Impact of Colonization of an Invasive Species on Genetic Differentiation in New Environments: A Study on American Artemia franciscana (Crustacea:Anostraca) in the United Arab Emirates
ASEMAlireza,SCHUSTERRolf,EIMANIFARAmin, LUHao,LIU Chunwei,WU Xiaofang,YAOLiping,MENG Xinyu,LIWeidong, andWANG Peizheng
1) College of Fisheries and Life Science, Hainan Tropical Ocean University, Sanya 572022, China
2) College of Ecology and Environment, Hainan University, Haikou 570208, China
3) Central Veterinary Research Laboratory, Dubai 597, United Arab Emirates
4) Independent Senior Research Scientist, Industrial District, 21601 Easton, Maryland 21601, USA
5) College of Ecology and Environment, Hainan Tropical Ocean University, Sanya 572022, China
Abstract Artemia franciscana, native to America, has recently colonized as non-indigenous population in Asia, Europe, North Africa, and Australia. We evaluated the effects of the colonization of A. franciscana on genetic differentiation in new environments in the United Arab Emirates (UAE). We used the COI marker to determine the genetic structure and origins of exotic populations in the UAE. Results confirmed the colonization of A. franciscana in two localities. Invasive populations of A. franciscana had significantly lower genetic variation than native populations in the Great Salt Lake and San Francisco Bay. Results showed that the studied populations could not have colonized directly from natural American habitats, and they possibly were from secondary introduction events of other non-indigenous populations. Genetic analysis yielded different demographic patterns for the studied invasive populations. The population in Al Wathba Wetland Reserve (AWWR) demonstrated demographic expansion, whereas in Godolphin Lakes(GL), it reached a demographic equilibrium. Neutrality tests showed an excess of recent and historical mutations in the COI gene pool of invasive AWWR Artemia in the new environment. The results suggest that different ecological conditions in new environments can exert selective pressures during the introduction of an exotic population, which can affect genetic variation.
Key words Artemia franciscana; non-native population; mtDNA-COI; genetic variation; biodiversity; UAE
1 Introduction
The introduction of an exotic species to natural environments can diminish the biodiversity and change biological community structure (Oldenet al., 2004; Lodgeet al., 2006).Although only nearly 1% of introduced exotics become invasive (Williamson, 2006), establishment of invasive species in new habitats might impose widespread ecological effects and economic damages (Pimentelet al., 2005).
Genetic structure of exotic species determines their successful colonization and dispersion (Lee, 2002). Genetic context analysis indicates that the capacity of a non-native species to adapt to new environmental conditions depends on the potential genetic diversity of the species(Lavergne and Molofsky, 2007). Exotic species experience a loss of genetic variation during colonization in non-native habitats because of the founder effect caused by genetic drift in a small population (Dlugosch and Parker, 2008).
The brine shrimp genusArtemiahas been widely used in larviculture (Sorgelooset al., 2001). AnnualArtemiacyst consumption by the aquaculture and fisheries industries increased rapidly from 60 t in 1980 to nearly 2000 t in 1994.At the beginning of the 21st century, the rapid development of aquaculture has increased the harvesting ofArtemiacysts up to 9000 tons in 2000-2001 (wet weight)from the Great Salt Lake, USA (Ben Naceuret al., 2010).About 1000 t ofArtemiacystswere consumed in China alone in 2016 (Van Stappen, 2019).
Since 1950, AmericanA.franciscanafrom the Great Salt Lake (GSL) and San Francisco Bay (SFB) in the USA were exported overseas for larviculture and fishery hatcheries (Van Stappen, 2008; Muñoz, 2009; Eimanifaret al.,2014). Because ofArtemiacysts have been harvested in a limited number of natural hypersaline lakes, growing importance ofArtemiain aquaculture needed to cultureArtemiain other saltwater sources. Given its high reproductive rateand adaptation (Amatet al., 2007; Sanchezet al.,2016),A.franciscanahas been selected to be recultured in non-indigenous natural and artificial environments for industrial aquaculture and fishery activities. As a result, it has a wide geographical distribution in the world, including inland salt lakes, coastal saltworks, salt ponds, and lagoons, and is used widely in fishery hatcheries. Unintentional escapes caused by normal use in hatcheries and/or transmission by migratory waterfowl should be considered as a secondary factor in the distribution ofA. franciscanain new habitats. At present,A.franciscanahas been colonized in numerous regions across Eurasia, especially in the Mediterranean (Amatet al., 2005; Muraet al., 2006;Van Stappen, 2008; Muñoz, 2009; Ben Naceuret al., 2010,Eimanifaret al., 2014; Scalone and Rabet, 2013; Horvathet al., 2018; Sajiet al., 2019; Eimanifaret al., 2020) and Australia (Asemet al., 2018).
TwoArtemiasites have been reported in the United Arab Emirates (UAE) (Aspinall and Hellyer, 1999; Sivakumaret al., 2018). Sajiet al. (2019) have documented the invasiveA.franciscanain Al Wathba Wetland Reserve (AWWR).Evidence thatArtemiahas been introduced intentionally into these localities for commercial activity is lacking (Sajiet al., 2019). In 1998, before the introduction of the greater flamingos in Godolphin Lakes (GL), cysts ofArtemiawere distributed in those water bodies (Sivakumaret al., 2018).Although previous studies have referred to theArtemiapopulation in GL asA.franciscana, phylogenetic proof to support that claim is lacking. Thus, the present study performed a phylogenetic analysis of populations from GL for the taxonomic identification ofArtemiain this locality. The mitochondrialCOIgene was sequenced and the genetic diversity and structure ofArtemiapopulations in the UAE were calculated to compare the evolutionary progress and genetic differentiation of invasive species in new environments.
2 Materials and Methods
2.1 Study Area and Sampling
In total, 82 cysts ofArtemiawere collected from two geographical localities in the UAE, including GL and AWWR,following the method described by Sajiet al. (2019) (Fig.1).Cysts ofArtemiawere collected by a sampling two conical nets with 180 μm in diameter mesh.
Total DNA was extracted from each decapsulatedArtemiacyst in accordance with the Chelex® 100 Resin method (Bio-Rad Laboratories, USA). Cyst samples were crushed and then incubated in a water bath at 56℃ for 3 h and finally at 80℃ for 10 min. Tubes were agitated every 30 min. The tubes were centrifuged at 10000 r min-1for 2 min, and the supernatant phase was stored at -80℃ to be used as a template for PCR (Eimanifaret al., 2014; Asemet al., 2016).
A fragment of the mitochondrial markercytochrome c oxidase subunit I(COI) was amplified using the universal primers LCO1490/HC02198 (Folmeret al., 1994). PCR was performed on a total volume of 20 μL containing 8 μL of ddH2O, 10 μL ofTaqpolymerase (2× EasyTaq® PCR SuperMix, Code#AS111 + dye, TransGen Biotech Co., Ltd.,CHN), 0.4 μL of DNA solution, and 0.8 μL of each primer.The PCR cycling program was as follows (Eimanifaret al.,2014; Asemet al., 2016): a cycle of 3 min at 94℃, followed by 35 cycles of 45 s at 94℃, 60 s at 45℃, and 60 s at 72℃, with a final step of 5 min at 72℃.
2.2 Sequence Alignment and Phylogenetic Analyses
Sequences were aligned using MEGA X with default setting (Kumaret al., 2018). For the taxonomic identification of the collected samples, theCOIreference sequences from bisexual species and parthenogenetic populations were downloaded from GenBank (Table 1) and utilized to draw phylogenetic tree. The phylogenetic tree was generated based on Bayesian Inference (BI) as performed in MrBayes 3.2.2 on XSEDE (Milleret al., 2010). The best fitting nucleotide substitution model was estimated using MrModeltest 2.2 (Nylander, 2004), and HKY + G was selected as the best-fit model.The sequences ofA.franciscanawere analyzed from two natural localities (SFB and GSL) of USA (Muñozet al.,2013) (Table 2). A median network was performed using the median-joining algorithm in the Network program ver.5.0.0.3 to detect the genealogical relationships and compare the UAE samples with AmericanA.franciscana(Bandeltet al., 1999).

Table 1 Species and GenBank accession numbers used in phylogenetic analysis (Pop., population)

Table 2 GenBank accession numbers of A. franciscana from GSL and SFB localities used for haplotype network analysis
For each population of UAE and AmericanA.franciscana, the number of polymorphic sites (S), the total number of mutations (Eta), the number of haplotypes (h), the haplotype diversity (Hd), the haplotype ratio (Hr), the nucleotide diversity (Pi), the average number of nucleotide differences (K), and neutrality tests (i.e., TajimaD, Fu and Li’sD*, Fu’sFs) were computed using DnaSP v.5.10 program (Librado and Rozas, 2009). Expected heterozygosity,FST(an overall population differentiation index), AMOVA (Analysis of Molecular Variance) and mismatch distribution were computed using Arlequin v.3.5(Excoffier and Lischer, 2010). Two parameters for population differentiation (GST,NST) were detected using the program Permut version 1.0 (Pons and Petit, 1996).
3 Results
The BI phylogenetic tree was utilized to studyArtemiasamples from GL and AWWR that were clustered in the clade ofA.franciscana(Fig.2). Results showed that both localities have been invaded by the exotic American speciesA.franciscana.COIsequences ofA.franciscanafrom UAE displayed 16 variable sites, where 8 sites were parsimony informative and 8 sites were singletons. TheCOIsequences of native AmericanA.franciscanarepresented 12 variable sites, where 10 sites were parsimony informa- tive and 2 sites were singletons, respectively.

Fig.2 COI phylogeny of Artemia samples analyzed using the Bayesian Inference (BI) approach. Numbers behind major nodes denote posterior probabilities. Daphnia tenebrosa (GenBank accession no. HQ972028) is used as an outgroup. P.P.,parthenogenetic population; URM, A. urmiana; TIB, A. tibetiana; SIN, A. sinica; FRA, A. franciscana; PER, A. persimilis;SAL, A. salina; H, haplotype.
Fig.3 shows the haplotype distribution network ofA.franciscanaamong native and invasive habitats. The 170COIsequences ofA.franciscanarepresented 14 distinct haplotypes. H1, H10, and H12 are the major types, and are 57.06% (97 ind.), 18.24% (31 ind.), and 12.94% (22 ind.)of the whole tested samples, respectively (as shown in Table 3). In addition, the majority of the sequences belong to H1, including 47.42% AWWR, 46.39% GL, 4.12% SFB,and 2.07% GSL sequences, respectively (Fig.3 and Table 4).
The haplotype distribution and frequency in each locality are presented in Table 5. The majority of haplotype frequencies of the native AmericanA.franciscanafrom SFB and GSL were located in H10 (83.87%) and H12 (95.45%),respectively. Two localities from the UAE possessed the greatest H1 haplotype frequency: AWWR (88.46%: 46 individuals out of 52) and GL (86.54%: 45 individuals out of 52).

Fig.3 Relationship of COI haplotype distribution among Artemia individuals from the Great Salt Lake (GSL), San Francisco Bay (SFB), Al Wathba Wetland Reserve (AWWR), and Godolphin Lakes (GL). Size of each circle is proportional to the frequency of individuals. Hatched circles indicate intermediate or unsampled haplotypes. Black dots/numbers between haplotypes represent the number of nucleotide substitutions.

Table 3 Haplotype information for the network of Artemia based on COI loci

Table 4 Distributions and frequencies of observed localities in each haplotype
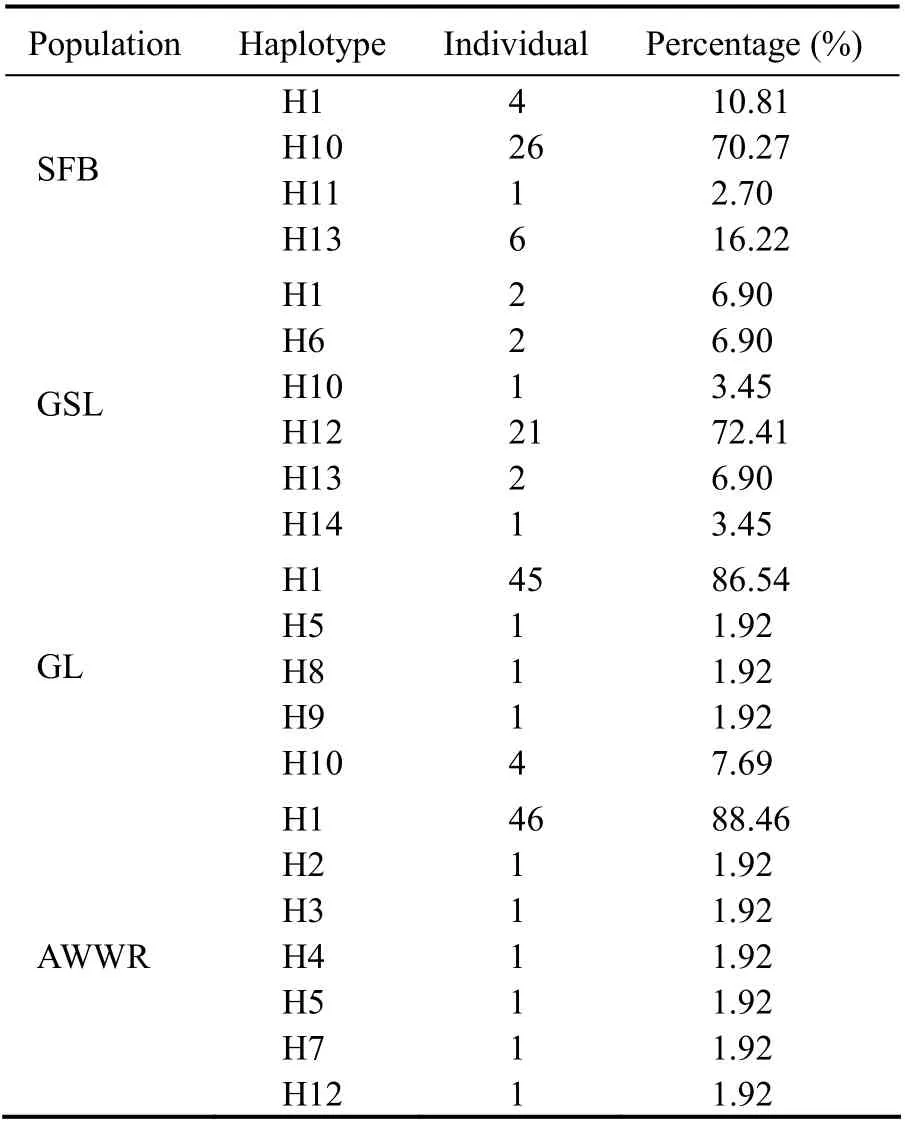
Table 5 Distributions and frequencies of observed haplotypes in each locality
The estimated genetic indices for the studied localities are displayed in Table 6. The highest-ranking amounts of Hr (0.206), Pi (0.0038 ± 0.002), and K (2.334 ± 1.013) were documented in GSL, whereas the SFB locality had the highest quantities of Hd (0.480 ± 0.087) and Ext Het (0.277± 0.136). The lowest genetic variations were observed in AWWR, except for Hr whose lowest value was noticed in GL (0.096). Neutrality tests yielded negative values with significant levels only for AWWR.
The mismatch distributions for invasive UAE and American populations ofA.franciscanashowed that GSL, SFB,and GL had a multimodal pattern, whereas the AWWR location revealed a unimodal pattern (Fig.4).
The lowest and non-significant values of the pairwise genetic differentiation index (FST) were between UAE populations (2.68%). The significant population differentiations were among UAE and American populations (P<0.01). The lowest and significant value ofFSTwas deter-mined between native American populations (SFB-GSL:58.39%) (Table 7).

Table 6 Population genetic indices for invasive and native American A. franciscana based on COI loci

Fig.4 Observed mismatch distributions and their curve fit to simulated model of demographic expansion. GSL, Great Salt Lake; SFB, San Francisco Bay; GL, Godolphin Lakes; AWWR, Al Wathba Wetland Reserve.
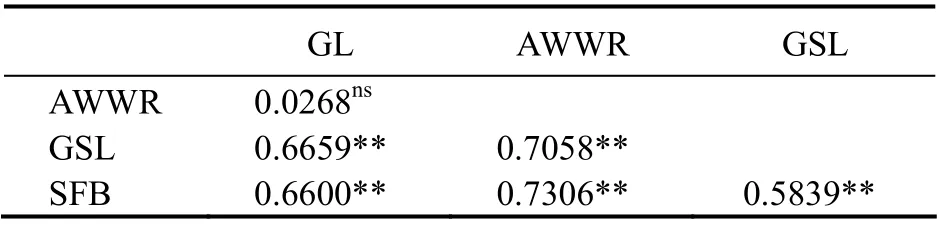
Table 7 Pairwise population matrix of FST values based on COI loci among studied populations
The permutation test indicates that the difference betweenGSTandNSTwas exclusively significant in UAE populations, whereNST(0.027) was higher thanGST(0.0001)(Table 8). AMOVA documented that more than a third of the genetic variation (37.20%) was within populations (Table 9).
The Prince reached the palace in safety, but was so dazzled at first by the Princess s beauty, which far surpassed his expectations, that he was quite dumb for a time
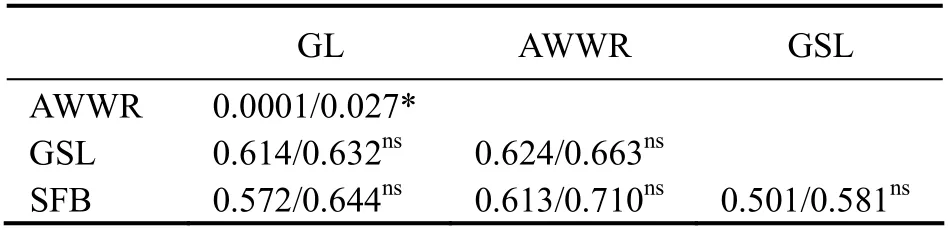
Table 8 Pairwise population GST/NST values based on COI loci among studied populations

Table 9 Molecular variation (within and among populations)for among and between populations of invasive and native A. franciscana (by AMOVA)
4 Discussion
The occurrence ofArtemiain UAE had been recorded in two geographical sites, AWWR (Aspinall and Hellyer,1999) and GL (Sivakumaret al., 2018). A molecular phylogenetic study confirmed that the population in Al Wathba Wetland belongs to an exotic AmericanA.franciscana(Sajiet al., 2019). Our results also confirmed the colonization of the same species in GL.
SFB and GSL are the two major sources ofArtemiathat are usually used to culture in saline ecosystems for industrial aquaculture and fishery activities to produceArtemiacysts and biomass (Eimanifaret al., 2014; Muñozet al.,2014; Asemet al., 2018; Sajiet al., 2019). Thus, these populations were considered in this analysis to determine the genetic alterations of colonized populations in new nonnative environments.
Mitochondrial DNA presents some exceptional characteristics, including rapid evolutionary rates, maternal origin, and lack of recombination (Boore, 1999; Milleret al.,2009). Therefore, mitochondrial markers are important for the apprehension of tracing and the explanation of the source of non-indigenous species in new habitats (Ashtonet al.,2008; Ficetolaet al., 2008; Mabuchiet al., 2008; Gaubertet al., 2009; Asemet al., 2018; Sajiet al., 2019).
Phylogeographic analysis using mitochondrialCOImarkers has clearly demonstrated that GSL and SFB are the origins of invasiveA.franciscanain the Mediterranean area(Muñozet al., 2014; Horvathet al., 2018). Genetic patterns of Asian populations showed that theywere colonized byA. franciscanafrom multiple origins, direct establishment from America and secondary introduction from Europe (Eimanifaret al., 2014). Sajiet al. (2019) studied the phylogeography ofA.franciscanain AWWR (Abu Dhabi; UAE). Regarding the haplotype distribution, the commercialized GSL source might be the origin of invasiveArtemiain Abu Dhabi. A recent study on the geographical origin of the invasive AmericanArtemiain Australia has shown different results. Genetic analyses have evidenced that populations from Port Hedland and St Kilda genetically originated from SFB population, whereasArtemiafrom Mulgundawa and Dampier were from secondary introduction events or were introduced from admixture sources containing GSL and SFB (Asemet al., 2018).Although Sajiet al. (2019) have suggested that GSL is the origin of AWWR (Abu Dhabi; UAE), our finding could not support any evidence that the origin of both localities in AWWR and GL of UAE were from native populations in America (GSL and/or SFB). This result can be ascribed to the fact that most of the sequences in the major haplotype (H1) contained 47.42% of AWWR and 46.39% of GL,and neither GSL nor SFB showed a significant contribution in H1. In addition, no meaningful relationship was found between American and invasive populations, sequences in other haplotypes (Fig.3 and Table 4). Conversely, the high and significantFSTvalues between American and exotic populations indicate that GSL and SFB cannot be an introduced source for UAE localities (Table 7).The paradox between our results and previous study on AWWE might be caused byusing short sequences ofCOIin the previous study (446 bpvs. 604 bp). Regarding the results of the present study, the geographical origin of both UAE populations might be due to the secondary introductions from otherArtemiaproduction sources, especially Eastern Asian sources, including the Mekong Delta (Vietnam) and Bohai Bay (China), where both of these cysts are commercially available in aquaculture markets (Van Stappenet al., 2007; Muñozet al., 2014; Asemet al., 2018;Leet al., 2018).
Golaniet al. (2007) showed that invasive populations generally possess lower genetic variation in new environments in comparison with source populations. An introduced population ofA.franciscanain Vinh Chau (Vietnam) displays a low intraspecific genetic variation and reduced haplotype diversity as compared with its original population from SFB (Kappaset al., 2004). By contrast, a comprehensive study on AsianA.franciscanadocumented that invasive populations had higher genetic diversity than American GSL populations and native Asian species(Eimanifaret al., 2014). Identical findings were found in some Mediterranean invasive populations (Hontoriaet al.,2012; Muñozet al., 2014). Asemet al. (2018) found an equilibrium population of exoticArtemiafrom Dampier in Australia without genetic variation. Other invasive populations in Australia presented different levels of genetic variation as compared with American native populations.Our results indicated that the genetic differentiation in both exotic UAE populations was low and that AWWR revealed lower variation than GL (Table 6).
The low genetic variation of exotic populations can be attributed to the founder effect (Kappaset al., 2004) or population bottleneck during colonization (Asemet al.,2018). Conversely, the higher genetic diversity can be attributed to adaptive ability and/or physiological plasticity in non-native populations (Dlugosch and Parker, 2008; Ruebhartet al., 2008; Vikaset al., 2012; Eimanifaret al., 2014;Muñozet al., 2014; Asemet al., 2018). We suggest that different ecological conditions in new environments can exert selective pressures during the introduction of an exotic population, which can affect genetic variation.
Our results revealed a negative and significant Tajima’sDvalue (-2.304) only for the exotic population in AWWR(Table 6), which demonstrated an excess of rare haplotypes followed by population expansion or purifying selection (Nei and Kumar, 2000; Swansonet al., 2001; Akeyet al., 2004; Crucianiet al., 2008; Levitan and Stapper,2009; Asemet al., 2019). Given the greater numbers of polymorphic sites (12 sites), mutations (12 mutations), and haplotypes (12 haplotypes) in comparison with other localities, Tajima’sDresult might be ascribed to the demographic expansion of theArtemiapopulation in AWWR.In addition, the unimodal mismatch distribution demonstrated the demographic expansion in this population (Fig.4).The negative values of Fu’s Fs test and Fu and Li’s D*test suggest the existence of rare recent mutations (Fu,1997; Ramos-Onsins and Rozas, 2002; Zhaoet al., 2008;Asemet al., 2019) and an excess of rare historical mutations in populations (Fu and Li, 1993; Fu, 1996; Zhaoet al., 2008; Asemet al., 2019), respectively. The results of both neutrality tests were negative and significant for the AWWR population. These findings can document the excess of novel and ancient mutations in theCOIgene pool of invasive AWWRArtemiaduring colonization in new environment.
The non-significant value of neutrality tests and multimodal mismatch distribution in GL suggests a demographic equilibrium. AmericanArtemiain GSL and SFB presented negative and non-significant values for neutrality tests, and a mismatch distribution was revealed as a multimodal pattern. These findings indicate that these populations reached a demographic equilibrium. By contrast,Asemet al. (2018) found that GSL represented a unimodal mismatch distribution and is under demographic expansion. This difference could be attributed to the use ofCOIsequence from GSL in different periods. Asemet al.(2019) suggested that the ecological variations could alter the genetic structure ofArtemiafrom Urmia Lake. Hence,the population genetics of native AmericanArtemiain GSL and SFB warrant a comprehensive study to estimate the genetic variation between these two major sources ofArtemiain long-term periods.
NSTshowed a higher and significant difference (0.027)thanGST(0.0001) between invasive populations in UAE.A significantly higherNSToverGSTgenerally points out the existence of phylogeographic structure (Pons and Petit,1996). The results reflect a differentiation in geographical structure between exotic populations from GL and AWWR and no phylogeographic differentiation between GSL and SFB populations and neither among UAE and American populations.
Overall, GL and AWWR are close localities (approximately at 120 km apart), but theirArtemiapopulationspresent a strong geographical structure. In addition, population genetics showed that these two populations have different demographic histories. Consequently, these findings can demonstrate the effect of colonization on genetic variation in new environments. Despite the lack of information about ecological conditions in these sites, previous studies recorded average salinities of 100 and 180 in GL and AWWR localities, respectively (Dhaheri and Saji, 2013;Sivakumaret al., 2018). In hypersaline environments, salinity is an important ecological parameter for the biology ofArtemiapopulationsincluding survival, growth, and reproduction. The existence of geographical structure and differentiation in UAE populations might be the result of different ecological conditions and the different adaptation processes in new environments.
The impact of exotic species in aquatic ecosystems has been considered less important than that of species from terrestrial habitats (Shea and Chesson, 2002). In addition,insufficient information is available about the effects of colonization on the biodiversity of hypersaline ecosystems,because the latter usually has a more limited number of species compared with marine or freshwater habitats, which limits their biodiversity. Therefore, the introduction of exotic species could represent a major threat to the biodiversity of native species/populations. For example, colonization of AmericanA.franciscanain Port Hedland and Dry Creek (Australia) has causedtheextinction of native parthenogenetic populations (Asemet al., 2018). Several studies have been focused on the biology of invasiveA.franciscanain non-native environments. The results suggest thatA.franciscanacan be employed as a suitable invasive model organism to investigate the biological effect of exotic species on genetic variation patterns and native communities.
In conclusion, this study provides evidence to demonstrate the impact of exotic economical species on genetic and geographical differentiation in new environments. Invasive species can present a different genetic pattern comparing with the same species in native populations. However, exotic populations in neighboring environments do not necessarily share a similar genetic pattern.
Acknowledgements
This project was funded by the Key Research and Development Program from Science and Technology Department of Hainan Province (No. ZDYF2019154). The help of Prof. Jim Clegg (University of California, USA) in English editing is greatly acknowledged.
 Journal of Ocean University of China2021年4期
Journal of Ocean University of China2021年4期
- Journal of Ocean University of China的其它文章
- Paleosalinity and Its Association with Organic Matter:A Case Study from the Eocene Shahejie Formation,Laizhou Bay Sag, Bohai Bay Basin (China)
- Studies on the Inversion Phenomenon of Physical Properties Observed in the Huagang Formation Reservoir in the Xihu Sag Based on the Water-Rock Reaction Experiments
- Effect of Sand Body Enrichment Under the Restriction of a Tectonic Transfer Zone: A Case Study on the Pinghu Formation in the Kongqueting Region on the Pinghu Slope
- Relationship Between Paleogene Reservoir Densification and Hydrocarbon Accumulation in the Xihu Depression
- Adjoint Method-Based Algorithm for Calculating the Relative Dispersion Ratio in a Hydrodynamic System
- Analysis of the Leading Modes of Autumn Precipitation over the Yangtze River Basin
