Synthesis,Structure,DNA Binding and Cleavage Activities of Novel Binuclear Copper(II)Complex with Hydrazide Derivative
TANG Qing,LIU Xia,TIAN Anni,HU Zhouping,YAN Jun,ZHANG Shouchun
(School of Chemistry and Chemical Engineering,Central South University,Changsha 410083,China)
Abstract:A water soluble binuclear Cu(II)complex[Cu2(BPH)2Cl4]·CH3OH(BPH is1-benzoyl-1-phenylhydrazine)was synthesized and characterized by elemental analysis,infrared spectroscopy and single crystal Xray crystallography.In the complex,two[Cu(BPH)Cl]units are bridged by two chloride ions,and each Cu(II)ion displays a five-coordinate slightly distorted square-pyramid structure.Multi-spectroscopic DNA interactions studies indicated that the complex may bind to DNA via groove binding modes.The cleavage effect of the complex on supercoiled pBR322 DNA was studied by gel electrophoresis method.The results show that the complex could cleave DNA effectively in the presence of H2O2 via an oxidative mechanism.The singlet oxygen and metal ions may play crucial roles in the DNA cleavage process.
Key words:hydrazide;binuclear copper complex;DNA binding;DNA cleavage
0 Introduction
Hydrazide is formed in the condensation of carboxylic acid and hydrazine with the general structure R-CO-NR1-NR2R3(R,R1,R2,and R3could be hydrogen,alkyl,aromatic groups,etc.).Hydrazide compounds have received much attention due to their remarkable biological and pharmacological properties[1-3].Moreover,hydrazide-based ligands have gained considerable interest owing to their excellent chelating ability with transition metal ions,and the obtained metal complexes also exhibited wide variety of biological activities,and the obtained metal complexes show great potential in new drug development[4-7].For example,Qurrat et al.found some Pd(II)complexes with hydrazide ligands that were approximately 300 times more potent a-glucosidase inhibitors than the standard,1-deoxynojirimycin(DNJ)[8].A series of Cu(II)-hydrazide complexes were prepared and exhibited an intrinsic binding with DNA and anti-inflammatory activity[9].
Copper is a bio-essential metal element and Cu2+has versatile coordination chemistry to form many complexes with numerous biological properties,such as antitumor,anti-candida,antimycobacterial,and antimicrobial activity etc.[10-14].DNA is usually considered as the target of many metal-based anticancer drugs like platinum complexes[15-16],and many copper complexes with anticancer activities could also interact with DNA by a variety of binding modes,and could efficiently cleave DNA via oxidative or hydrolytic mechanism[17].For example,Krikavova reported five copper(II)quinolinonato-7-carboxamido complexes,and found that all the complex exhibit potent cytotoxicity and the ability to cleave pUC-19 DNA by an oxidative mechanism in the presence of a reducing agent[18].Zhang et al.synthesized mononuclear and binuclear copper complexes,and found these complexes could effectively inhibit the proliferation and induce apoptosis via ROS-triggered pathway of He-La cells[19].Some copper(II)complexes with hydrazide and heterocyclic bases ligands were found to possibly bind to DNA with a binding constant K values ranging from 2.62×10-4to 4.38×10-4,and induce apoptotic cell death in MDA-MB-231 cell line[20].
In this paper,we report the synthesis and characterization of a novel binuclear copper complex with hydrazide derivative,[Cu2(BPH)2Cl4]·CH3OH.The interaction between the complex and DNA is studied by spectroscopy and gel electrophoresis.
1 Experimental section
1.1 Materials and methods
Benzoyl chloride,phenylhydrazine,copper(II)chloride,methanol and dichloromethane are analytical grade.All medicines and solvents were purchased from commercial sources and were used without further purification.All water used in the experiments was deionized water.Plasmid pBR322 DNA and calf thymus DNA(CT-DNA)were commercially obtained from MBI Fermentas,whereas ethidiumbromide(EB)and tris(hydroxylmethyl)aminomethane(Tris)were purchased from Energy Chemical.Infrared spectrum was carried on a Bruker VEC-TOR22 spectrometer using KBr tablet.Electronic absorption spectrum was measured on a Shimadzu UV-2600 spectrometer.Fluorescence spectrum was obtained on a Hitachi F4600 luminescence spectrometer.The circular dichroism(CD)spectrum was carried on a JASCO J-815 automatic recording spectropolarimeter.1-Benzoyl-1-phenylhydrazine(BPH)was synthesized according to the reported procedures[21].
1.2 Synthesis of[Cu2(BPH)2 Cl4]·CH3 OH
The methanol solution of CuCl2·2H2O(0.017 g,0.1 mmol)was added dropwise to the methanol solution of ligand BPH(0.021 g,0.1 mmol).After ultrasound at room temperature for 5 minutes,the resulting solution was allowed to stand and slowly evaporated.The blue crystal of the complex suitable for X-ray diffraction analysis was obtained after standing for a few days.The yield(based on Cu):0.03 g(80%).Elemental analysis(%)Calcd,for C27H28Cu2N4O3∶C,45.67;H,3.70;N,7.61.Found:C,45.73;H,3.66;N,7.58.IR(KBr/pellet,cm-1)∶ν(N-H)3 205 m;ν(C-H)3 106 w;ν(C=O)1 620 s;ν(C-N)1 423 s,δ(NH)1 117 s cm-1.
1.3 X-ray Crystallography
Bruker APEX-II diffractometer(Mo-Kα,λ=0.710 73Å)was used for X-ray analysis of singlecrystal.The structure was determined by Olex2 direct method and optimized through Olex2 least square method.Details of the crystal parameters,data collection and refinements of the complex are listed in Table 1.All the non-hydrogen atoms were refined anisotropically on F2by full-matrix least-squares minimization.
The crystallographic data(excluding structural factors)of the structures presented in this paper have been deposited with the Cambridge Crystallographic Data Centre,CCDC,12 Union Road,Cambridge CB21EZ,Cambridge,UK.Copies of the data can be obtained free of charge on quoting the depository numbers CCDC-2067264 for the complex.

Tab.1 Crystal data and structure refinement for[Cu2(BPH)2Cl4]·CH3 OH
1.4 DNA-binding Studies
Absorption Spectra:The CT-DNA reserve solution was dissolved in the buffer solution(50 mM NaCl/5 mM Tris-HCl,pH=7.2).The UV absorption ratio of CT-DNA solution at 260 nm and 280 nm ranged from 1.8 to 1.9,indicating that CT-DNA had not deteriorated.The concentration of CT-DNA was determined by using a molar extinction coefficient of 6 600 M-1cm-1at 260 nm.[Cu2(BPH)2Cl4]·CH3OH concentration was 2.5×10-5M.The electron spectra of the complex was carried out,where the molar ratios of DNA to the complex were 0.0,0.1,0.2,0.3,0.4,0.5 and 0.6.The values of the intrinsic equilibrium DNA binding constant(Kb)were calculated for the complex using the equation[22]:
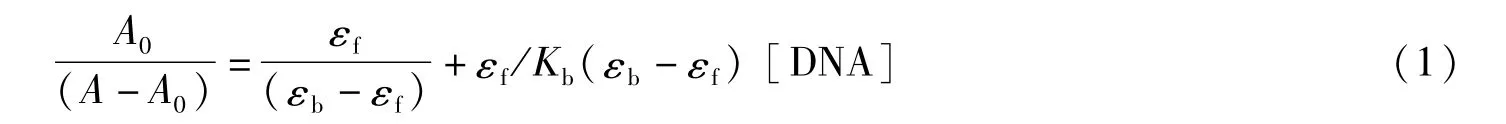
Where,εbandεfrepresent the extinction coefficients of[Cu2(BPH)2Cl4]·CH3OH in the bonded and free from,respectively.A0denotes the initial absorbance of the free complex and A denotes the absorbance of[Cu2(BPH)2Cl4]·CH3OH in the presence of DNA.The curve of A0/(A-A0)vs.1/[DNA]is linear,and the binding constant(Kb)is the ratio of the slope to the intercept.
Fluorescence Spectra:The solution of[Cu2(BPH)2Cl4]·CH3OH was added to the EB-DNA solution for fluorescence titration experiment.The fluorescence quenching results were taken down when the molar ratios of the complex to EB-CT-DNA were 0.0,0.2,0.4,0.6,0.8,1.0,1.2,1.4 and 1.6.The sample excitation wavelengths were 530 nm,and the emission was recorded from 550 to 750 nm.According to the classical Stern-Volmer equation[23]:

In the formula,I0and I are fluorescence intensity of CT-DNA solution in the absence and presence of quenching agent([Cu2(BPH)2Cl4]·CH3OH),respectively.Ksvis the linear Stern-Volmer quenching constant and[Q]is the molar concentration of the complex.
CD Spectra:The circular dichroism spectrum was performed on the samples,which were incubated with the ratio of complex to CT-DNA of 0.0,0.1,0.2 and 0.4 for 12 h at 37℃.And concentrations of[Cu2(BPH)2Cl4]·CH3OH and CT-DNA solution were 1×10-4M and 3×10-4M,respectively.
1.5 DNA Cleavage Experiments
Gel electrophoresis:The DNA cleavage activity of[Cu2(BPH)2Cl4]·CH3OH was detected by agarose gel electrophoresis.Plasmid pBR322 DNA(1μL,200 ng·μL-1)was treated with the complex in proper concentration of H2O2.The complex sample was dissolved in Tris-HCl buffer and then continued to be diluted in Tris-HCl buffer to a concentration of 10-4M with a concentration gradient from5 to 25μM.The sample was incubated at 37 C for 1 h.Then termination buffer(2μL)was added to the solution to stop DNA cleavage.The sample was electrophoresis for 1 h in a TAE buffer solution(40 mM Tris acetate/1 mM EDTA,pH=8.4)at 100 V on a 1% agarose gel containing ethidium bromide(1μg·mL-1).In the mechanism study,we selected the best concentration of the complex(20μM),added DMSO,L-histidine,KI,EDTA respectively,and then repeated the electrophoresis step.The bands were observed and photographed under ultraviolet light.
2 Results and discussion
2.1 Crystal structures of[Cu2(BPH)2Cl4]·CH3OH
The X-ray structure of[Cu2(BPH)2Cl4]·CH3OH is shown in Figure 1,and the selected bond lengths and bond angles of the complex are given in Table 2.
The structure of[Cu2(BPH)2Cl4]·CH3OH consists of two Cu(II)centers which are bridged by two chloride ions[Cl(3)and Cl(4)].Each Cu(II)ion is five-coordinated with two atoms(N and O)from the BPH ligand and three chloride ions.The Cu(1)ion is situated in a distorted square-pyramidal.In the basal plane,the Cu(II)ion is bonded to two atoms[N(1)and O(1)]of the BPH ligand,a bridged chloride ion Cl(3)and another chloride ion Cl(1)with bond lengths Cu(1)-N(1)=1.997(3)Å,Cu(1)-O(1)=1.981(4)Å,Cu(1)-Cl(3)=2.263(3)Åand Cu(1)-Cl(1)=2.228(3)Å,respectively.The apical position of the square-pyramidal is occupied by another bridged chloride ion Cl(4)with the bond length Cu(1)-Cl(4)=2.746(5)Å.Interestingly,the bond lengths of Cu(1)with two bridged chloride ions are different.The Cu(1)-Cl(3)bond length(2.263(3)Å)in the basal plane is shorter than Cu(1)-Cl(4)bond length(2.746(5)Å)in the apical position,indicating that the coordination ability of Cu(1)with Cl(3)is stronger than that of Cu(1)with Cl(4).The bond angles of O(1)-Cu(1)-Cl(3)(170.75(6)°)and N(1)-Cu(1)-Cl(1)(164.89(7)°)are contracted from the ideal value of 180°for a regular square-planar structure,indicating distortion in the basal plane.The Cu(2)ion is also coordinated in a distorted square-pyramidal.The Cu(2)-NBPH,Cu(2)-OBPH,and Cu(2)-Cl bond lengths are similar with the corresponding bonds of Cu(1)ion.

Tab.2 Selected bond lengths(Å)and angles(°)of[Cu2(BPH)2 Cl4]·CH3 OH
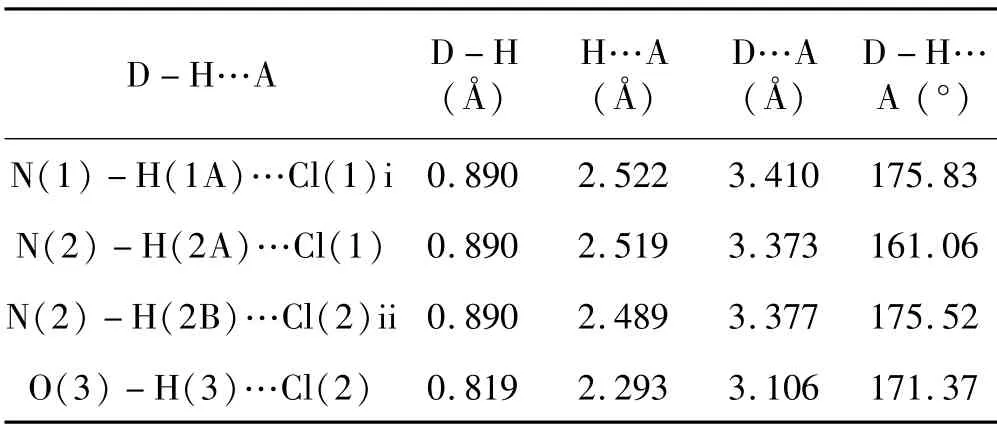
Tab.3 Hydrogen-bonding geometries for[Cu2(BPH)2Cl4]·CH3OH

Fig.1 Crystal structure of[Cu2(BPH)2 Cl4]·CH3 OH
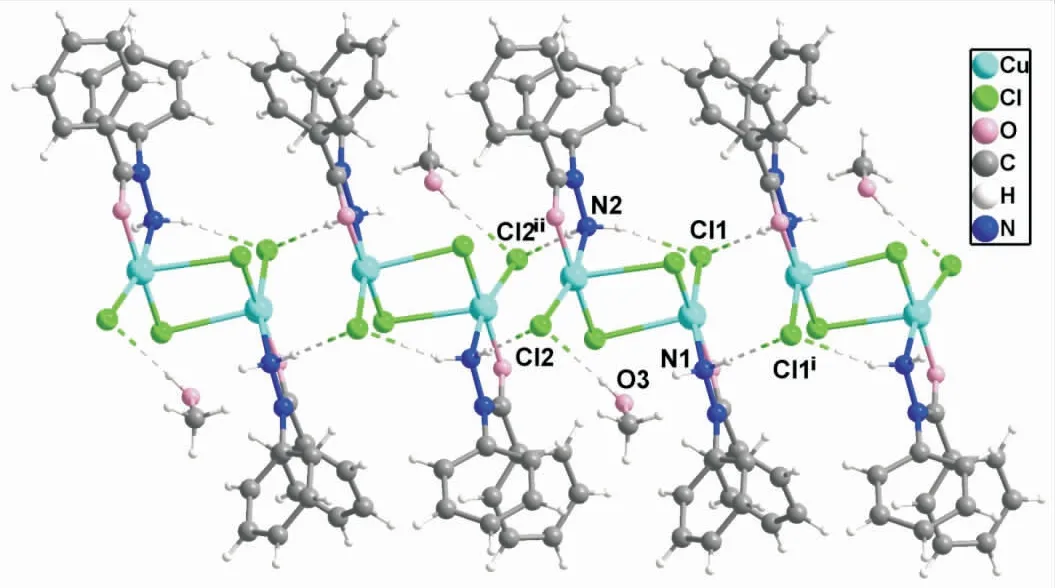
Fig.2 The stacking diagram of the complex
The crystal packing of[Cu2(BPH)2Cl4]·CH3OH is shown in Figure 2.There are hydrogen bonding interactions in the packing of the complex,and the most relevant hydrogen bonding interactions are given in Table 3.The intramolecular hydrogen bond for N(2)-H(2A)…Cl(1),the separation is 2.519Å and the angle is about 161.06°.Two adjacent molecules are connected by intermolecular hydrogen bonds N(1)-H(1A)…Cl(1)iand N(2)-H(2B)…Cl(2)ii.In addition,there is also a hydrogen bond between the hydrogen atom of the solvent methanol molecule and Cl(2),with the separation of 2.293Å and the angle of 171.37°.This hydrogen bond may be responsible for the excellent solubility of the complex in proton-containing solvents such as water and methanol[24-25].
2.2 Spectroscopic study on DNA interaction
The interaction of[Cu2(BPH)2Cl4]·CH3OH with CT-DNA was studied by electron absorption spectra,fluorescence spectra and CD spectra.
Electron absorption spectroscopy is one of the commonly used techniques to study the interaction between complexes and DNA.The absorption spectra of[Cu2(BPH)2Cl4]·CH3OH in the absence and presence of CT-DNA are shown in Figure 3.Increasing concentration of CT-DNA results in some hyperchromism and a slight blueshift(253→252 nm).These changes are typical of complexes that bind to DNA by groove modes,which may involve loosening of the DNA double helix and greater exposure of the base pairs[26].In addition,the intrinsic binding constant(Kb)of the complex was obtained as 9.3×103M-1.The Kb value is lower than that observed for the classical intercalator EB(ca.7×105M-1),but it has similar magnitudes to the classic groove binding reagent DAPI(8.90×103M-1).Hence,it is likely that the complex associates with CT-DNA via groove binding[27].
Fluorescence quenching experiments were carried out to further study the interaction between[Cu2(BPH)2Cl4]·CH3OH and CT-DNA.Ethidium bromide(EB)itself is weak in fluorescence and can fluoresce strongly in the presence of DNA due to its intercalation with DNA.However,this enhanced fluorescence can be quenched or partially quenched by the addition of other competitors which can replace bound EB or damage the secondary structure of DNA.Therefore,EB can be a powerful tool for studying the interaction between complexes and DNA.

Fig.4 Fluorescence emission spectra of the EBCT-DNA system in the absence(…)and presence(—)of the complex

Fig.5 Fluorescence quenching diagram of interaction between[Cu2(BPH)2 Cl4]·CH3 OH and EB-DNA
CT-DNA fluorescence quenching spectrum of[Cu2(BPH)2Cl4]·CH3OH is shown in Figure 4.As the complex was gradually added,the fluorescence intensity of EB-DNA gradually decreased,indicating that the complex could compete with EB in binding DNA.Figure5 shows the fluorescence quenching diagram of the interaction between the complex and EB-DNA.According to the classical Stern-Volmer equation,the apparent binding constant(Ksv)of the complex was calculated as 4.7×104M-1.This suggests that the complex may bind to DNA in non-intercalated ways,such as groove binding.Furthermore,the binding energy of the complex in the fluorescence spectrum is smaller than that of classical intercalation,which is consistent with the results of electron absorption studies[28].
CD spectroscopy is a sensitive and effective technique for evaluating whether nucleic acids undergo conformational changes due to interactions with complex.The CD spectra of free DNA shows positive bands at 275 nm due to base accumulation,and negative bands at 245 nm owing to the loss of B-type DNA helicity.Figure 6 displays the CD spectra of CT-DNA treated with the complex.The strength of both the positive(about 276 nm)and negative(about 246 nm)bands was decreased when[Cu2(BPH)2Cl4]·CH3OH was added into DNA.The results show that the complex could loosen the DNA helix and lead to the loss of helicity[29].This change indicates a non-intercalated binding pattern and supports the grooves binding nature,because intercalation can increase the intensity of the two bands while the electrostatic interaction has little effect on the CD spectrum.
Based on these observations,we speculated that the copper complex could interact with DNA through groove binding.The complex acts on the large or small furred region of the double helix DNA,thus causing changes in the DNA structural microenvironment.
2.3 DNA Cleavage Activity
The DNA cleavage activity of[Cu2(BPH)2Cl4]·CH3OH was evaluated by gel electrophoresis analysis to detect the presence of different DNA isoforms:supercoiled DNA(Form I),nicked circular(Form II)and linear DNA(Form III)(Figure7 and Figure 8),and the amounts of supercoiled,nicked and linear DNA were quantified and listed in the Table 4 and Table 5.

Tab.4 The percentage of DNA cleavage at concentration gradient of[Cu2(BPH)2 Cl4]·CH3 OH

Fig.6 CD spectra of 1×10-4 M CT-DNA in the absence and presence of increasing amounts of[Cu2(BPH)2 Cl4]·CH3OH(1×10-5 M)at the ratio=0,0.1,0.2,0.4
Figure 7 shows the effects of[Cu2(BPH)2Cl4]·CH3OH on DNA cleavage at different concentrations.Lanes 1-3 were used as the control group,and lanes 4-8 were used as the concentration gradient group.The pBR322 DNA had no obvious transformation from Form I to Form II or Form III(lanes 1-3)when only DNA,the complex,or H2O2were present.With the increase of complex concentration,Form I decreased gradually and finally disappeared completely(Figure 7,lanes7 and 8).The cleavage efficiency of the complex reached 100%at a concentration of 20μM in converting Form I to Form II(56.75%)and Form III(43.25%)(Figure 7,lane 7).Increasing the concentration of the complex to 25μM,some smears corresponding to multi-fragmented DNA were observed(Figure 7,lane 8),which suggested that the optimum concentration showing most efficient cleavage was about 20μM.In addition,the cleavage reactions are dependent on the activation of H2O2as an inductive agent,indicating that the processes may be oxidative[30].
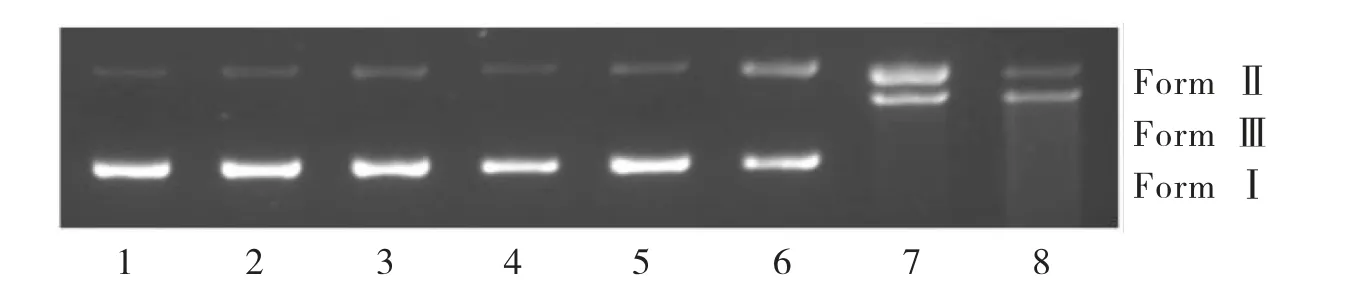
Fig.7 Cleavage of pBR322 DNA by[Cu2(BPH)2Cl4]·CH3OH in Tris-HCl buffer solution(pH=7.24)at 310 K
To elucidate the possible mechanism of DNA cleavage,several reactive oxygen species(ROS)inhibitors were used in the DNA cleavage experiment,including hydroxyl radical trapping agent(DMSO),singlet oxygen quenching agent(L-histidine),H2O2scavenger(KI),and metal chelating agents(EDTA)as shown in Figure 8.In parallel,DNA,DNA+H2O2,DNA+complex and DNA+H2O2+complex were used as controls(Figure 8 lanes 1-4).The inhibitory effect of DMSO and KI on the DNA cleavage reaction was not obvious(Figure 8 lanes 5 and 7).However,from lanes 6 and 8 of Figure 8,we could observe that only a small part of From I was converted to Form II,and no Form III appeared.In other words,L-histidine and EDTA had significant inhibitory effect in the DNA cleavage process,while L-histidine and EDTA are singlet oxygen quenching agent and metal chelating agents respectively.It can be inferred from the above results that the active component of singlet oxygen and metal ions play major roles in the cleavage reaction[31-32].
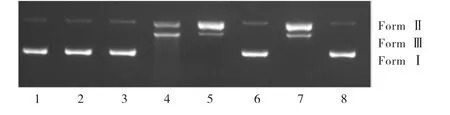
Fig.8 Cleavage of pBR322 DNA by[Cu2(BPH)2Cl4]·CH3OH with different inhibitors in Tris-HCl buffer solution(pH=7.24)at 310 K
Based on the above observations,we propose the mechanisms of DNA cleavage as follows:Initially,the Cu(II)complex binds to DNA through groove binding interaction and results in changes in DNA structural microenvironment.Then,the Cu-DNA reacts with H2O2to produce singlet oxygen.Finally,singlet oxygen can cause structural changes in DNA,such as the shedding of glycosides or the oxidation of bases,which can cause permanent damage to the DNA,resulting in metabolic abnormalities and initiating DNA chain breakage[33-34].

Tab.5 The percentage of DNA cleavage of[Cu2(BPH)2 Cl4]·CH3 OH with different inhibitors
3 Conclusion
A novel binuclear copper complex was synthesized and characterized.The interaction between[Cu2(BPH)2Cl4]·CH3OH and CT-DNA was through groove binding.The complex can efficiently cleavage pBR322 DNA in the presence of H2O2via oxidative mechanism,and singlet oxygen and metal ions were proposed to be active substances involved in DNA fragmentation.These results may be valuable in understanding the mode of the complex with DNA as well as laying a foundation for the rational design of novel and powerful agents for DNA cleavage.

