase,)、P21-活化激酶1P21-activated kinases 1,腺苷活化蛋白激酶α1亚基(protein kinase AMP-activated catalytic subunit alpha 1,)等多个产羔数调节基因[17]。WANG等[18]在相同饲养管理条件下,对在5个连续分娩记录中表现出稳定的产羔数差异的12只山羊进行基因组重测序,结果发现细胞分裂周期蛋白25同源蛋白C(cell division cycle 25C,)、核酸内切酶G(endonuclease G,)和Nanos同源基因3(nanos C2HC-type zinc finger 3,)的变异与产羔数性状显著相关,螺旋卷曲过程对生殖能力有潜在的调控作用。这些候选基因的发现拓展了人们对繁殖力遗传基础的认知,为山羊产羔数性状的遗传机制的解析提供重要线索。【本研究切入点】川中黑山羊是我国优良地方山羊品种,具有生长速度快,产肉性能高,产羔率高等优良特性[19-20]。我们在川中黑山羊和的前期研究中未检测到与绵羊多羔性状相关突变的存在,但高繁川中黑山羊与单胎藏山羊之间检测到5个新的碱基突变,其中4个导致氨基酸改变,有2个碱基突变,其中1个导致氨基酸突变,两个品种的15核苷酸序列则完全一致[21]。说明山羊和中氨基酸改变的突变与绵羊中不同,这些突变是否与山羊产羔数相关尚有待进一步研究。从卵巢转录组和miRNA水平也不能完全解析川中黑山羊多羔性状的分子遗传机制[22-23]。【拟解决的关键问题】本研究选择高、低繁殖力的川中黑山羊为研究对象,利用基因组重测序技术扫描其低杂合性、高遗传分化区域,挖掘高、低繁殖力山羊关键基因遗传变异,并对候选靶基因进行GO富集与KEGG通路分析,以期筛选调节川中黑山羊产羔数的关键候选基因,为山羊产羔数性状遗传机制的阐释及后续山羊分子育种工作提供理论基础。
1 材料与方法
1.1 样品的采集
本研究根据第一、二胎产羔记录,于2019年9月选择饲养于四川省乐至县天龙育种场,胎产羔数差异显著的高繁殖力(high fecundity,HF)和低繁殖力(low fecundity,LF)三岁龄川中黑山羊各6只,其中HF组山羊连续两胎胎产羔数4—6只,LF组连续两胎胎产羔数1只(表1)。每只山羊采集其颈静脉血液2 mL于EDTA-K2抗凝管中,试剂盒提取基因组DNA后用于基因组重测序研究。
1.2 山羊血液基因组DNA的提取及质量检测
使用1%琼脂糖凝胶电泳与紫外分光光度计分别鉴定山羊基因组DNA的完整性、浓度和纯度,12管质检合格的HF组和LF组DNA样品各抽取1.5 μg分别建库。利用Agilent 2100检测文库中插入片段的长度,片段符合预期后,送至Illumina HiSeq PE150平台进行双端测序,整个流程委托北京诺禾致源生物信息科技有限公司完成。

表1 12只川中黑山羊的胎产羔数信息
编号HF1—6为高繁殖力山羊,编号LF1—6为低繁殖力山羊
The symbols from HF1 to HF6 were high fecundity goats, and the symbols from LF1 to LF6 were low fecundity goats
1.3 测序数据的变异检测与注释
原始数据经初步质控处理,将接头序列、未测出碱基超过10%和低质量碱基数超过50%的paired reads去除,获得净数据。利用BWA[24]软件将clean reads比对至山羊参考基因组ARS1[25],比对结果经SAMTOOLS软件去除重复[26]。利用GATKv3.2-2进行SNP鉴定与分型[27],过滤后获得的高质量SNPs用ANNOVAR[28]软件工具进行功能注释。
1.4 群体选择分析
利用滑动窗口法(sliding windows)对HF、LF组川中黑山羊进行全基因组扫描,按照窗口内SNPs个数不超过20,确定选择消除窗口大小(100 kb)。计算每个窗口的固定系数(Fst)值并转换为值,选择top 5% 最大t值窗口为选择区域。通过计算窗口内SNPs位点的杂合性(Hp),进而对Selective sweep进行评估。分别计算窗口的Hp值并转化为值,选择top 5%最小值窗口作为选择区域。在计算得到每个窗口Hp和Fst值的基础上,选择最小值和最大值均达到Top 5%的区域为候选区域,并对该区域进行基因注释。
1.5 候选基因的功能注释
基于Bio-Mart在线数据库获得山羊候选区域内的基因注释信息(基因类型和ENSEMBL号)后,利用g:Profiler网站将山羊基因符号ID转换为小鼠同源基因。利用g:Profiler[29]网站和KOBAS[30]在线数据库分别进行Gene ontology(GO)富集分析和Kyoto Encyclopedia of Genes and Genomes(KEGG)通路分析。
1.6 SNP位点的验证
为进一步鉴定调节山羊产羔数目的关键遗传标记,据重测序变异分析报告对19个繁殖候选基因的同义与非同义SNPs进行定位筛选。候选位点经Primer Premier 5.0 软件设计引物(表2),12个山羊样本的扩增产物送至成都擎科生物有限公司进行Sanger测序。
2 结果
2.1 高、低繁殖力组山羊基因组测序数据质控
分别完成12只山羊血液样本基因组重测序工作,共获得431.50 Gb 净数据,错误分布率均为0.03%,测序质量较高(Q20≥96.9%,Q30≥91.69%),所有样本比对率在99.61%—99.73%之间,平均测序深度10.70×,1×覆盖度在94.91%—95.32%之间,4×覆盖度在88.70%—93.13%之间。所有原始序列数据已上传至国家基因组数据中心基因组序列档案库(https://bigd.big.ac.cn/gsa.),注册号为CRA003846。

表2 SNP位点验证引物
2.2 变异检测与注释
基于严格的阈值,两组山羊共检测到16 707 324个高质量SNPs位点,其中LF组8 935 907个SNPs位点,HF组7 771 417个SNPs位点,且LF组的各类SNPs 均多于HF组(表3)。在这些SNPs中,HF组和LF组山羊外显子SNPs平均各有53 425(0.68%)和60 588(0.69%)个,基因上游1 kb区域分别有44 949个和50 798个,基因下游1 kb区域分别分布44 201个和50 349个,两组基因间区SNPs占比最多,分别占总SNPs的70.01%和70.09%;其次是分布在内含子区域SNPs,HF和LF组分别含有2 145 836个和2 473 891个;位于剪切位点的SNPs数目最少。在编码区SNPs 中,同义突变和错义突变分别占外显子总SNPs的58.66%和40.77%,且两组相差不大。此外,HF组和LF组在终止密码子处分别检测到300和323个SNPs,在剪切位点处分别检测到214个和237个SNPs,这些位点可能会影响转录剪接,从而影响蛋白质产物及功能。

表3 川中黑山羊SNPs检测及注释结果统计
2.3 群体选择分析
经不同大小窗口调试发现,窗口大小为100 kb时,SNPs数目小于20个并逐渐趋于稳定,因此,选择100 kb作为选择信号检测最佳窗口长度,使用50%的重叠区为滑动尺,计算川中黑山羊HF组与LF组之间的Fst值,并标准化作图,选取LF和HF之间的阈值为1.9,共筛选到579个窗口,注释623个基因(图1)。
分别计算川中黑山羊HF组与LF组染色体窗口内SNP位点的值,Z转换后并标准化作图,选择top 5%最小值为阈值,HF群体的阈值为-2.05,筛选到579个窗口(图2),共注释617个候选基因。 LF群体的阈值为-2.08,筛选到579个窗口(图3),注释到610个基因。

图1 高繁组和低繁组川中黑山羊1-29号常染色体平均固定系数ZFst的分布
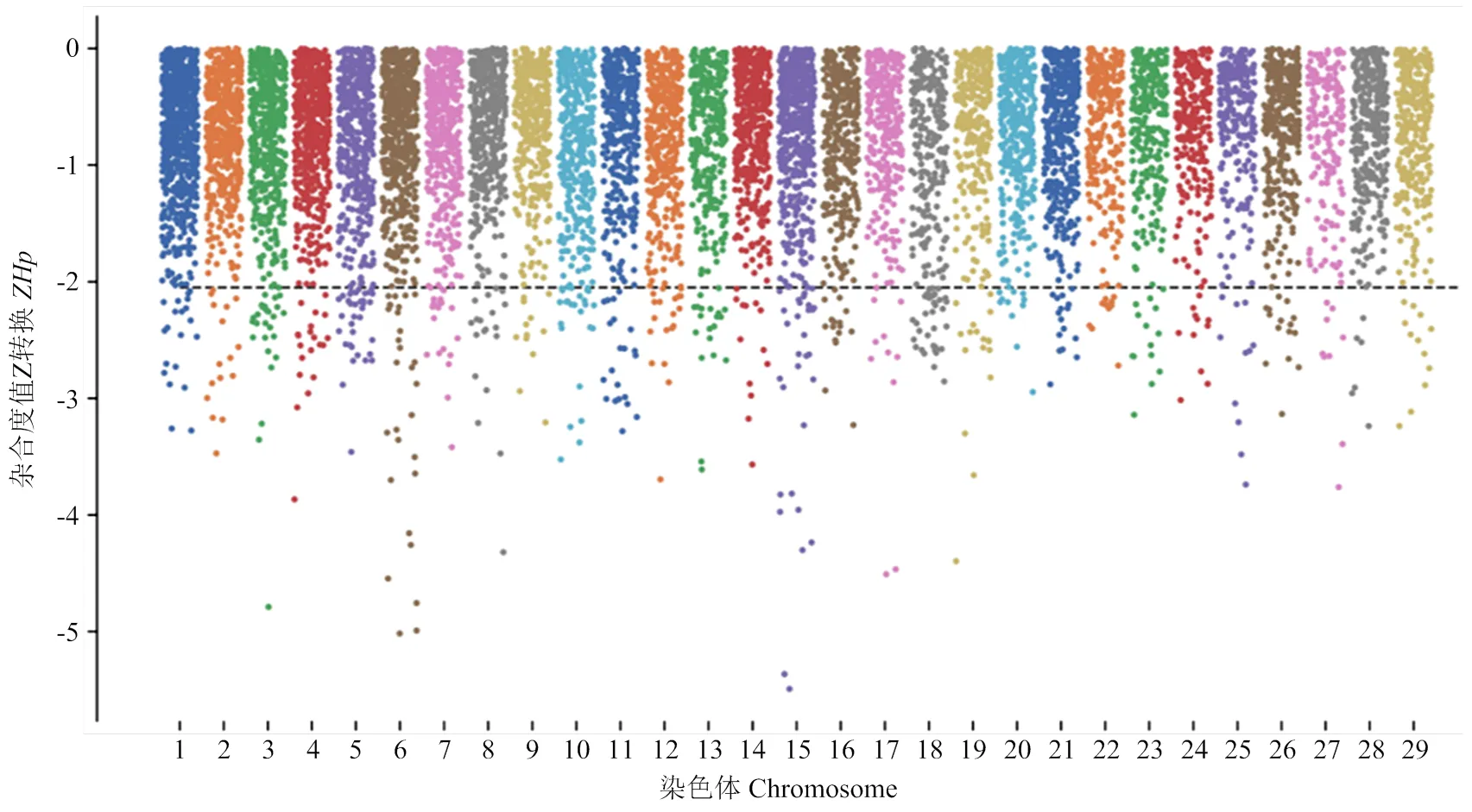
图2 高繁组川中黑山羊1-29号常染色体平均杂合度ZHp的分布
为进一步筛选川中黑山羊产羔性状关键调控基因,本研究初步扫描了具有低杂合性、高遗传分化的区域,即候选窗口值均达到top 5%最大值和top 5%最小值标准。其中HF组中鉴定65个候选窗口,共注释84个候选基因(图4-A),LF组山羊共鉴定了42个候选窗口,注释了59个候选基因(图4-B)。因两组候选基因中存在、等13个交集基因,所以HF组和LF组共注释了130个差异候选基因。
2.4 高、低繁殖力山羊候选基因基因GO富集分析
进一步揭示与产羔数性状的候选靶基因功能,本研究对两群体差异表达基因(130个)进行生物功能分析,-value经Bonferroni校正后,设置corrected-value ≤0.05为阈值,选择候选靶基因中显著富集的GO terms。

图3 低繁组川中黑山羊1-29号常染色体平均杂合度ZHp的分布

图4 川中黑山羊高繁组(A)与低繁组(B)受选择区域
GO结果显示,川中黑山羊群体中筛选的130个强选择性候选基因,共富集到193条显著性GO功能条目。其中归类为生物学过程的GO条目最多,共113条,主要行使生长发育和代谢功能,其次是细胞组成,共50条GO条目,分子功能本体条目最少,其富集的30条GO条目主要参与跨膜信号受体活性、神经肽受体活性和蛋白多糖生物合成的过程等。
值得注意的是,19个基因与繁殖紧密相关,如繁殖、繁殖过程、胚胎发育和生殖过程调控(表4)。其中HF组特异性强选择性基因为11个,包含腺苷酸环化酶10(adenylate cyclase 10,)、多巴胺受体D1(dopamine receptor D1,)、硫酸肝素6-邻磺酸转移酶1(heparan sulfate 6-O-sulfotransferase 1,)、胰岛素样生长因子结合蛋白7(insulin like growth factor binding protein 7,)、促黑激素同源框2(msh homeobox 2,)、头蛋白(noggin,)、肾单位肾痨4(nephronophthisis 4 (juvenile) homolog,)、妊娠相关血浆蛋白A(pregnancy- associated plasma protein-A,、睾丸发育相关蛋白(testis development-Related Protein,)、木糖转移酶1(xylosyltransferase 1,)和催乳素释放激素受体(prolactin releasing hormone receptor,),HF和LF组共享窗口基因3个,如醛酮还原酶家族成员B3(aldo-keto reductase family 1,member B3,)、组蛋白脱乙酰酶4(histone deacetylase 4,)和mu阿片受体(opioid receptor mu 1,),以及LF组特异性候选基因5个,如膜联蛋白5(annexin A5,)、内皮素A型受体(endothelin receptor type A,)、FA核心复合体连接酶(FA complementation group L,)、胰岛素样生长因子1(insulin-like growth factor1,)和速激肽前体1(tachykinin precursor 1,)。同时,大多数GO术语,如G蛋白偶联受体活性、激素反应和神经肽信号通路等,都包含这19个候选基因。

表4 川中黑山羊繁殖相关GO条目
2.5 高、低繁殖力山羊候选基因KEGG分析
KEGG通路分析结果显示,川中黑山羊差异候选基因集共富集112条代通路,选择前20条显著的代谢通路作为展示(图5)。KEGG代谢通路结果表明,14个HF山羊候选基因中,9个基因显著富集在代谢途径(、)、糖胺聚糖-硫酸乙酰肝素/肝素的生物合成(、)、神经活性配体-受体相互作用()、钙离子信号通路()、cAMP信号通路()和叶酸生物合成()通路(corrected-value ≤0.05)中,这些通路可能在山羊产羔数性状的调控中极具关键作用。
2.6 SNP位点的验证
为筛选多羔性状关键遗传标记,本研究扫描LF和HF组外显子SNPs分布。19个繁殖候选基因中,共有2个同义突变(,)与2个非同义突变(G529A,),且仅定位于HF候选基因中。其中G529A多态性导致第177位丙氨酸突变为苏氨酸,多态性导致翻译提前终止。重要的是,4个SNPs仅有3个与高通量测序结果呈现相同趋势(,G529A,),的外显子SNP测序结果与重测序变异报告不一致,未发现该突变位点(图6)。
3 讨论
山羊产羔数是复杂的表型特征,其不仅由排卵率、胚胎或胎儿成活率、子宫容受性等多个次级性状调控,也受遗传、季节、降雨、气温和营养等因素影响[31-33]。GWAS虽是检测复杂性状的遗传学基础的最有力的工具[34],但深入挖掘GWAS的统计能力需要巨大的样本量和适当的对照设计[35]。因此,大样本、家系信息详尽等要求限制了GWAS在山羊群体中的应用。基因组重测序仍是目前分析山羊复杂性状的强有力手段。该技术不仅可发掘出特定性状的多个候选基因,其公布的变异数据也为其他性状研究提供思考。例如最新发现的山羊产羔数候选基因、等[36-37],遗传标记信息均是来自先前的山羊基因组重测序报道。

图5 川中黑山羊选择信号KEGG通路分析
本研究对高产山羊品种川中黑山羊不同产羔数群体进行了全基因组测序,获得了大量遗传变异,为山羊高繁殖力遗传机制解析提供了重要的遗传资料。为阐明川中黑山羊产羔性状的选择特征,设置top 5%为阈值[13-14],在川中黑山羊群体的低杂合性、高遗传分化的区域共注释130个候选基因。
通过GO富集进一步发现了19个候选基因可能参与川中黑山羊产羔性状的调节,参与繁殖、繁殖过程、胚胎发育等生殖过程调控。在这19个候选基因中,11个候选基因在HF组中特异性选择,可能与川中黑山羊的多羔性状有关。这些基因的功能主要与生殖细胞发育及胚胎发育等生殖过程相关。其中编码的可溶性腺苷酸环化酶(SAC),对男性生殖至关重要[38]。SAC调控cAMP/PKA通路与AMPK的活性,其中cAMP/PKA通路在颗粒细胞FSH的功能调节和E2产生过程中发挥重要作用[39],AMPK的活性影响卵母细胞成熟[40]。推测SAC通过调节生殖激素分泌水平进而调控动物繁殖过程,其表达的丰度制约了雌性个体排卵率。与与家禽产卵相关[41-42],可能在山羊群体的排卵机制中发挥特殊作用。可能是卵巢早衰病因学的候选基因[43],该基因突变也被认为与特发性低促性腺激素减退症有关[44]。牛繁殖候选基因下调促进了细胞凋亡,阻碍了细胞增殖,增加了孕酮和雌二醇的产生[45],其还与[46]功能类似,对胚胎着床、妊娠的建立与维持至关重要[47]。抑制IGFBP7表达可显著降低植入胚胎数与妊娠率[47],这意味着除排卵数外,早期胚胎的丢失也是山羊窝产羔数目减少的诱因。目前并未发现功能突变与山羊早期胚胎丢失有关,其调控机制还需深入研究。在卵巢中表达并与BMP蛋白相互作用,参与正常胚胎的发育调控[48]。PAPAA蛋白的缺乏会下调小鼠卵巢类固醇生成和损害女性生育能力[49],其mRNA水平与妊娠期间血清水平分别是识别卵母细胞能正常发育至胚胎和评估子宫内膜容受性的潜在重要标志[50-51]。PAPAA蛋白具有成为预测高繁殖力个体诊断工具的潜力。和是参与精子发育的关键基因[52-53]。是一种木糖基转移酶,参与对着床期囊胚的附着和生长有直接作用的蛋白多糖肝素/硫酸肝素合成过程[54]。这些证据均表明了这11个繁殖调控基因可能是调控山羊多羔性状的关键基因,在改良山羊繁殖性能方面具有较大的应用价值,但其在产羔调节网络中的具体功能还需进一步研究。

A:PRLHR,B:MSX2,C:DRD1,D:ADCY10
LF组差异表达基因(和)在川中黑山羊排卵、植入后胚胎发育等生殖过程可能呈现负调控作用。研究表明,可抑制应激条件引发的牛卵丘细胞的凋亡[55],对牛卵母细胞发育可能具有增益作用。其5′侧翼区杂合SNPs虽不影响其在卵巢中的表达,但对山羊的多产性具有显著影响[56],且编码区同义突变还可通过调控基因表达影响与受体结合的亲和力[57],进一步影响动物繁殖。因此,后续拟定分析部分SNPs是否与山羊排卵率相关,对于解析川中黑山羊产羔性状遗传基础可能具有理论指导意义。启动子区域内的SNPs可下调其基因的表达[58],进而导致胎盘介导的妊娠并发症及女性的反复流产[59-60]。EDNRA作为一种内皮素受体,不仅参与山羊毛色调控[61],还在肥胖引起的小鼠排卵缺失过程中发挥重要作用[62]。突变可破坏DNA损伤修复过程,可能是引发人类卵巢早衰的潜在病因[63-64],具体的生殖功能调控机制还有待探索。基因编码速激肽家族成员P(SP)和神经激肽A(Neurokinin A,NKA)[65],参与调控哺乳动物生殖激素分泌[66]。速激肽信号对排卵前促黄体素分泌激增具有促进作用,也被认为是调节排卵的一个负面因素,其功能障碍导致雌鼠排卵障碍[67]。此外,川中黑山羊LF组次优选择基因与美姑山羊的繁殖性状相关[68]。这些发现均为产羔数调控网络解析提供了新视角。
HF与LF组间存在多个共享窗口基因,正如之前崂山奶山羊与大足黑山羊的相关报道[13-14],川中黑山羊19个繁殖候选基因中也存在3个HF和LF组的交集基因()。其中调控前列腺素水平[69],参与睾酮的生成[70],存在于卵母细胞和颗粒细胞中,体外成熟液中其激动剂的添加可提高卵母细胞的囊胚率[71],然用其激动剂孵育精子,囊胚率的结果相反[72]。这些基因的功能与动物繁殖紧密相关,其功能的深入挖掘对山羊繁殖学研究是极为重要的,但该相关报道并未对共享窗口基因功能定位做深入剖析,这3个共享基因在川中黑山羊产羔性状调控机制中发挥主控或微效调控作用还有待探查。
进一步的功能分析表明,神经活性配体-受体相互作用、糖胺聚糖-硫酸乙酰肝素/肝素的生物合成、钙离子信号通路、cAMP信号通路和叶酸生物合成通路,在动物繁殖活动中也发挥重要功能。例如硫酸肝素蛋白聚糖是在GDF9信号通路中起着重要作用,并参与排卵周期卵泡中卵母细胞信号和卵丘细胞功能的形成[73]。钙离子信号通路参与调控哺乳动物卵母细胞发育[74-75]。cAMP通路调节卵巢类固醇P4、E2的产生[76],参与卵泡颗粒细胞分化和成熟[77]。神经活性配体-受体相互作用途径不仅调节家禽卵泡发育和卵子生产[78-79],还参与软体动物性腺发育和产卵[80-81]。推测这些通路可能参与川中黑山羊卵母细胞发育及排卵过程,并且对排卵率至关重要。此外,叶酸(维生素B9)是机体健康和发育的最佳营养必需物质。叶酸缺乏会引发贫血、生殖健康和胎儿发育受损[82-83]。多个HF特异性强选择候选基因在这些通路中显著富集,暗示这些通路可能是川中黑山羊产羔数性状关键调控途径,对其多羔性状调节起促进作用。
本研究还特别分析19个候选基因外显子区域的SNPs,以期进一步筛选山羊多羔机制关键分子标记。发现导致氨基酸序列变化的外显子突变只在HF组中,并且仅定位于(G529A)与(A281T)候选基因中。测序结果表明,外显子G529A与A281T突变在人工选择条件下被强烈选择,表现出较高的遗传分化,改变了、的翻译,可能在山羊繁殖力中发挥关键作用,有望作为高产标记用于山羊分子育种。后续在川中黑山羊群体中扩群验证G529A与A281T突变位点,将成为本课题组下一步工作的方向与重点。
总而言之,本研究在川中黑山羊群体中发现了许多新的变异体。这些可能是调控生殖的关键的基因。其中、、、、、和可能是调控山羊多羔性状的关键基因,同时这些候选基因大部分显著富集在神经活性配体-受体相互作用、糖胺聚糖-硫酸乙酰肝素/肝素的生物合成、钙离子信号通路、cAMP信号通路和叶酸生物合成等潜在的生殖相关通路中。外显子G529A与突变可能是川中黑山羊多羔性状的关键遗传标记,在改良山羊繁殖性能方面可能具有较大的应用价值。
4 结论
本研究筛选到19个与产羔性状相关的新候选基因,包括11个HF组特异性候选因(和),5个LF组特异性候选因(和)和3个LF组和HF组共享窗口基因(和)。
[1] DE LIMA L G, DE SOUZA N O B, RIOS R R, DE MELO B A, DOS SANTOS L T A, DE MORAES SILVA K, MURPHY T W, FRAGA A B. Advances in molecular genetic techniques applied to selection for litter size in goats (): a review. Journal of Applied Animal Research, 2020, 48(1): 38-44. doi:10.1080/09712119.2020.1717497.
[2] MULSANT P, LECERF F, FABRE S, SCHIBLER L, MONGET P, LANNELUC I, PISSELET C, RIQUET J, MONNIAUX D, CALLEBAUT I, CRIBIU E, THIMONIER J, TEYSSIER J, BODIN L, COGNIÉ Y, CHITOUR N, ELSEN J M. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(9): 5104-5109. doi:10.1073/pnas.091577598.
[3] GALLOWAY S M, MCNATTY K P, CAMBRIDGE L M, LAITINEN M P E, JUENGEL J L, JOKIRANTA T S, MCLAREN R J, LUIRO K, DODDS K G, MONTGOMERY G W, BEATTIE A E, DAVIS G H, RITVOS O. Mutations in an oocyte-derived growth factor gene () cause increased ovulation rate and infertility in a dosage- sensitive manner. Nature Genetics, 2000, 25(3): 279-283. doi:10. 1038/77033.
[4] MARTINEZ-ROYO A, JURADO J J, SMULDERS J P, MARTÍ J I, ALABART J L, ROCHE A, FANTOVA E, BODIN L, MULSANT P, SERRANO M, FOLCH J, CALVO J H. A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Animal Genetics, 2008, 39(3): 294-297. doi:10.1111/j.1365-2052.2008.01707.x.
[5] HANRAHAN J P, GREGAN S M, MULSANT P, MULLEN M, DAVIS G H, POWELL R, GALLOWAY S M. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and belclare sheep (). Biology of Reproduction, 2004, 70(4): 900-909. doi:10.1095/biolreprod.103.023093.
[6] NICOL L, BISHOP S C, PONG-WONG R, BENDIXEN C, HOLM L E, RHIND S M, MCNEILLY A S. Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep. Reproduction (Cambridge, England), 2009, 138(6): 921-933. doi:10.1530/rep-09-0193.
[7] AHLAWAT S, SHARMA R, MAITRA A. Screening of indigenous goats for prolificacy associated DNA markers of sheep. Gene, 2013, 517(1): 128-131. doi:10.1016/j.gene.2012.12.015.
[8] TEJANGOOKEH H M, SHAHNEH A Z, ZAMIRI M J, DALIRI M, KOHRAM H, JAVAREMI A N. Study of BMP15 gene polymorphism in Iranian goats. African Journal of Biotechnology, 2009, 8(13), 2929-2932.
[9] SUPAKORN C, PRALOMKARN W. Sheep FecB gene polymorphism role in Thai meat goat proliferation rate//Proceedings of 9th World Congress Genetics Applied to Livestock Production, Leipizig Germany, 2010.
[10] HUA G H, CHEN S L, AI J T, YANG L G. None of polymorphism of ovine fecundity major genes FecB and FecX was tested in goat. Animal Reproduction Science, 2008, 108(3/4): 279-286. doi:10.1016/ j.anireprosci.2007.08.013.
[11] HE Y Q, MA X K, LIU X Y, ZHANG C X, LI J. Candidate genes polymorphism and its association to prolificacy in Chinese goats. Journal of Agricultural Science, 2010, 2(1): 88–92. doi:10.5539/jas. v2n1p88.
[12] 李恒, 字向东. 全基因组测序在山羊上的研究进展. 中国畜牧杂志, 2021, 57(10): 29-34. doi:10.19556/j.0258-7033.20200910-01.
LI H, ZI X D. Research progress on whole-genome sequencing on goat. Chinese Journal of Animal Science, 2021, 57(10): 29-34. doi:10.19556/j.0258-7033.20200910-01. (in Chinese)
[13] LAI F N, ZHAI H L, CHENG M, MA J Y, CHENG S F, GE W, ZHANG G L, WANG J J, ZHANG R Q, WANG X, MIN L J, SONG J Z, SHEN W. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (). Scientific Reports, 2016, 6: 38096. doi:10.1038/srep38096.
[14] GUANG-XIN E, ZHAO Y J, HUANG Y F. Selection signatures of litter size in Dazu black goats based on a whole genome sequencing mixed pools strategy. Molecular Biology Reports, 2019, 46(5): 5517-5523. doi:10.1007/s11033-019-04904-6.
[15] ISLAM R, LIU X X, GEBRESELASSIE G, ABIED A, MA Q, MA Y H. Genome-wide association analysis reveals the genetic locus for high reproduction trait in Chinese Arbas Cashmere goat. Genes & Genomics, 2020, 42(8): 893-899. doi:10.1007/s13258-020-00937-5.
[16] E G X, ZHOU D K, YANG B G, DUAN X H, NA R S, HAN Y G, ZENG Y. Association analysis of sixty-seven single nucleotide polymorphisms with litter size in Dazu Black goats. Animal Genetics, 2020, 51(1): 151-152. doi:10.1111/age.12879.
[17] WANG J J, ZHANG T, CHEN Q M, ZHANG R Q, LI L, CHENG S F, SHEN W, LEI C Z. Genomic signatures of selection associated with litter size trait in Jining gray goat. Frontiers in Genetics, 2020, 11: 286. doi:10.3389/fgene.2020.00286.
[18] WANG K, LIU X F, QI T, HUI Y Q, YAN H L, QU L, LAN X Y, PAN C Y. Whole-genome sequencing to identify candidate genes for litter size and to uncover the variant function in goats (). Genomics, 2021, 113(1): 142-150. doi:10.1016/j.ygeno.2020.11.024.
[19] ZI X D, MU X K, LU J Y, MA L, WANG Y. Polymorphisms of growth hormone(GH) and insulin-like growth factor I(IGF-I) genes in prolific Lezhi Black Goat: Possible association with litter size. Journal of Southwest University for Nationalities (Natural Science Edition), 2014, 40(3): 344-349.
[20] LÜ M J, LI H, ZI X D. Assessment of estrous synchronization protocols and pregnancy specific protein B concentration for the prediction of kidding rate in Lezhi black goats. Small Ruminant Research, 2021, 195: 106299. doi:10.1016/j.smallrumres.2020.106299.
[21] YANG C X, ZI X D, WANG Y, YANG D Q, MA L, LU J Y, NIU H R, XIAO X. Cloning and mRNA expression levels of GDF9, BMP15, and BMPR1B genes in prolific and non-prolific goat breeds. Molecular Reproduction and Development, 2012, 79(1): 2. doi:10. 1002/mrd.21386.
[22] ZI X D, LU J Y, MA L. Identification and comparative analysis of the ovarian microRNAs of prolific and non-prolific goats during the follicular phase using high-throughput sequencing. Scientific Reports, 2017, 7: 1921. doi:10.1038/s41598-017-02225-x.
[23] ZI X D, LU J Y, ZHOU H, MA L, XIA W, XIONG X R, LAN D L, WU X H. Comparative analysis of ovarian transcriptomes between prolific and non-prolific goat breeds via high-throughput sequencing. Reproduction in Domestic Animals, 2018, 53(2): 344-351. doi:10. 1111/rda.13111.
[24] LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009, 25(14): 1754- 1760. doi:10.1093/bioinformatics/btp324.
[25] BICKHART D M, ROSEN B D, KOREN S, SAYRE B L, HASTIE A R, CHAN S, LEE J, LAM E T, LIACHKO I, SULLIVAN S T, BURTON J N, HUSON H J, NYSTROM J C, KELLEY C M, HUTCHISON J L, ZHOU Y, SUN J J, CRISÀ A, PONCE DE LEÓN F A, SCHWARTZ J C, HAMMOND J A, WALDBIESER G C, SCHROEDER S G, LIU G E, DUNHAM M J, SHENDURE J, SONSTEGARD T S, PHILLIPPY A M, VAN TASSELL C P, SMITH T P L. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nature Genetics, 2017, 49(4): 643-650. doi:10.1038/ng.3802.
[26] LI H, HANDSAKER B, WYSOKER A, FENNELL T, RUAN J, HOMER N, MARTH G, ABECASIS G, DURBIN R. 1000 genome project data processing subgroup. The sequence alignment/ map format and SAMtools. Microbiology Spectrum, 2009, 25(16): 2078-2079. doi:10.1093/bioinformatics/btp352.
[27] MCKENNA A, HANNA M, BANKS E, SIVACHENKO A, CIBULSKIS K, KERNYTSKY A, GARIMELLA K, ALTSHULER D, GABRIEL S, DALY M, DEPRISTO M A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Cell Reports, 2010, 20(9): 1297-1303. doi:10.1101/ gr.107524.110.
[28] YANG H, WANG K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nature Protocols, 2015, 10(10): 1556-1566. doi:10.1038/nprot.2015.105.
[29] RAUDVERE U, KOLBERG L, KUZMIN I, ARAK T, ADLER P, PETERSON H, VILO J. G: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Research, 2019, 47(W1): W191-W198. doi:10.1093/ nar/gkz369.
[30] BU D C, LUO H T, HUO P P, WANG Z H, ZHANG S, HE Z H, WU Y, ZHAO L H, LIU J J, GUO J C, FANG S S, CAO W C, YI L, ZHAO Y, KONG L. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Research, 2021, 49(W1): W317-W325. doi:10.1093/nar/gkab447.
[31] MOKHTARI M S, ASADI FOZI M, GUTIERREZ J P, NOTTER D R. Genetic and phenotypic aspects of early reproductive performance in Raeini Cashmere goats. Tropical Animal Health and Production, 2019, 51(8): 2175-2180. doi:10.1007/s11250-019-01915-0.
[32] ĐURIČIĆ D, BENIĆ M, ŽAJA I Ž, VALPOTIĆ H, SAMARDŽIJA M. Influence of season, rainfall and air temperature on the reproductive efficiency in Romanov sheep in Croatia. International Journal of Biometeorology, 2019, 63(6): 817-824. doi:10.1007/s00484-019- 01696-z.
[33] ASTUTI D A, KHOTIJAH L, MAIDIN M S, NUGROHO P. Reproductive profile of etawah crossbred does fed Flushing diet containing different kinds of plant oil and animal fat. Pakistan Journal of Biological Sciences, 2020, 23(5): 650-657. doi:10.3923/pjbs.2020. 650.657.
[34] MCCARTHY M I, ABECASIS G R, CARDON L R, GOLDSTEIN D B, LITTLE J, IOANNIDIS J P A, HIRSCHHORN J N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Reviews Genetics, 2008, 9(5): 356-369. doi:10. 1038/nrg2344.
[35] HONG E P, PARK J W. Sample size and statistical power calculation in genetic association studies. Genomics & Informatics, 2012, 10(2): 117-122. doi:10.5808/gi.2012.10.2.117.
[36] LIU N, CUI W B, CHEN M Y, ZHANG X L, SONG X Y, PAN C Y. A 21-bp indel within thegene is significantly associated with litter size in goat. Animal Biotechnology, 2021, 32(2): 213-218. doi:10.1080/10495398.2019.1677682.
[37] JIANG E H, KANG Z H, WANG X Y, LIU Y, LIU X F, WANG Z, LI X C, LAN X Y. Detection of insertions/deletions (InDels) within the goat Runx2 gene and their association with litter size and growth traits. Animal Biotechnology, 2021, 32(2): 169-177. doi:10.1080/10495398. 2019.1671858.
[38] BALBACH M, FUSHIMI M, HUGGINS D J, STEEGBORN C, MEINKE P T, LEVIN L R, BUCK J. Optimization of lead compounds into on-demand, nonhormonal contraceptives: Leveraging a public- private drug discovery institute collaboration. Biology of Reproduction, 2020, 103(2): 176-182. doi:10.1093/biolre/ioaa052.
[39] CHEN H, CHAN H C. Amplification of FSH signalling by CFTR and nuclear soluble adenylyl cyclase in the ovary. Clinical and Experimental Pharmacology & Physiology, 2017, 44(Suppl 1): 78-85. doi:10.1111/1440-1681.12756.
[40] JAYARAJAN V, APPUKUTTAN A, ASLAM M, REUSCH P, REGITZ-ZAGROSEK V, LADILOV Y. Regulation of AMPK activity by type 10 adenylyl cyclase: Contribution to the mitochondrial biology, cellular redox and energy homeostasis. Cellular and Molecular Life Sciences, 2019, 76(24): 4945-4959. doi:10.1007/s00018-019-03152-y.
[41] WANG C, LI S J, LI C, FENG Y P, PENG X L, GONG Y Z. Molecular cloning, expression profile, polymorphism and the genetic effects of the dopamine D1 receptor gene on duck reproductive traits. Molecular Biology Reports, 2012, 39(9): 9239-9246. doi:10.1007/ s11033-012-1797-3.
[42] LIU Z, YANG N, YAN Y, LI G, LIU A, WU G, SUN C. Genome-wide association analysis of egg production performance in chickens across the whole laying period. BMC Genetics, 2019, 20(1): 67. doi:10.1186/ s12863-019-0771-7.
[43] BARONCHELLI S, VILLA N, REDAELLI S, LISSONI S, SACCHERI F, PANZERI E, CONCONI D, BENTIVEGNA A, CROSTI F, SALA E, BERTOLA F, MAROZZI A, PEDICINI A, VENTRUTO M, POLICE M A, DALPRÀ L. Investigating the role of X chromosome breakpoints in premature ovarian failure. Molecular Cytogenetics, 2012, 5(1): 32. doi:10.1186/1755-8166-5-32.
[44] FESTA A, UMANO G R, MIRAGLIA DEL GIUDICE E, GRANDONE A. Genetic evaluation of patients with delayed puberty and congenital hypogonadotropic hypogonadism: Is it worthy of consideration? Frontiers in Endocrinology, 2020, 11: 253. doi:10.3389/fendo.2020. 00253.
[45] LI J, LIU J, CAMPANILE G, PLASTOW G, ZHANG C, WANG Z, CASSANDRO M, GASPARRINI B, SALZANO A, HUA G, LIANG A, YANG L. Novel insights into the genetic basis of buffalo reproductive performance. BMC Genomics, 2018, 19(1): 814. doi:10. 1186/s12864-018-5208-6.
[46] NALLASAMY S, KAYA OKUR H S, BHURKE A, DAVILA J, LI Q X, YOUNG S L, TAYLOR R N, BAGCHI M K, BAGCHI I C. Msx homeobox genes act downstream of BMP2 to regulate endometrial decidualization in mice and in humans. Endocrinology, 2019, 160(7): 1631-1644. doi:10.1210/en.2019-00131.
[47] LIU Z K, WANG R C, HAN B C, YANG Y, PENG J P. A novel role of IGFBP7in mouse uterus: regulating uterine receptivity through Th1/Th2 lymphocyte balance and decidualization. PLoS ONE, 2012, 7(9): e45224. doi:10.1371/journal.pone.0045224.
[48] GERHART J, SCHEINFELD V L, MILITO T, PFAUTZ J, NEELY C, FISHER-VANCE D, SUTTER K, CRAWFORD M, KNUDSEN K, GEORGE-WEINSTEIN M. Myo/Nog cell regulation of bone morphogenetic protein signaling in the blastocyst is essential for normal morphogenesis and striated muscle lineage specification. Developmental Biology, 2011, 359(1): 12-25. doi:10.1016/j.ydbio. 2011.08.007.
[49] NYEGAARD M, OVERGAARD M T, SU Y Q, HAMILTON A E, KWINTKIEWICZ J, HSIEH M, NAYAK N R, CONTI M, CONOVER C A, GIUDICE L C. Lack of functional pregnancy- associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female Fertility1. Biology of Reproduction, 2010, 82(6): 1129-1138. doi:10.1095/biolreprod.109.079517.
[50] KORDUS R J, HOSSAIN A, CORSO M C, CHAKRABORTY H, WHITMAN-ELIA G F, LAVOIE H A. Cumulus cell pappalysin-1, luteinizing hormone/choriogonadotropin receptor, amphiregulin and hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta- isomerase 1 mRNA levels associate with oocyte developmental competence and embryo outcomes. Journal of Assisted Reproduction and Genetics, 2019, 36(7): 1457-1469. doi:10.1007/s10815-019- 01489-8.
[51] YU M, WANG J, LIU S, WANG X Q, YAN Q. Novel function of pregnancy-associated plasma protein A: promotes endometrium receptivity by up-regulating N-fucosylation. Scientific Reports, 2017, 7: 5315. doi:10.1038/s41598-017-04735-0.
[52] WON J, DE EVSIKOVA C M, SMITH R S, HICKS W L, EDWARDS M M, LONGO-GUESS C, LI T S, NAGGERT J K, NISHINA P M. NPHP4is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Human Molecular Genetics, 2010, 20(3): 482-496. doi:10.1093/hmg/ ddq494.
[53] MAO S, WU F, CAO X, HE M, LIU N, WU H, YANG Z, DING Q, WANG X. Tdrp deficiency contributes to low sperm motility and is a potential risk factor for male infertility. American Journal of Translational Research, 2016, 8(1): 177-187.
[54] FARACH M C, TANG J P, DECKER G L, CARSON D D. Heparin/heparan sulfate is involved in attachment and spreading of mouse embryos. Developmental Biology, 1987, 123(2): 401-410. doi:10.1016/0012-1606(87)90398-8.
[55] 潘阳阳, 王萌, 芮弦, 王立斌, 何翃闳, 王靖雷, 马睿, 徐庚全, 崔燕, 樊江峰, 余四九. IGF-1调控RBM3表达抑制低温应激诱导牦牛卵丘细胞凋亡. 中国农业科学, 2020, 53(11): 2285-2296.
PAN Y Y, WANG M, RUI X, WANG L B, HE H H, WANG J L, MA R, XU G Q, CUI Y, FAN J F, YU S J. RNA-binding motif protein 3(RBM3) expression is regulated by insulin-like growth factor(IGF-1) for protecting yak() cumulus cells from apoptosis during hypothermia stress. Scientia Agricultura Sinica, 2020, 53(11): 2285-2296. (in Chinese)
[56] THOMAS N, VENKATACHALAPATHY T, ARAVINDAKSHAN T, RAGHAVAN K C. Molecular cloning, SNP detection and association analysis of 5' flanking region of the goat IGF1gene with prolificacy. Animal Reproduction Science, 2016, 167: 8-15. doi:10.1016/j. anireprosci.2016.01.016.
[57] CHENG Y Y, LIU S C, WANG G, WEI W Z, HUANG S, YANG R, GENG H W, LI H Y, SONG J, SUN L D, YU H, HAO L L. Porcine IGF1synonymous mutation alter gene expression and protein binding affinity with IGF1R. International Journal of Biological Macromolecules, 2018, 116: 23-30. doi:10.1016/j.ijbiomac.2018.05.022.
[58] INAGAKI H, OTA S, NISHIZAWA H, MIYAMURA H, NAKAHIRA K, SUZUKI M, NISHIYAMA S, KATO T, YANAGIHARA I, KURAHASHI H. Obstetric complication-associatedpromoter polymorphisms may affect gene expression via DNA secondary structures. Journal of Human Genetics, 2019, 64(5): 459-466. doi:10.1038/s10038-019-0578-4.
[59] ARANDA F, UDRY S, PERÉS WINGEYER S, AMSHOFF L C, BOGDANOVA N, WIEACKER P, LATINO J O, MARKOFF A, LARRAÑAGA G. Maternal carriers of the ANXA5 M2 haplotype are exposed to a greater risk for placenta-mediated pregnancy complications. Journal of Assisted Reproduction and Genetics, 2018, 35(5): 921-928. doi:10.1007/s10815-018-1142-4.
[60] DRYLLIS G, GIANNOPOULOS A, ZOI C, POULIAKIS A, LOGOTHETIS E, VOULGARELIS M, ZOI K, KOUSKOUNI E, DINOU A, STAVROPOULOS-GIOKAS C, KREATSAS G, KONSTANTOPOULOS K, POLITOU M. Correlation of single nucleotide polymorphisms in the promoter region of the(annexin A5) gene with recurrent miscarriages in women of Greek origin. The Journal of Maternal-Fetal & Neonatal Medicine, 2020, 33(9): 1538-1543. doi:10.1080/14767058.2018.1521799.
[61] DI GERLANDO R, MASTRANGELO S, MOSCARELLI A, TOLONE M, SUTERA A M, PORTOLANO B, SARDINA M T. Genomic structural diversity in local goats: Analysis of copy-number variations. Animals, 2020, 10(6): E1040. doi:10.3390/ani10061040.
[62] HOHOS N M, ELLIOTT E M, GIORNAZI A, SILVA E, RICE J D, SKAZNIK-WIKIEL M E. High-fat diet induces an ovulatory defect associated with dysregulated endothelin-2 in mice. Reproduction (Cambridge, England), 2021, 161(3): 307-317. doi:10.1530/rep-20- 0290.
[63] YANG Y, GUO T, LIU R, KE H, XU W, ZHAO S, QIN Y.gene mutations in premature ovarian insufficiency. Human Mutation, 2020, 41(5): 1033-1041. doi:10.1002/humu.23997.
[64] YANG Y, ZHAO S, QIN Y. Response to “Should FANCL heterozygous pathogenic variants be considered as potentially causative of primary ovarian insufficiency? ”. Human Mutation, 2020, 41(9): 1700-1701. doi:10.1002/humu.24073.
[65] FERGANI C, NAVARRO V M. Expanding the role of tachykinins in the neuroendocrine control of reproduction. Reproduction (Cambridge, England), 2016, 153(1): R1-R14. doi:10.1530/rep-16-0378.
[66] LEÓN S, FERGANI C, TALBI R, SIMAVLI S, MAGUIRE C A, GERUTSHANG A, NAVARRO V M. Characterization of the role of NKA in the control of puberty onset and gonadotropin release in the female mouse. Endocrinology, 2019, 160(10): 2453-2463. doi:10. 1210/en.2019-00195.
[67] LEÓN S, FERGANI C, TALBI R, MAGUIRE C A, GERUTSHANG A, SEMINARA S B, NAVARRO V M. Tachykinin signaling is required for induction of the preovulatory luteinizing hormone surge and normal luteinizing hormone pulses. Neuroendocrinology, 2021, 111(6): 542-554. doi:10.1159/000509222.
[68] GUO J Z, TAO H X, LI P F, LI L, ZHONG T, WANG L J, MA J Y, CHEN X Y, SONG T Z, ZHANG H P. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Scientific Reports, 2018, 8: 10405. doi:10.1038/s41598-018- 28719-w.
[69] PASTEL E, POINTUD J C, LOUBEAU G, DANI C, SLIM K, MARTIN G, VOLAT F, SAHUT-BARNOLA I, VAL P, MARTINEZ A, LEFRANÇOIS-MARTINEZ A M. Aldose reductases influence prostaglandin F2α levels and adipocyte differentiation in male mouse and human species. Endocrinology, 2015, 156(5): 1671-1684. doi:10. 1210/en.2014-1750.
[70] ZHANG Q, PEI L G, LIU M, LV F, CHEN G H, WANG H. Reduced testicular steroidogenesis in rat offspring by prenatal nicotine exposure: Epigenetic programming and heritability via nAChR/ HDAC4. Food and Chemical Toxicology, 2020, 135: 111057. doi:10.1016/j.fct.2019.111057.
[71] OLABARRIETA E, TOTORIKAGUENA L, AGIRREGOITIA N, AGIRREGOITIA E. Implication of mu opioid receptor in thematuration of oocytes and its effects on subsequent fertilization and embryo development in mice. Molecular Reproduction and Development, 2019, 86(9): 1236-1244. doi:10.1002/mrd.23248.
[72] OLABARRIETA E, TOTORIKAGUENA L, ROMERO- AGUIRREGOMEZCORTA J, AGIRREGOITIA N, AGIRREGOITIA E. Mu opioid receptor expression and localisation in murine spermatozoa and its role in IVF. Reproduction Fertility and Development, 2020, 32(4): 349-354. doi:10.1071/rd19176.
[73] WATSON L N, MOTTERSHEAD D G, DUNNING K R, ROBKER R L, GILCHRIST R B, RUSSELL D L. Heparan sulfate proteoglycans regulate responses to oocyte paracrine signals in ovarian follicle morphogenesis. Endocrinology, 2012, 153(9): 4544-4555. doi:10. 1210/en.2012-1181.
[74] TIWARI M, PRASAD S, SHRIVASTAV T G, CHAUBE S K. Calcium signaling during meiotic cell cycle regulation and apoptosis in mammalian oocytes. Journal of Cellular Physiology, 2017, 232(5): 976-981. doi:10.1002/jcp.25670.
[75] STEWART T A, DAVIS F M. An element for development: Calcium signaling in mammalian reproduction and development. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2019, 1866(7): 1230-1238. doi:10.1016/j.bbamcr.2019.02.016.
[76] LU N S, LI M J, LEI H L, JIANG X Y, TU W L, LU Y, XIA D. Butyric acid regulates progesterone and estradiol secretion via cAMP signaling pathway in porcine granulosa cells. The Journal of Steroid Biochemistry and Molecular Biology, 2017, 172: 89-97. doi:10. 1016/j.jsbmb.2017.06.004.
[77] JOZKOWIAK M, HUTCHINGS G, JANKOWSKI M, KULCENTY K, MOZDZIAK P, KEMPISTY B, SPACZYNSKI R Z, PIOTROWSKA- KEMPISTY H. The stemness of human ovarian granulosa cells and the role of resveratrol in the differentiation of MSCs-A review based on cellular and molecular knowledge. Cells, 2020, 9(6): E1418. doi:10.3390/cells9061418.
[78] ZHANG T, CHEN L, HAN K P, ZHANG X Q, ZHANG G X, DAI G J, WANG J Y, XIE K Z. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Animal Reproduction Science, 2019, 208: 106114. doi:10.1016/j.anireprosci. 2019.106114.
[79] CHEN X, SUN X, CHIMBAKA I M, QIN N, XU X, LISWANISO S, XU R, GONZALEZ J M. Transcriptome analysis of ovarian follicles reveals potential pivotal genes associated with increased and decreased rates of chicken egg production. Frontiers in Genetics, 2021, 12: 622751. doi:10.3389/fgene.2021.622751.
[80] XU R Y, PAN L Q, YANG Y Y, ZHOU Y Y. Characterizing transcriptome in female scallopfarreri provides new insights into the molecular mechanisms of reproductive regulation during ovarian development and spawn. Gene, 2020, 758: 144967. doi:10. 1016/j.gene.2020.144967.
[81] HUANG D X, ZHANG B, HAN T, LIU G B, CHEN X, ZHAO Z H, FENG J Q, YANG J W, WANG T M. Genome-wide prediction and comparative transcriptomic analysis reveals the G protein-coupled receptors involved in gonadal development of. Genomics, 2021, 113(1): 967-978. doi:10.1016/j.ygeno.2020.10.030.
[82] NADERI N, HOUSE J D. Recent developments in folate nutrition// Advances in Food and Nutrition Research. Amsterdam: Elsevier, 2018: 195-213. doi:10.1016/bs.afnr.2017.12.006.
[83] BROWN L L, COHEN B E, EDWARDS E, GUSTIN C E, NOREEN Z. Physiological need for calcium, iron, and folic acid for women of various subpopulations during pregnancy and beyond. Journal of Womens Health (Larchmt), 2021, 30(2): 207-211. doi:10.1089/jwh. 2020.8873.
Screening of Key Regulatory Genes for Litter Size Trait Based on Whole Genome Re-Sequencing in Goats ()
LI Heng1, ZI XiangDong1, WANG Hui2, XIONG Yan1, LÜ MingJie1,LIU Yu1, JIANG XuDong1
1Key Laboratory of Animal Science of National Ethnic Affairs Commission, Southwest Minzu University, Chengdu 610041;2Key Laboratory of Qinghai-Tibetan Plateau Animal Genetic Resource Reservation and Utilization of Ministry of Education, Southwest Minzu University, Chengdu 610041
【Objective】The purpose of this study was to analyze the genome of different fecundity populations of goats (s) and to explore the key regulatory genes involved in the regulation of litter size traits of Chuanzhong black goats (CBGs), and to provide the theoretical reference for analyzing the genetic mechanism of litter size traits and molecular genetic improvement of fecundity in goats. 【Method】The high fecundity (HF) CBG does (= 6) that produced 4-6 kids per doe kidding and low fecundity (LF) does (= 6) that produced only one kid per doe kidding were chosen in this study. The jugular blood samples were collected to extract genomic DNA. The 350 bp double-terminal sequencing library was constructed, and then 12 whole genome libraries were resequenced by IlluminaHiSeqPE150 platform. The clean data from sequencing were mapped to goat reference genome ARS1 by using BWA software, and two whole-genome scanning analysis methods (and) were used to comprehensively analyze the high-quality SNPs obtained to identify candidate regions. GO analysis and KEGG pathway analysis were performed on the G:Profiler and KOBAS online databases, respectively, to screen candidate genes for regulating the number of kids in CBGs. To further identify the key genetic markers that regulate the number of kids, the synonymous and non-synonymous single nucleotide polymorphisms (SNPs) of reproductive candidate genes were mapped and screened according to the variation analysis report of genome resequencing. The amplified products of 12 goat samples were sequenced by Sanger sequencing to verify the resequencing results.【Result】A total of 431.50 Gb clean data were obtained from the genome resequencing study of 12 CBGs. Through mutation detection and annotation, 7 771 417 SNPs were detected in HF group and 8 935 907 SNPs were detected in LF group, and all types of the LF group SNPs were more than those in HF group. The windows that reach the maximumvalue of top 5% and the minimumvalue of top 5% were set as candidate regions. A total of 130 strong selection signals were annotated in the regions with low heterozygosity and high genetic differentiation, of which 84, 59 and 13 genes were annotated in HF group, LF group and shared window, respectively. GO enrichment analysis and KEGG pathway showed that 19 candidate genes were involved in the regulation of reproduction, reproduction and embryonic development of CBG, including 11 HF group-specific candidate genes (,,,,,,,,,, and), and five strong selection signal genes (,,,, and) in LF group, and three window genes (,and) in HF group shared with LF group. The most GO terms, such as G-protein-coupled receptor activity, hormone response and neuropeptide signal pathway, contained these 19 candidate genes. In addition, nine of the 14 HF candidate genes were significantly enriched in metabolic pathway, neuroactive ligand-receptor interaction, glycosaminoglycan-heparan sulfate/heparin biosynthesis, calcium signal pathway, cAMP signal pathway and folate biosynthesis KEGG pathways (<0.05). Among the 19 reproductive candidate genes, there were two synonymous mutations (G771T,G529,,andgene mutations could be detected, and this result was consistent with the results of genome resequencing, in whichG529A polymorphism led to alanine mutation to threonine, and【Conclusion】A total of 11 HF group-specific candidate genes were found in this study, which were speculated to be the key regulatory genes for fecundity trait. The mutations ofgene exon G529A andexon A281T might be the key genetic markers for regulating prolificacy traits in goats, which had great application value in improving reproductive performance of goats.
Chuanzhong black goat; genome resequencing; fecundity; candidate genes

10.3864/j.issn.0578-1752.2022.23.015
2021-08-17;
2022-10-12
国家自然科学基金(31902154)、西南民族大学中央高校基本科研业务专项(2021PTJS26)
李恒,E-mail:lih199501 @sina.com。通信作者字向东,E-mail:zixd@sina.com
(责任编辑 林鉴非)

