Construction of miRNA-mRNA regulatory network and drug prediction for ulcerative colitis associated colorectal cancer
Qing-Song Liu ,Shuang-Lan Chen ,Yi Zhang* ,Bin Li ,Zi-Yan Xie ,Shuai Xiong
1Affiliated Hospital of Chengdu University of Traditional Chinese Medicine,Chengdu 610075,China.
Abstract Objective:To construct the miRNA-mRNA regulatory network,and identify more reliable therapeutic targets and potential drugs in ulcerative colitis associated colorectal cancer.Methods: Two datasets were downloaded from the GEO,and the differently expressed analysis were conducted by R software limma package.Functional enrichment analysis was performed using R software.The targets of differently expressed miRNAs were predicted by FunRich software,and the miRNA-mRNA regulatory network was constructed by Cytoscape software.The cMAP and TCMSP databases were used to predict small molecule drugs and traditional Chinese medicine respectively.Results: A total of 79 differently expressed miRNAs and 8865 differently expressed mRNAs were identified.Then the miRNA-mRNA regulatory network was constructed.Among DE miRNAs in the network,hsa-miR-520e,hsa-miR-199b-5p,hsa-miR-140-5p may be the most significant due to their large number of connecting nodes in ulcerative colitis associated colorectal cancer.The integrated differently genes were mainly concentrated in protein processing in endoplasmic reticulum,ferroptosis and other signaling pathways.In addition,10 kinds of small molecule drugs and 6 kinds of traditional Chinese medicine were screened as therapeutic agents for ulcerative colitis associated colorectal cancer.Conclusion:hsa-miR-520e,hsa-miR-199b-5p,hsa-miR-140-5p can be used as therapeutic targets for ulcerative colitis associated colorectal cancer.The pathogenesis of ulcerative colitis associated colorectal cancer may be related to the protein processing in endoplasmic reticulum/ferroptosis signaling pathway,and it is predicted that 10 kinds of small molecule drugs,such as Isoflupredone,and 4 traditional Chinese medicines,such as Baiqucai (Celandine),Guanhuangbai (Cortex phellodendri amurensis),Huangbai(Phellodendron amurense)and Bajiaolian (Dysosma Versipellis),can be used as therapeutic drugs for ulcerative colitis associated colorectal cancer.
Key words:Ulcerative colitis associated colorectal cancer,miRNA-mRNA regulatory network,Small molecule drugs,traditional Chinese medicine
Background
Ulcerative colitis associated colorectal cancer(UC-CRC) is one of the most serious complications of ulcerative colitis (UC) [1].UC-CRC and colorectal cancer (CRC) have different clinical characteristics and carcinogenesis mechanism,but the specific mechanism of carcinogenesis has not been revealed[2].MicroRNA (miRNA) is an endogenous RNA with 20 to 24 nucleotides in length,which has many important and complex regulatory functions in cells[3].The effect of miRNA on mRNA is expressed by cutting off the mRNA molecule and inhibiting the translation [4].Previous studies have shown that RNA plays a key role in the occurrence and development of UC-CRC,for example,miR-370-3p attenuates UC-CRC in mice by inhibiting inflammation and epithelial-mesenchymal transformation [5].miR-193a-3p promotes ulcerative colitis carcinogenesis through up-regulation of IL17RD [6].
In this study,miRNA-mRNA regulatory network was constructed by using miRNA and mRNA associated with UC-CRC in order to explore the potential molecular mechanism of the disease,and to search for new therapeutic drugs for UC-CRC by using small molecular compounds and natural drugs prediction.
Methods
Data download and screen differentially expressed
miRNA data set GSE68306 [7] and mRNA data set GSE3629 [8] were downloaded from the GEO database.As to GSE68306,it includes 9 UC and 20 UC-CRC samples.GSE3629 includes 43 UC samples and 16 UC-CRC samples.The package of limma analysis in R software(version:4.0.2)was used to identify the disorder genes in UC tissue and UC-CRC tissue.In order to control the error detection rate,the miRNA cutoff standard was set to |logFC| >0.585 andp<0.05,and the mRNA cutoff standard was set to|logFC| >2 andp<0.05.The R package of ggplot 2 was used to draw volcanic maps.
Construction of a miRNA-mRNA regulatory network
Differential miRNAs in GSE68306 were fed into FunRich software (version:3.1.4) to predict their target genes,and then crossed with the differential mRNA in GSE3629 data set.In this way,we can predict the regulatory relationship between differentially expressed miRNAs and differentially expressed genes.Cytoscape (version:3.8.0) was used to visualize the miRNA-mRNA regulatory network.
Enrichment analysis and protein-protein interaction (PPI) network analysis
Use R software to perform gene ontology (GO)functional enrichment analysis and kyoto encyclopedia of genes and genomes pathways (KEGG) analysis on the differential mRNAs.Both useadj.P<0.05 as the screening criteria and draw GO Bar and KEGG Bubble charts.The top five terms were visualized if there were more than five terms.
Using STRING database to predict gene interaction,set the minimum required interaction score,high confidence (0.700),downloading the protein interaction relationship and importing Cytoscape software to construct the PPI network.
Related small-molecule compounds and herbs screening
The Connectivity Map (cMAP,https://portals.broadinstitute.org/cMAP) was a Gene expression profiling database of small molecule drugs,gene expression and disease,which plays an important role in The field of drug research.According to miRNA-mRNA regulatory network,differential mRNAs were divided into up-regulation and down-regulation groups,which were imported into cMAP database to calculate similarity enrichment scores from-1 to 1.Small molecules generated from up-regulated genes suggested therapeutic goals,while down-regulated genes predicted inhibitors of therapy for the disease.The top 10 small molecule compounds were selected according to theP valueand entered into Traditional Chinese Medicine Database and Analysis Platform (TCMSP,https://tcmspw.com/tcmsp.php) to predict the corresponding natural drugs.Small molecule compounds and natural drugs predicted by cMAP and TCPSP database may provide references for the use of UC-CRC natural products.
Results
Preliminary screening of differentially expressed miRNAs and mRNAs
GSE68306 and GSE3629 were analyzed by the limma package in R software.Finally,32 down-regulated miRNAs and 47 up-regulated miRNAs were obtained.4438 down-regulated mRNAs and 4427 up-regulated mRNAs were screened (Figure 1A and Figure 1B).
Analysis of miRNA-mRNA regulatory network
The predicted regulatory table of miRNA-mRNA was derived from FunRich Software,and Cytoscape was used to visual analysis of regulatory networks (Figure 2).The figure includes 13 miRNAs and 357 mRNAs,in which the red node represents up-regulation and the green node represents down-regulation,the number of genes around miRNAs regulation or the number of miRNAs around a surrounding gene was defined as degrees,which represented the center of the miRNA-mRNA regulatory network,and the more important of the network was the greater the degree of its base.Obviously,four miRNAs,which include hsa-miR-520e (Degree=82),hsa-miR-199b-5p(Degree=64),hsa-miR-140-5p (Degree=62),are at the core of the regulatory network (Table 1).

Table 1 The degree of Hub miRNA
Functional and pathway enrichment analysis
GO and KEGG pathway enrichment analysis were performed for differential mRNAs in miRNA-mRNA network.GO was divided into three parts (Figure 3A):Molecular Function (MF),Biological Process (BP),and Cellular Component (CC).BP of GO analysis were associated with Ras protein signal transduction,cardiac epithelial to mesenchymal transition,endocardial cushion development,RNA splicing,Rap protein signal transduction.MF were associated with GDP binding,Rho Gtpase binding,Rac GTPase binding,small GTPase binding,GTPase activity.CC were associated with focal adhesion,cell-substrate junction,nuclear speck,midbody,recycling endosome.The results of the KEGG pathway enrichment analysis were associated with Renal cell carcinoma,Protein processing in endoplasmic reticulum,Ras signaling pathway,mRNA surveillance pathway,Ferroptosis(Figure 3B).
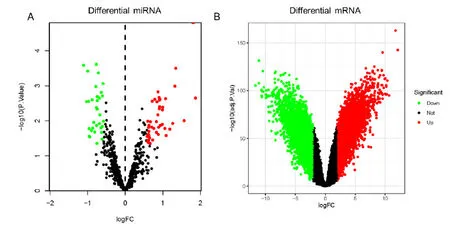
Figure 1 The volcano of Differential miRNA and Differential mRNA expression

Figure 2 miRNA-mRNA regulatory network

Figure 3 Enrichment analysis of gene ontology and kyoto encyclopedia of genes and genomes pathways pathway
Construction of PPI Network
PPI network was constructed for of differential mRNAs in miRNA-mRNA network according to STRING database (Figure 4).Many of these differentially expressed genes have high node degrees:RAC1(degree=31),APP (degree=26),HNRNPA2B1(degree=23),HSPA5 (degree=21),PABPN1 (degree=19),PAPOLA (degree=18),SRRM1 (degree=18),PCBP1 (degree=18),PAX6 (degree=17),CDC40(degree=16).These nodes may play an important role in the carcinogenesis of UC.

Figure 4 The network of protein-protein interaction
Related small-molecule compounds and herbs screening
The cMAP database was used for small molecule drug screening based on differentially expressed genes in the miRNA-mRNA regulatory network.Select the top 10 small molecule compounds according toP<0.005(Table 2).Among these compounds,isoflupredone,3-acetamidocoumarin,isoxicam,trihexyphenidyl,canadine,podophyllotoxinmay perturb the development of the carcinogenesis of UC.whilebetonicine,gliclazide,zimeldine,camptothecin,alpha-estradiolmight provide potentially therapeutic goals for UC-CRC.Then,four kinds of traditional Chinese medicines (Celandine,Cortex phellodendri amurensis,Phellodendron amurense and DysosmaVersipellis) were obtained by inputting small molecular compounds into TCMSP database,which may have potential anti UC-CRC effect.Besides,bitter almond and black sesamemay promote the change from UC to UC-CRC,which will be confirmed by further study.

Table 2 Screening of small molecule drug and traditional Chinese medicine
Discussion
Currently,the prediction of novel miRNA tumor markers for tumor auxiliary diagnosis,therapeutic response and prognosis has gradually become a research hotspot [9],and the miRNA-mRNA regulatory network regulates the biological pathways and processes of tumor genesis and development through complex relationships.UC-CRC is one of the most serious complications of UC,which is difficult to cure,seriously reduces the quality of life of patients,and even endangers their lives [10].Therefore,early detection,early diagnosis and early treatment are of great significance to improve the prognosis of UC-CRC.
In this study,we used UC as the control group and UC-CRC as the experimental group to obtain the differentially expressed miRNAs and their target genes.Then,miRNA-mRNA regulatory and PPI networks were constructed.Next,GO and KEGG pathways were enriched by differentially expresses genes in miRNA-mRNA regulatory network.finally,the differentially expressed genes in miRNA-mRNA regulatory networks were predicted for their potential small molecule drugs and Traditional Chinese medicine.
So far,the regulatory mechanism of hsa-miR-520e,hsa-miR-199b-5p and hsa-miR-140-5p in UC-CRC has not been found.However,studies have shown that they have a certain relationship with the occurrence and development of tumors.For example,hsa-miR-520e plays a key role in hereditary non-polyposis colorectal cancer [11].hsa-miR-199b-5p is a diagnostic biomarker of tumor [12,13].The functional and clinical significance of Mir-140-5p suggest that it is a key regulator of CRC progression and metastasis,and may serve as a novel therapeutic molecule to treat CRC [14,15].KEGG pathways enrichment results showed that these miRNA were involved in Renal cell carcinoma,Protein processing in endoplasmic reticulum,Ras signaling pathway,mRNA surveillance pathway,Ferroptosis.MUC2 mucin is one of the most abundant secreted proteins in intestinal goblet cells[16].Accumulation of misfolded non-glycosilated MUC2 precursors in goblet cells and activation of the unfolded protein response (UPR) was observed in both inflammatory and non-inflammatory colon tissues of UC [17],but in fact endoplasmic reticulum stress and UPR can participate in the occurrence and development of tumors [18].Protein processing in endoplasmic reticulum is closely associated with iron death,and it has been shown that phosphorylated NF-κBp65 protects against iron death by inhibiting er stress [19].Iron death is an iron-dependent form of cell death characterized by the accumulation of oxidized lipids in membranes [19].Iron chelating agent significantly reduced reactive oxygen species in colon tissues of UC patients and improved clinical symptoms and endoscopic manifestations [20].There are few studies on iron death in colorectal cancer with ulcerative colitis,but its activation leads to non-apoptotic destruction of some tumor cells [21],which provides ideas for the study of UC-CRC related mechanisms.
The cMAP database showed thatisoflupredone,3-acetamidocoumarin,isoxicam,trihexyphenidyl,canadine,podophyllotoxinmay perturb the development of the carcinogenesis of UC.whilebetonicine,gliclazide,zimeldine,camptothecin,alpha-estradiolmight provide potentially therapeutic goals for UC-CRC.The small molecule drugs were imported into TCMSP database to obtain 6 traditional Chinese medicines includingbitter almond,black sesame,celandine,Celandine,Cortex phellodendri amurensis,Phellodendron amurenseandDysosma Versipellis.Among these Herbs,celandine,Celandine,Cortexphellodendriamurensis,Phellodendron amurense and Dysosma Versipelliscould improve the quality of life of patients and support traditional chemotherapy or radiotherapy,suggesting that natural compounds have more advantages as anticancer drugs.Traditional Chinese medicine has significant curative effect and advantages in the treatment of UC.ccording to the clinical manifestations and disease development of UC-CRC,it can be classified into the category of"long toxic dysentery" by referring to Yan Yonghe in Song Dynasty's "Jisheng Fang,Daybimen,Dysentery Treatment" [22].Celandinedampness anddetoxification,Cortex phellodendri amurensispurges heat to check dysentery,Phellodendron amurenseandDysosma Versipellisclear heat and detoxify,in line with the pathogenesis of "long toxic dysentery".However,the limitation of this study is that it is currently limited to the theoretical level.To obtain further accurate verification,it depends on the further experimental verification,which will be the direction of our future efforts.
Conclusion
In this study,the miRNA-mRNA regulatory network of UC-CRC was constructed and revealed that hsa-miR-520e,hsa-miR-199b-5p and hsa-miR-140-5p could be used as therapeutic targets of UC-CRC.It was also found thatisoflupredone,3-acetamidocoumarin,isoxicam,trihexyphenidyl,canadine,podophyllotoxinand 4 Chinese herbs,incloudscelandine,Celandine,Cortexphellodendriamurensis,Phellodendron amurense and Dysosma Versipelliscould be potential therapeutic drugs for UC-CRC.Bioinformatics can help effectively reveal the pathophysiological molecular mechanism of diseases for more indepth research,we can perfect the clinical information of UC-CRC and expand the sample size of UC-CRC population through clinical follow-up in the next stage,and also we can excavate more small molecule drugs and traditional Chinese Medicine of anti UC-CRC through in biological information and vitro experiments;finally,establishing a reliable animal model of UC-CRC will be essential to all research areas of UC-CRC.
- Medical Data Mining的其它文章
- NiuHuangJiangYa capsule for hypertension:a Bayesian dose-response analysis of multiple N-of-1 trials
- The research on rule of Acupoints and Massage Manipulations selection for Postischemic Stroke Constipation based on association rule and entropy clustering analysis
- Efficacy of acupuncture as an adjuvant to speech and language therapy for aphasia after ischemic stroke:a protocol of systematic review and meta-analysis
- Molecular mechanism of Mimenghua Granules in treating dry eye
- Based on Network Pharmacology to Explore the Action Mechanism of Caulis Sinomenii-Caulis Piperis Kadsurae Herb Pair in the Treatment of Rheumatoid Arthritis

