Laetmonice iocasica sp.nov.,a new polychaete species(Annelida:Aphroditidae) from seamounts in the tropical Western Pacific,with remarks on L.producta Grube,1877*
Xuwen WU ,Pat HUTCHINGS ,Anna MURRAY ,Kuidong XU ,
1 Laboratory of Marine Organism Taxonomy and Phylogeny, Shandong Province Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
2 Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai 519082, China
3 Australian Museum, Australian Museum Research Institute, Sydney NSW 2000, Australia
4 Biological Sciences, Macquarie University, North Ryde 2109, Australia
5 University of Chinese Academy of Sciences, Beijing 100049, China
Abstract Laetmonice Kinberg,1856 is a remarkable genus characterized by having harpoon notochaetae in the polychaete family Aphroditidae.We describe a new species of Laetmonice,Laetmonice iocasica sp.nov.,found from seamounts on the Caroline Ridge in the tropical Western Pacific.The new species is readily distinguished from congeners,particularly those distributed in the Indo-Pacific Ocean by possessing 45 segments with 18 pairs of elytra,and the tuberculated harpoon notochaetae in the elytrigerous segments,which are replaced by tuberculated notochaetae without recurved fangs on segments 4 and 5.Laetmonice iocasica sp.nov.is closely related to L.producta Grube,1877,but differs in both morphology and the genetic distance of the mitochondrial cytochrome oxidase subunit I (COI) sequences.Laetmonice producta Grube,1877 contained five varieties reported in various marine areas,which have been raised to species level.However,the records of L.producta from the Sagami Bay and Suruga Bay in Japan and in the south-eastern Australia remain obscure and probably represent a different species.The data indicate that L.producta,which was originally described from Kerguelen Islands in the Southern Ocean and later commonly found on the Antarctic shelf,is probably distributed only at high latitudes of the Southern Hemisphere.
Keyword:Polychaeta;taxonomy;biodiversity;deep-sea
1 INTRODUCTION
Members in the family Aphroditidae are predominantly deep-water scale-worms,and few species occur intertidally (Hutchings and McRae,1993).Within the family,the genusLaetmoniceKinberg,1856 is characterized by the remarkable harpoon notochaetae which are distally tapering with several recurved fangs on lateral margins (Hutchings,2000).Up to date,27 species have been included in the genus,in which 24 species were originally described by early taxonomists more than a century ago (Read and Fauchald,2021a).It is not surprising that as many as five generic names have been considered as synonymies ofLaetmonicebecause the early descriptions are brief and not all important characters are mentioned (Hutchings and McRae,1993;Barnich and Fiege,2003).
Species ofLaetmoniceare highly diverse in the Indonesian Archipelago and Australian waters.Hutchings and McRae (1993) made an important revision ofLaetmonicemainly based on the types of the Indo-Pacific and Australian species and provided a checklist of 18 species distributed in China,Philippines,Indonesia,New Guinea,and Australia including two new species.In contrast,only a few species ofLaetmoniceare reported in the Atlantic.Barnich and Fiege (2003) made a comprehensive work on the Mediterranean species ofLaetmoniceand only two valid species were found in the East Atlantic,Mediterranean Sea,and the northern part of the Red Sea.The most recent new record ofLaetmoniceis from Barnich et al.(2013),who described a new species associated with cold-water corals in the eastern Gulf of Mexico.
Based on the examination of polychaete specimens collected from the seamounts located on the Caroline Ridge in the tropical Western Pacific in 2019,we identified and described a new species of Aphroditidae:Laetmoniceiocasicasp.nov.For species comparison,we evaluated the taxonomic status of the speciesLaetmoniceproductaGrube,1877 and its varieties and subspecies,which were recorded in the Indian,Pacific,Atlantic,and Southern Oceans.The speciesLaetmoniceproductawas commonly found at high latitudes of the Southern Ocean (Grube,1877;Parapar et al.,2013),and it is confusing thatL.productawas reported from the Sagami Bay and Suruga Bay in Japan and the southeastern Australia (Hutchings and McRae,1993;Imajima,2003).In this study,we provided some remarks on the speciesL.producta,particularly its records in Japan and Australia.
2 MATERIAL AND METHOD
2.1 Specimen collection and morphological examination
Specimens were collected from two adjacent seamounts (provisionally named as M5 and M6)located on the Caroline Ridge in the tropical Western Pacific (Fig.1).A total of 3 specimens were obtained from three dives (140°11ʹE–140°14ʹE,10°04ʹN–10°07ʹN,water depth of 888–980 m) by the ROVFaxian(Discoveryin Chinese) on board the R/VKexue(Sciencein Chinese) in June 2019.Photos were taken in situ by the imaging equipment of the ROV.The specimens were photographed on board using a Canon EOS camera,then preserved in 80% ethanol solution and later deposited in the Marine Biological Museum of Chinese Academy of Sciences(MBMCAS),Institute of Oceanology,Chinese Academy of Sciences (IOCAS).Details of prostomium,pygidium,segments,parapodia and chaetae were observed through a Zeiss Discovery.V20 stereomicroscope.Digital photographs were taken using an AxioCam 512 digital camera mounted on the microscope.Images of different focal planes were stacked by the software Helicon Focus 7.The character choice and terminology mainly followed Hutchings and McRae (1993).
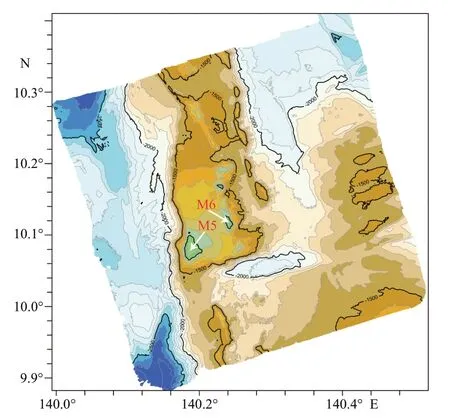
Fig.1 Sampling sites of Laetmonice iocasica sp.nov.
2.2 DNA extraction and sequencing
Genomic DNA was extracted from each of the three specimens using the TIANamp Marine Animal DNA Kit (Tiangen Bio.Co.,Beijing,China).Approximately 650 bp of cytochrome c oxidase subunit I (COI),400 bp of 16S,1 700 bp of 18S,1 000 bp of 28S,and 900 bp of ITS (ITS1-5.8S-ITS2)genes were amplified using the following primers:LoboF1 and LoboR1 for COI (Lobo et al.,2013);Ann16SF and Ann16SR for 16S (Sjölin et al.,2005);18SA and 18SB for 18S (Medlin et al.,1998;Nygren and Sundberg,2003),28S D1_F and D3 for 28S(Brown et al.,1999;Vonnemann et al.,2005);ITS18SFPOLY and POLY_28R for ITS (Barfuss,2012).The 25-μL reaction contained 12-μL 2X Es TaqMasterMix (CWBio Co.,Ltd.,Beijing,China),1 μL of each primer (10 mmol/L),2 μL of template DNA (50 ng/μL),and 9-μL dH2O.Thermal cycling protocols followed Wu et al.(2019).Amplified DNA was sequenced using the ABI 3730 DNA Analyzer sequencing facility of the Shanghai Sangon Biological Engineering and Technical Service Company,Shanghai,China.
2.3 Barcoding and phylogenetic analyses
Three partial sequences of COI were obtained from each of the three specimens in this study and were deposited in GenBank.To examine species delimitation,all available COI sequences ofLaetmonicespecies were downloaded from GenBank and used in the barcoding and phylogenetic analyses(Table 1).The outgroup comprised species ofAphroditaLinnaeus,1758,PalmyraSavigny in Lamarck,1818,NereisLinnaeus,1758,andGlyceraLamarck,1818.The former two genera represented the other species of the family Aphroditidae available in GenBank.Sequences were aligned using ClustalW method provided in MEGA-X.Pairwise distances of COI were calculated using MEGA-X based on the Kimura 2-parameter andp-distance models (Kumar et al.,2018).
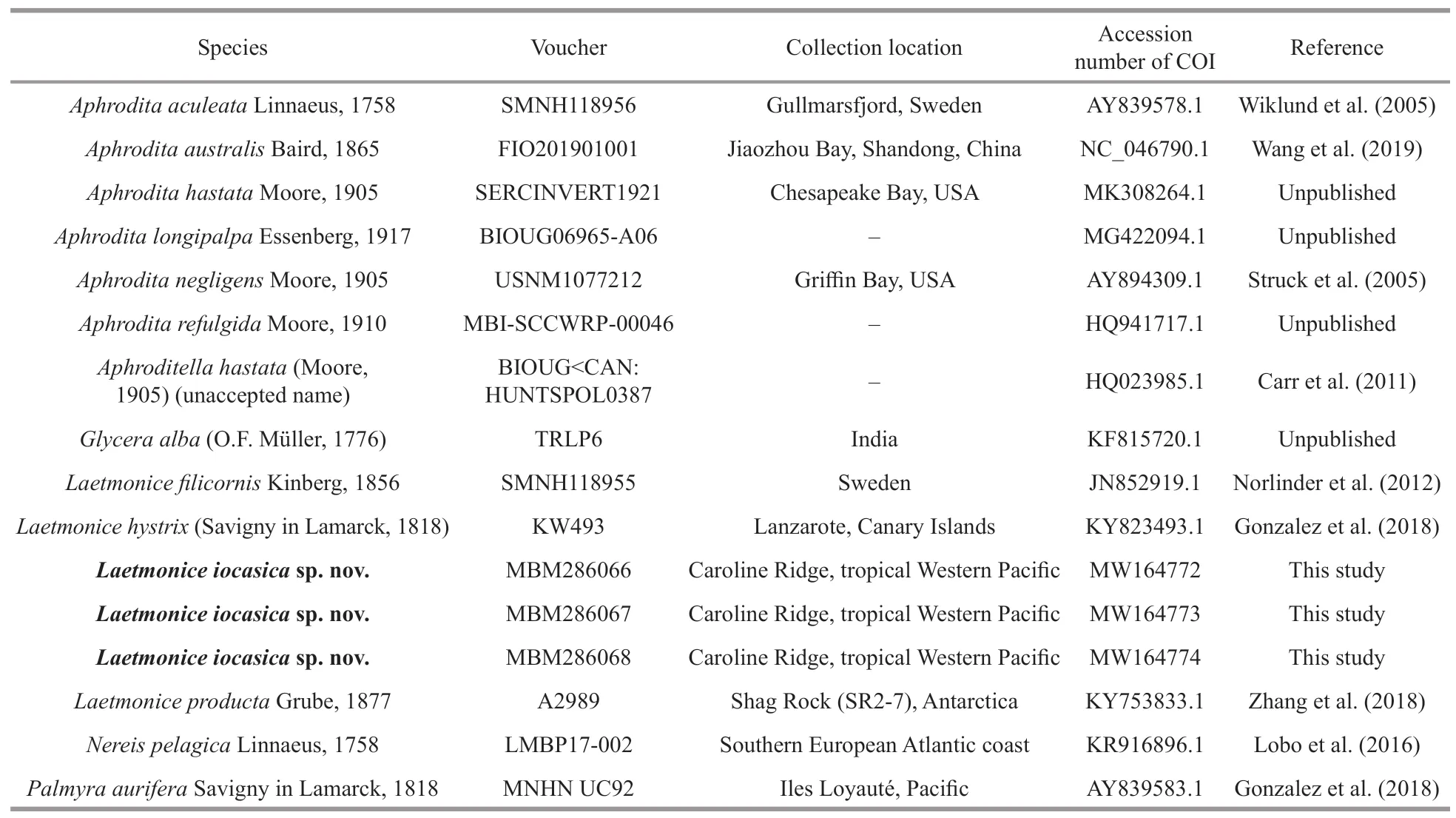
Table 1 List of species and corresponding accession numbers of COI used in this study
Maximum likelihood (ML) analysis was carried out using IQ-TREE (Nguyen et al.,2015) in PhyloSuite v1.2.2 (Zhang et al.,2020) under the model automatically selected by IQ-TREE (“Auto”option in IQ-TREE) for 5 000 ultrafast (Minh et al.,2013) bootstraps.Bayesian inference (BI) analysis was carried out using MrBayes 3.2.6 (Ronquist et al.,2012) under GTR+I+G model (2 parallel runs,four chains of 3 000 000 generations with sampling every 1 000 generations),in which the initial 25% of sampled data were discarded as burn-in trees.
3 RESULT
3.1 Taxonomy
Family Aphroditidae Malmgren,1867
Genus Laetmonice Kinberg,1856
Laetmoniceiocasica sp.nov.
Figs.2–5;Table 2
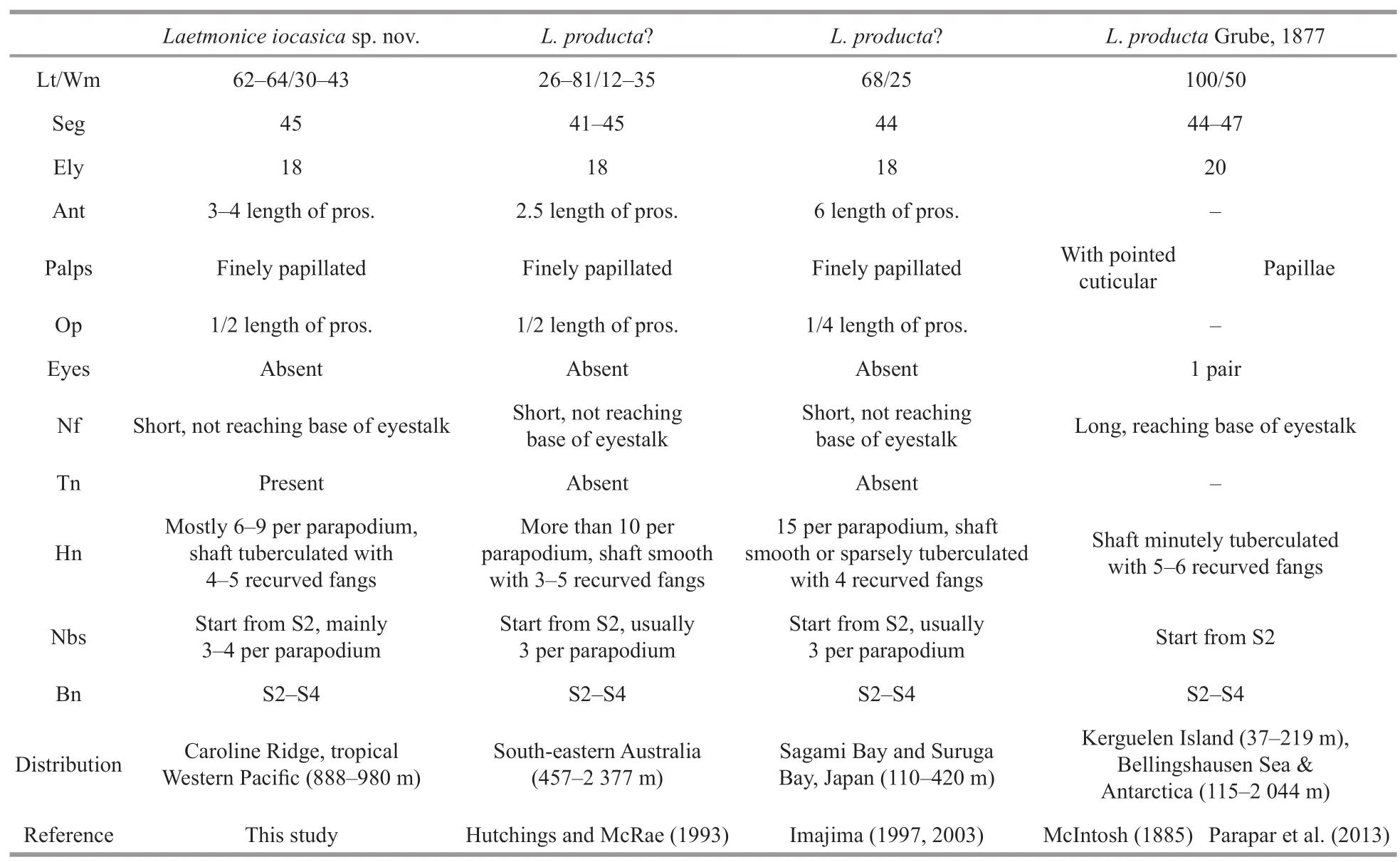
Table 2 Morphological comparison of Laetmonice iocasica sp.nov.and L.producta Grube,1877
Material examined
Holotype:MBM286066,Dive 215,140°10.8ʹE,10°4.9ʹN,946 m,2 June 2019.Paratypes:MBM286067,1 specimen,Dive 215,140°11ʹE,10°4.8ʹN,888 m,2 June 2019;MBM286068,1 specimen,Dive 218,Dive 215,140°14ʹ18″E,10°07ʹ25″N,980 m,6 June 2019.
Diagnosis
Body with 45 segments,maximum length 63.5 mm,maximum width 42.6 mm.Elytra 18 pairs,located on segments 2,4,5,7,9,11,13,15,17,19,21,23,25,28,31,34,37,and 40.Dorsum without felt.Ventral surface finely papillated.Palps stout and finely papillated,extending to segment 10.Ocular peduncles without eyes.Nuchal flaps small,not reaching base of ocular peduncles.Facial tubercle with distinct papillae.Bipinnate neurochaetae on segments 2–4.Neurochaetae with basal spur and distal filamentous fringe located from segment 2 to posterior end,usually 3 or 4 per parapodium,a gap between basal spur and distal fringe.Harpoon notochaetae present on elytrigerous segments after segment 5;replaced by 3–7 stout golden brown tuberculated notochaetae on segments 4 and 5.Harpoon notochaetae with shafts covered by fine tubercles,distally with 4 or 5 recurved fangs.
Description(based on type material)
Holotype (Fig.2a &d),well-preserved with 45 segments,length about 62.2 mm,maximum width 35.1 mm (including chaetae) and 21.2 mm (excluding chaetae).Two paratypes (MBM286067 and MBM286068) both with 45 segments (Fig.3e,f,i),length 63.4 and 63.5 mm,maximum width 30.4 and 42.6 mm (including chaetae),21.2 and 25.2 mm(excluding chaetae),respectively.Body ovate to elongate,dorsoventrally flattened (Fig.2b,c,e,f).Dorsum with no felt covering (Fig.3c &e).Ventral surface brown to cream-colored,evenly covered by fine papillae (Fig.3b,f,i).Base of parapodium including position between notopodium and neuropodium also covered by papillae (Fig.4a–h,k,l).

Fig.2 Three specimens of Laetmonice iocasica sp.nov.holotype (a,d) and paratypes (b,e,MBM 286067;c,f,MBM286068)
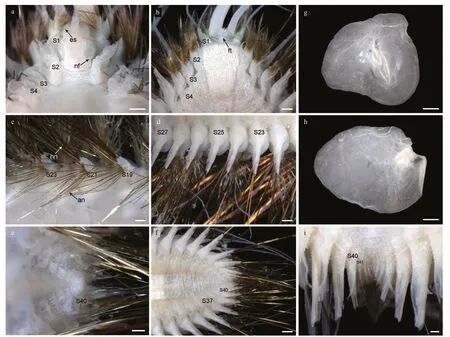
Fig.3 Laetmonice iocasica sp.nov.holotype (c–f) and paratype (a,b,g–i,MBM286067)
Prostomium rounded,longer than wide;anterolateral with a pair of cylindrical ocular peduncles (eyestalks),eyes absent (Fig.3a).Ceratophore of median antenna long,basally articulated (Fig.3a);style slender with a filamentous tip,3–4 times as long as prostomium.Palps stout and finely papillated,tapered with fine tips,extending to segment 10.A pair of prominent nuchal flaps,arising from posterolateral part of prostomium,not reaching base of ocular peduncles (Fig.3a).Facial tubercle located below base of palps,covered by distinct papillae (Fig.3b).
Elytra 18 pairs,attached to elytrophores on segments 2,4,5,7,9,11,13,15,17,19,21,23,25,28,31,34,37,and 40 (Fig.3a,c,e),completely covering dorsum;elytra soft,opaque,rounded to triangular,without tubercles or papillae on surface and margins(Fig.3g &h).Dorsal cirri present on segments without elytra;cirrophores short and cylindrical (Fig.4a,b,g,h),styles slender and smooth with filamentous tips,more than 3 times length of parapodia.
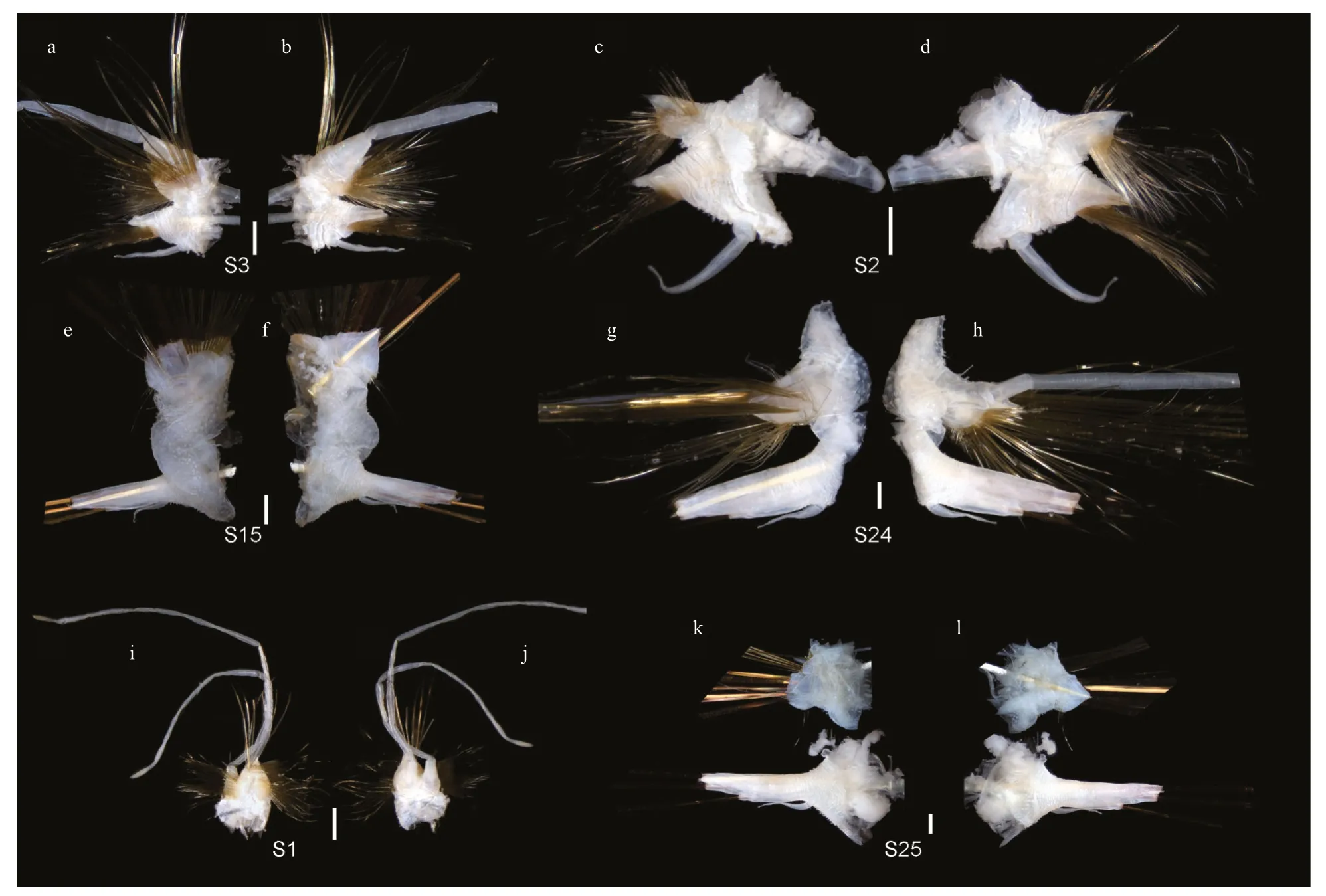
Fig.4 Parapodia of Laetmonice iocasica sp.nov.,holotype
First or tentacular segment with triangular,flattened uniramous parapodia,inserted anterolaterally to prostomium (Figs.3a,b,4i,j);3 tufts of fine,stiffgolden acicular chaetae,fanning around margin of parapodia (Figs.4i &5c).Two pairs of long dorsal and ventral tentacular cirri,consisting of cylindrical tentaculophores and slender styles with long terminal filaments (Fig.4i).
Following segments with biramous parapodia.Segment 2–4 with notopodia conical,as long as neuropodia (Fig.4a–d).Neuropodium conical,with two tiers of neurochaetae (Fig.4a–d);lower tier with numerous golden bipinnate neurochaetae (Fig.5a),upper tier with 1–2 stout neurochaetae,with lateral spur on subdistal position and row of numerous filamentous hairs on distal recurved surface (Fig.5g,l,m).Neuropodia from segment 5 to posterior end elongated,cylindrical with an inflated base (Fig.4e–h,k,l);3–4 golden yellow neurochaetae with basal spur and distal filamentous fringe,inserting on distal margin of neuropodia (Fig.3d &f).Ventral cirri short and smooth,tapered,attached on ventral bases of neuropodia on anterior 4 segments,posteriorly gradually shifting to middle position of ventral surface of neuropodia (Figs.3b,d,f,i,4a–h,k,l).
Elytrigerous segments with tuft of 18 or more golden yellow acicular notochaetae on uppermost position (Figs.3c,5d,e).Lateral to acicular notochaetae with tuft of usually 6–9 stout golden-brown harpoon notochaetae,inserting on anterior margin of acicular lobes (Figs.3c,e,4e,k),directed latero-posteriorly(Fig.3d–f).Harpoon notochaetae elongated,hollow,basally flattened (Fig.5f);shafts covered by fine tubercles (Fig.5k),distally tapering with 4 or 5 recurved fangs on lateral margins (Fig.5f &j).Harpoon notochaetae replaced on segment 4 and 5 by 3–7 stout golden brown tuberculated notochaetae (Fig.5h);this notochaetae covered by fine tubercles (Fig.5i),distally tapering to blunt tips (Fig.5h).Anterior to harpoon notochaetae or tuberculated notochaetae with small tuft of yellow capillary notochaetae.Notopodial acicula yellow,tapering with blunt tip,projecting from triangular acicular lobe (Fig.4d,f,l).Ventral to base of notopodium with tuft of short,fine capillary notochaetae (Figs.4e,f,k,l,5b).
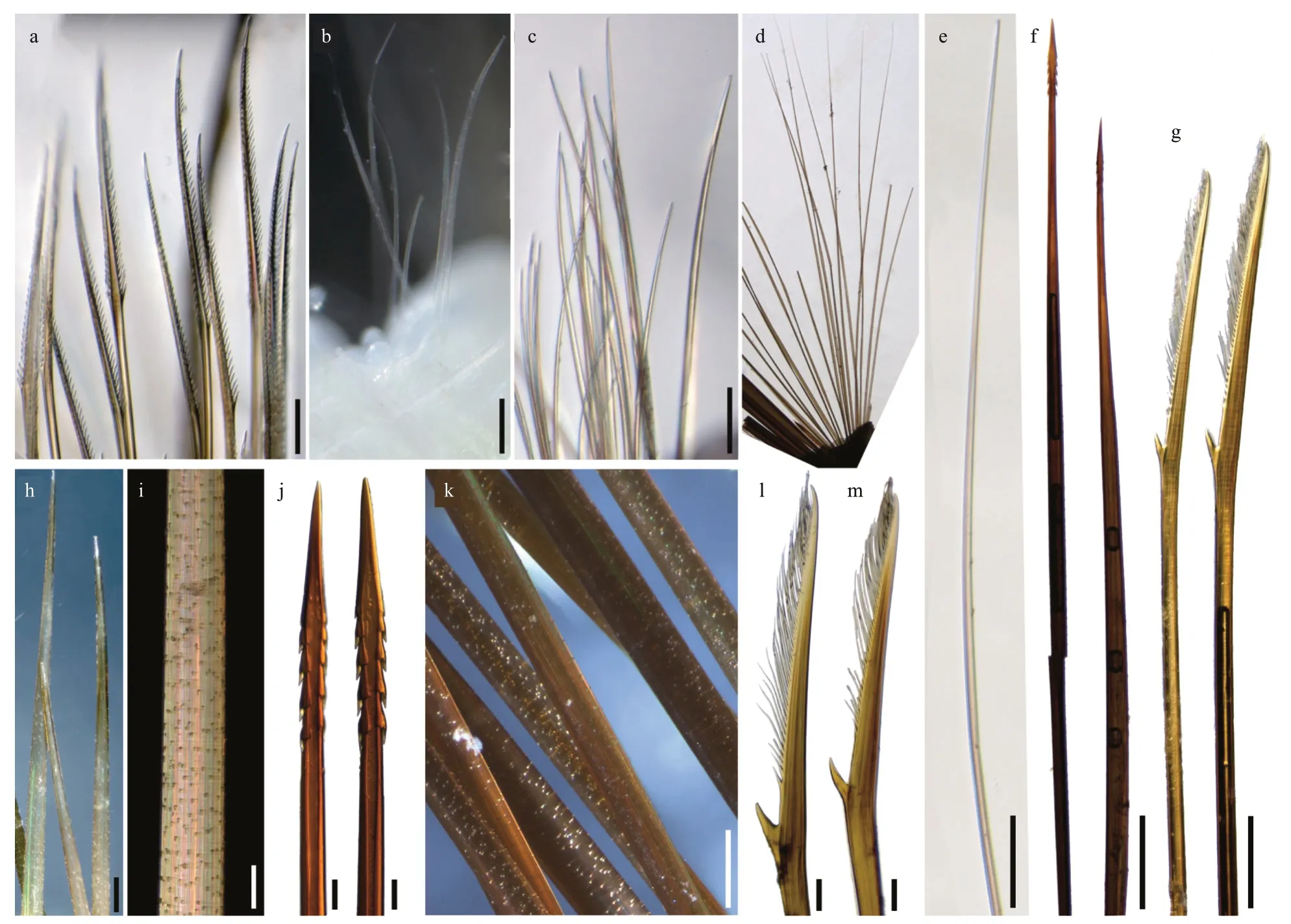
Fig.5 Chaetae of Laetmonice iocasica sp.nov.,holotype (a,c,d,k,m) and paratypes (b,e–h,j,l:MBM 286067;i:MBM286068)
Cirrigerous segments mainly including three tufts of notochaetae (Fig.4g).Uppermost position with a fan of golden yellow,smooth acicular notochaetae(Figs.4a,g,5c).Ventral to acicular notochaetae with a fan of yellow unidentate notochaetae.Both acicular notochaetae and unidentate notochaetae located on anterior side of acicular lobe (Fig.4a &g).Acicular notochaetae stouter than unidentate notochaetae(Fig.4a &g).Posterior to acicular lobes with a fan of capillary notochaetae (Fig.4b &h).
Etymology
The specific name is derived from the composite of IOCAS (abbreviation of Institute of Oceanology,Chinese Academy of Sciences) and the Latin suffi x-icus(belonging to),in celebration of the 70thanniversary of the establishment of the IOCAS in 1950.
Distribution and habitat
Seamounts on the Caroline Ridge in the tropical Western Pacific (888–980-m depth).In situ,the animals crawled slowly on rocky bottoms covered by fine sands (Fig.2a–c).
Remarks
The number of pairs of elytra is one of the most important characters for separating species ofLaetmonice,ranging from 12 pairs inL.batheiaHorst,1916 to 20 pairs inL.productaGrube,1877 (Hutchings and McRae,1993).Laetmoniceiocasicasp.nov.is a large-sized animal (up to 64 mm) with as many as 45 segments and 18 pairs of elytra.Among all theLaetmonicespecies,four species have large numbers of segments (>40 segments) and elytra (18–20 pairs),includingL.productasensuGrube (1877) reported from Kerguelen Islands,L.productasensuHutchings and McRae (1993) from the south-eastern Australia,L.productasensuImajima (2003) from the Sagami Bay and Suruga Bay in Japan,andL.britannicasensuMcIntosh (1900) from Achill Head of Ireland.LaetmoniceproductasensuGrube (1877) differs by having 20 pairs of elytra and a pair of eyes,whileL.iocasicasp.nov.has 18 pairs of elytra and no eyes.Furthermore,the nuchal flaps are long reaching the base of eyestalk inL.productasensuGrube (1877)but short in the new species (Table 2).The genetic distance and phylogenetic analyses further supported the separation of the new species andL.producta(Fig.6;Table 3).Laetmoniceiocasicasp.nov.is characterized by the harpoon notochaetae which are covered by fine tubercles on the shafts,while the harpoon notochaetae are smooth or tuberculated inL.productasensuHutchings and McRae (1993) andL.productasensuImajima (2003).Moreover,the harpoon notochaetae inL.iocasicasp.nov.andL.productasensuHutchings and McRae (1993) are replaced on segments 4 and 5 by stout notochaetae without recurved fangs,which is tuberculated in the new species but smooth in the latter (Table 2).The speciesL.britannicais characterized by reticulate cordate structures on the elytra (Grube,1877) which are absent inL.iocasicasp.nov.
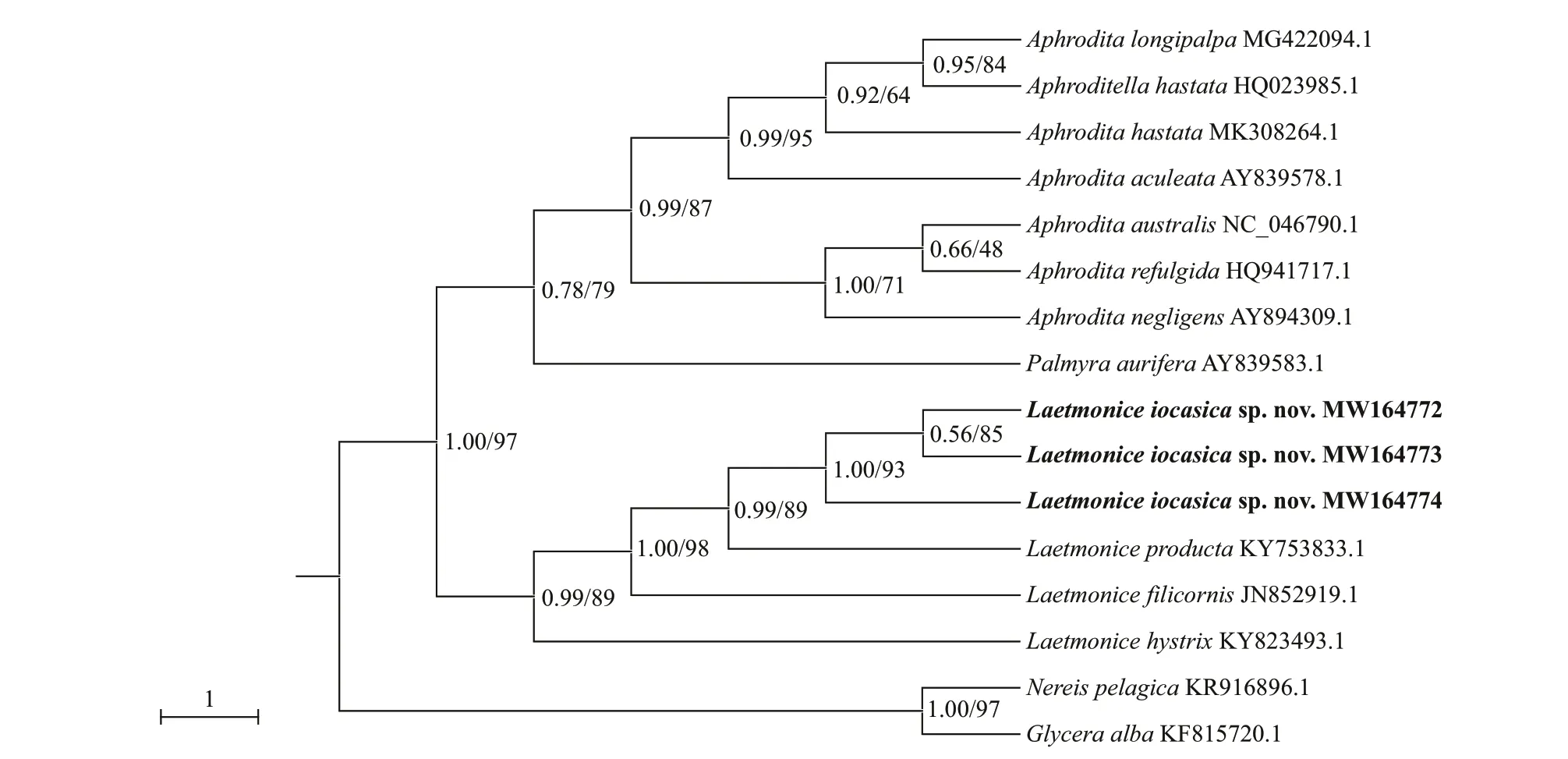
Fig.6 Bayesian inference (BI) tree reconstructed based on COI sequences of Laetmonice and the other species of Aphroditidae
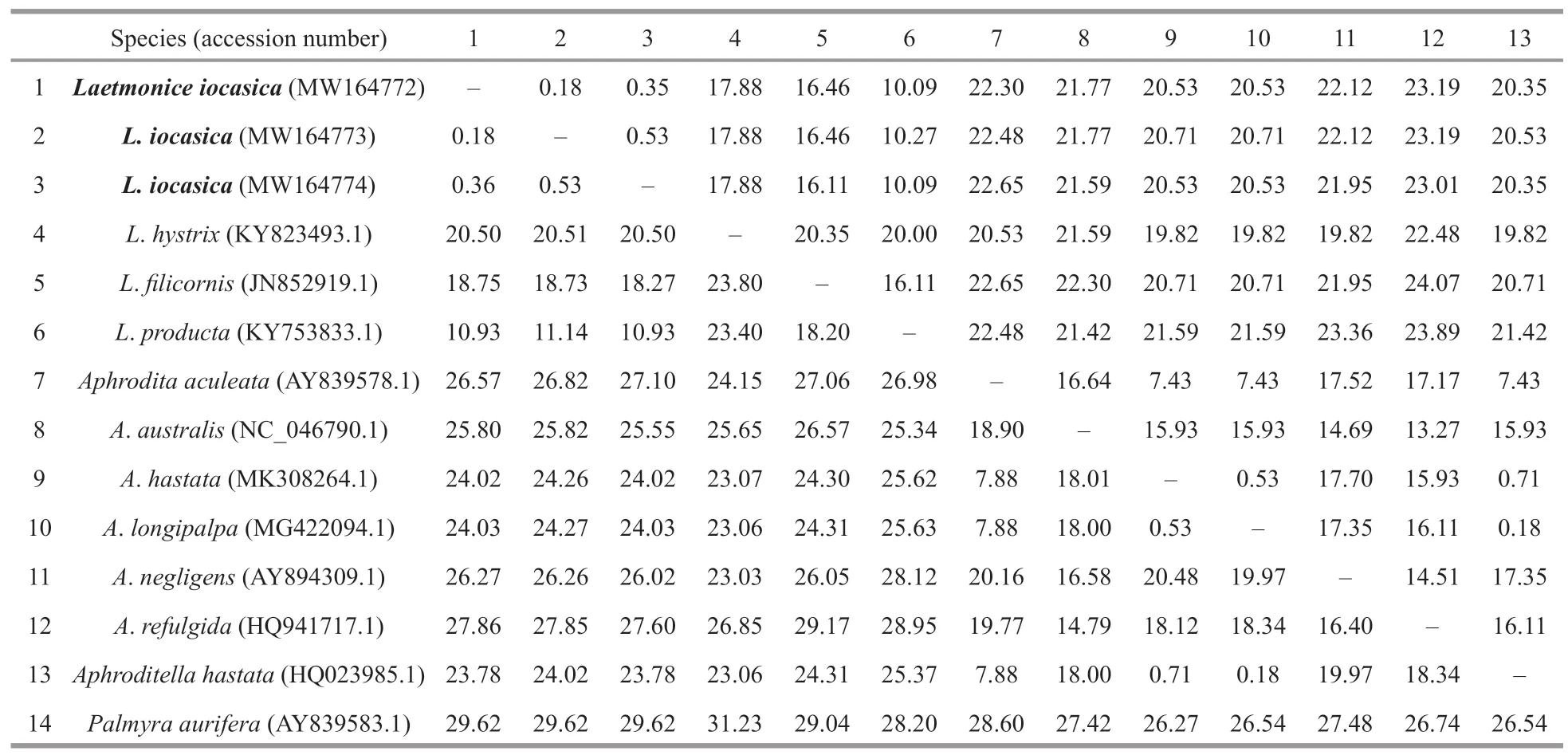
Table 3 Pairwise distances within and among species of Laetmonice and the other genera of Aphroditidae based on Kimura 2-parameter (below diagonal) and p-distance model (above diagonal) (%)
Laetmoniceproducta sensu Hutchings and McRae (1993)
Laetmoniceproducta.Hutchings and McRae(1993):333–336,Figs.45a–f,46a–j,59E,Tables 6,8,9;Gunton et al.(in press).
Material examined
N2017_V03_014,Australia,Tasmania,Flinders Commonwealth Marine Reserve,149°6.08ʹE,40°27.83ʹS,CSIRO Four Meter Beam Trawl,2 298–2 486 m,May 20,2017.
Description
Large-bodied specimen,body shape elongate,more than twice as long as maximum width.Dorsal felt of fine notochaetae absent,18 pairs elytra with purple coloration on inner halves.Prostomium with pair of large ocular peduncles,without eye pigment(pigment may be present);facial tubercle welldeveloped,with long conical papillae;small nuchal flaps present.Palps extending to segment 11,margins finely papillate.Median antenna with ceratophore halflength of the prostomium;antennal ceratostyle longer than prostomium,slender,clavate-tipped,three times length of prostomium.Notochaetae of three kinds:~15 smooth,golden,unidentate acicular chaetae;around ten long,stout,yellow brown harpoon-like chaetae with 3–5 recurved fangs below tips,shafts smooth or tuberculate;tuft of short,fine mud-covered capillary chaetae ventrally.Neurochaetae in two tiers:superior tier of yellow acicular chaetae with basal spur and subdistal fringe of long hairs and bare tips,inferior tier with numerous golden bipinnate neurochaetae.Ventrum covered with small papillae.
Distribution
South-east Australia,from Sandy Cape,Tas.to Broken Bay,New South Wales (457–2 486-m depth).
Remarks
Laetmoniceproductadisplays much morphological variability according to Hutchings and McRae (1993)and has a broad distribution according to McIntosh(1885,1900),who described several subspecies(“varieties”),ranging from the Azores to Antarctic waters.The variety that occurs in Australian waters is assigned toL.productaby Hutchings and McRae(1993),who examined material from southeast Australia,but these authors suggest a reappraisal of the validity of the subspecies.Live specimens are usually pale,with a longitudinal purple stripe middorsally.
As stated in the remarks above,L.productasensu Hutchings and McRae (1993) closely resemblesL.iocasicasp.nov.in most characters.The two species differ in the details of harpoon notochaetae(tuberculated inL.iocasicavs.tuberculated or smooth inL.productasensu Hutchings and McRae) and the size of antennae (3–4 length of prostomium vs.2.5 length of prostomium).Despite the small morphological differences,we suggest they belong to separated species due to their distinct distribution and habitats (Hutchings and Kupriyanova,2018).All the material cited in Hutchings and McRae (1993) were collected from soft sediments in the southeastern Australia,whereasL.iocasicasp.nov.was sampled on rocky seamounts in the tropical Western Pacific,which are more than 5 000 km apart from the former.In addition,previous studies from off the Australian coast have shown the endemism of the invertebrate seamount fauna (De Forges et al.,2000).Although some widespread species in the deep sea have been reported (Meyer et al.,2008;Georgieva et al.,2015;Guggolz et al.,2020;Zhou et al.,2020),there is no evidence that any species on seamounts can also be found on soft sediments on the seafloor,especially in this case of no obvious ocean current connecting the two locations which are far away.
3.2 Barcoding and phylogenetic analyses
Three new sequences of COI were obtained from each of the examined specimens.The accession numbers ofL.iocasicasp.nov.are MW164772,MW164773,and MW164774,and their lengths are 614 bp.Each gene of 16S (MW288699),18S(MW288700),28S (MW288698),and ITS(MW288701) was sequenced successfully from a single specimen and uploaded to GenBank for the supplementary of this study.The alignment sequences of COI comprised 565 nucleotide positions.Pairwise distances based on Kimura 2-parameter andpdistance models were calculated within and among species ofLaetmoniceand the other genera of Aphroditidae (Table 3).Genetic distances among the species ofLaetmoniceare 10.93%–23.80% (Kimura 2-parameter) and 10.09%–20.35% (p-distance),while the intraspecific distance withinL.iocasicasp.nov.was very low (0.18%–0.53%,based on both models).The genetic distance ofLaetmoniceproductaGrube,1877 (Table 1) andL.iocasicasp.nov.was in the range of 10.93%–11.14% (Kimura 2-parameter) and 10.09%–10.27% (p-distance),supporting the separation of both species.
Two phylogenetic trees were generated using BI and ML analyses.The topologies were completely consistent,both with high support values in most nodes (ML>70%,BI>0.95),thus only the BI tree was displayed with BI and ML support values marked at the nodes (Fig.6).All available sequences from species ofLaetmonicewere separated in the phylogenetic trees,and the monophyly ofLaetmonicewas supported.Laetmoniceiocasicasp.nov.andL.productaclustered together with high node support,as consistent with their high morphological similarity.
All the available sequences from species ofAphroditaformed a clade with high support.Genetic distances among the species ofAphroditaare 7.88%–20.16% (Kimura 2-parameter) and 7.43%–17.70%(p-distance),excluding those betweenA.hastataMK308264.1,A.longipalpaMG422094.1,andAphroditellahastataHQ023985.1 (Table 3).The sequence information ofAphroditellahastataHQ023985.1 should be revised asAphroditahastataHQ023985.1 because the genusAphroditellahas been synonymized withAphrodita(Pettibone,1966).The sequenceA.longipalpaMG422094.1 was clustered withA.hastataMK308264.1 andAphroditellahastataHQ023985.1,with Kimura 2-parameter distances ranging from 0.18% to 0.71%,indicating these sequences likely belong to the same species.We suggest that future studies check the type specimens ofA.longipalpaandA.hastataand look for precise sequences in subsequent study.
4 DISCUSSION
LaetmoniceproductaGrube,1877 was originally described from the area of Kerguelen Islands in the Southern Ocean.Later,McIntosh (1885,1900)describedL.productadredged off Kerguelen and five varieties ofL.producta.These areL.productavar.assimilisMcIntosh,1885 from South of Halifax,Canada,L.productavar.benthalianaMcIntosh,1885 from North Pacific and southern Indian Ocean,L.productavar.britannicaMcIntosh,1900 off Achill Head of Ireland,L.productavar.willemoesiMcIntosh,1885 from Azores to the Antarctic Ocean,and northeastern shores of Australia and New Zealand,andL.productavar.wyvilleiMcIntosh,1885 from southern Atlantic Ocean and southern Indian Ocean.Hartman(1965) gathered all the geographical information of the related species and suggested the distribution of the subspeciesL.productaproductais restricted from the Kerguelen Islands to the South Georgia Islands and the Antarctic Peninsula.The subspecies was among the most common polychaetes on the Antarctic continental shelf (Parapar et al.,2013).
It is worth mentioning that many of these varieties and subspecies exhibit distinct morphological differences such as the number of segments as well as elytra (Hutchings and McRae,1993).For example,L.productavar.assimilis,L.productavar.willemoesiandL.productavar.benthalianagenerally have 33–35 segments and 15 pairs of elytra,while the others have 43–47 segments and 18–20 pairs of elytra.These differences are suffi cient to raise the subspecies to species level.So far,all varieties and subspecies have been raised to species level,includingL.productavar.assimilisthat was synonymized withL.filicornis(Hartman,1959;Kongsrud et al.,2013;Read and Fauchald,2021b).Among these,L.wyvilleiandL.benthaliana,were also recorded in the Antarctica(Stiller,1996).
Laetmoniceproductais characterized by 44–47 segments with 20 pairs of elytra,nuchal flaps reaching base of eyestalk,a pair of eyes,palps minutely papillated,and harpoon notochaetae with shafts minutely tuberculated based on the material collected off Kerguelen according to McIntosh (1885) (Table 2).Thus,the populations ofL.productasensuHutchings and McRae (1993) andL.productasensuImajima (2003) likely represent a different species,considering that both populations have 18 pairs of elytra (Table 2).Thus,we agree with Hartman (1965)thatLaetmoniceproductais probably restrictedly distributed in the Southern Ocean and the Antarctic.The other species related toL.producta,including the species from Japan and Australia,probably belong to a complex of species.A revision of these species is needed based on the examination of the types and voucher specimens.
5 DATA AVAILABILITY STATEMENT
Sequence data that support the findings of this study have been deposited in the GenBank.
6 ACKNOWLEDGMENT
We are grateful for the crew of the R/VKexueand the ROVFaxianteam,for their eff ort in sampling the precious material.We also thank Mr.Shaoqing WANG for photographing the fresh specimens onboard.
 Journal of Oceanology and Limnology2021年5期
Journal of Oceanology and Limnology2021年5期
- Journal of Oceanology and Limnology的其它文章
- Screening of stable internal reference genes by quantitative real-time PCR in humpback grouper Cromileptes altivelis*
- Morphology and multifractal features of a guyot in specific topographic vicinity in the Caroline Ridge,West Pacific*
- Geochemical characteristics and geological implication of ferromanganese crust from CM6 Seamount of the Caroline Ridge in the Western Pacific*
- Deep-sea coral evidence for dissolved mercury evolution in the deep North Pacific Ocean over the last 700 years*
- Physical oceanography of the Caroline M4 seamount in the tropical Western Pacific Ocean in summer 2017*
- Characteristics and biogeochemical effects of oxygen minimum zones in typical seamount areas,Tropical Western Pacific*
