苹果果实大小相关的ARF-Aux/IAA互作组合筛选
周喆,卞书迅,张恒涛,张瑞萍,高启明,刘珍珍,阎振立
苹果果实大小相关的ARF-Aux/IAA互作组合筛选
周喆,卞书迅,张恒涛,张瑞萍,高启明,刘珍珍,阎振立
中国农业科学院郑州果树研究所,郑州 459000
【】通过转录组学和生物信息学,在苹果全基因组中对可能互作的MdARF和MdIAA进行鉴定,为明确相关基因功能和解析生长素调控苹果果实大小的分子机理奠定基础。对野生大果型‘皇家嘎啦’和过表达的转基因小果型‘皇家嘎啦’进行不同发育时期和不同组织材料的转录组测序,对测序结果进行基因的功能注释及差异表达分析。利用转基因小果和野生型大果的转录组数据,筛选在果实发育中表达的苹果MdARFs和MdIAAs基因家族成员,通过逐一在两个家族间计算基因时空表达的相关系数,筛选可能互作的MdARF-MdIAA组合,将从拟南芥基因组中下载的23个ARFs和34个Aux/IAAs、番茄基因组中下载的21个ARFs和25个Aux/IAAs,分别与互作候选MdARFs和MdIAAs进行比对,并进一步构建系统发育树。使用MEME和TBtools对苹果候选互作对中的MdARFs和MdIAAs蛋白进行Motif分析。利用STRING蛋白互作预测数据库进行同源映射,构建苹果中的蛋白-蛋白互作网络,进一步的确认候选互作对,最终得到苹果中通过互作参与果实发育可能性最高的MdARF-MdIAA组合。分别对野生型‘皇家嘎啦’和miR172OX转基因‘皇家嘎啦’盛花期后两周的全果和盛花期后4周的果皮、果肉和果核进行转录组测序,共生成178.19 Gb的数据量,各项指标均表明,3个生物学重复在所有组织类型上均具有高度一致性。在转录组数据中,共鉴定到38个和27个在至少一个文库中的FPKM值大于2,在苹果果实发育时期表达。通过计算Pearson相关系数对表达的MdARFs和MdIAAs两两进行相关性分析,其中8对MdARF-MdIAA的相关系数大于0.9或小于-0.9,作为初步筛选的候选互作组合。将8对组合中的MdARFs和MdIAAs分别与拟南芥和番茄中的ARFs和IAAs进行序列比对并构建系统进化树后发现,MdARF6和MdARF19与起转录激活作用的AtARFs同属一个分支。而MdARF2、MdARF4和MdARF9则与起转录抑制作用的AtARFs具有较近的亲缘关系。Motif分析结果显示,候选MdARF、MdIAA蛋白中均包含Motif 2和Motif 5。Motif 2和Motif 5分别对应IAA蛋白中的保守结构域Motif IV和Motif III。互作蛋白在拟南芥中进行同源映射校验后,最终得到两对MdARF-MdIAA组合可用于进一步的功能验证。苹果MdARF和MdIAA家族成员,在果实发育时期,有8对组合在表达量上存在显著的相关性,进一步同源映射确认互作后,最终确定MdARF4-MdIAA17和MdARF4-MdIAA19两对互作组合,极有可能通过互作传递生长素信号参与调控苹果果实发育。
苹果;ARF;Aux/IAA;互作筛选
0 引言
【研究意义】苹果(×)是世界范围内栽培最为广泛的果树之一。苹果果实的大小是衡量其品质和商品价值的重要指标。果实的最终大小,主要由果实内细胞数量和细胞大小共同决定[1-2]。植物激素作为协调细胞增殖和细胞膨大的关键因子,对苹果果实生长有着至关重要的影响[3-5]。在众多参与调节果实大小的植物激素中,生长素(auxin)起着非常关键的作用[6]。在果实发育时期,其内部生长素信号的传递,主要是通过生长素响应因子(auxin response factor,ARF)和生长素/吲哚乙酸蛋白(Auxin/Indole-3-Acetic Acid,Aux/IAA)之间的相互作用完成[7-8]。目前,相关的ARF和Aux/IAA互作鉴定主要集中在模式植物中,且ARF和Aux/IAA互作关系在调控果实发育中的研究鲜有报道。【前人研究进展】生长素在植物中广泛存在,参与植物器官发育、胁迫响应、果实成熟等各类生物学过程[9]。对植物施用外源生长素,极短的时间内即可观察到、(Gretchen Hagen3)、(Small Auxin Up RNA)等一系列生长素相关基因的表达水平发生改变[3,10-11]。ARF和Aux/IAA作为两个功能上相关的基因家族,是感应植物体内生长素浓度变化的核心元件[12]。两者通过互作形成复合体,将接收到的信号以促进或抑制上下游相关基因表达的形式释放,最终引起植物形态发育上的差异[13]。ARF与Aux/IAA调控生长素信号转导的模式为,当植物体内的生长素浓度处于较低水平时,Aux/IAA蛋白与ARF蛋白结合,阻止ARF激活生长素相关基因的转录;当生长素浓度上升时,Aux/IAA与生长素受体TIR结合并被降解,释放ARF来调控相关生长素响应基因的表达[13-14]。ARF家族蛋白在结构上主要包含一个N端B3型DNA绑定结构域(DNA binding domain,DBD)、一个C端的二聚绑定下游生长素响应基因的结构域(dimerization domain,CTD)和一个非保守的中间区域(middle region,MR)。Aux/IAA家族蛋白包含I、II、III、IV四个保守结构域,其中结构域I是一个起阻遏作用的富亮氨酸重复结构域(LxLxL),结构域II可以与生长素受体TIR1(Transport Inhibitor Resistant 1)结合引起Aux/IAA的泛素化降解,结构域III、IV是与ARF的CTD同源的结构域,参与Aux/IAA与ARF的二聚化[15-19]。Aux/IAA和ARF蛋白都是在转录水平对下游基因进行调控。Aux/IAA家族成员通常对下游生长素相关基因的表达起抑制作用,而ARF蛋白主要通过DBD结构域识别下游基因启动子区域的生长素响应元件(AuxREs)来调控其表达,调控作用取决于其MR的氨基酸序列[14]。起转录激活作用的ARF,其MR富含谷氨酰胺、亮氨酸和丝氨酸残基,例如拟南芥的ARF5、6、7、8、19[12];而阻遏转录活性的ARF,其MR则富含谷氨酸、亮氨酸、丝氨酸和脯氨酸残基,例如拟南芥AtARF1、2、4、9[20]。到目前为止,已有15种植物的ARF基因家族(包括苹果)和30种植物的Aux/IAA基因家族通过全基因组分析得到了鉴定。而全基因组范围内对两个基因家族间互作可能性的鉴定主要集中在拟南芥、番茄和水稻中[20]。其中拟南芥中共鉴定到213对可能的互作关系,在番茄的21个ARF和24个Aux/IAA之间进行了互作筛选,水稻中的8个ARF和15个Aux/IAA之间可能存在相互作用[8,21-22]。拟南芥中的ARF-Aux/ IAA互作复合体主要在子叶下胚轴和根部发育中起作用;番茄中的SlARF5-SlIAA3、SlARF7A-SlIAA8和SlARF4-SlIAA15可能通过互作调控果实发育[22-25]。在苹果中,MdARF13可以与MdIAA121互作,生长素可以通过MdARF13- MdIAA121介导的信号路径调控花青苷合成[26]。【本研究切入点】苹果中全基因组范围内ARF和Aux/IAA互作筛选尚未开展,且ARF和Aux/IAA的互作在果实发育中扮演的角色尚未可知。【拟解决的关键问题】课题组前期通过在‘皇家嘎啦’中过表达miRNA172p获得了果实显著变小的转基因苹果(miR172OX)[27]。通过对miR172OX和野生型的‘皇家嘎啦’(WT)不同发育时期的果实和组织进行转录组测序,发现ARF和Aux/IAA家族的基因表达量变化差异显著且存在相关关系。本研究拟通过对转录组中鉴定到的ARF和Aux/IAA进行成对表达分析,筛选到可能存在的果实发育过程中的互作组合。
1 材料与方法
试验于2019—2020年于中国农业科学院郑州果树研究所进行。
1.1 试验材料
本试验选择野生型‘皇家嘎啦’(WT)和:转基因‘皇家嘎啦’(miR172OX)两个基因型的苹果进行转录组测序。两个基因型的苹果均定植在新西兰植物和食品研究所。分别选取盛花后2周的全果(WF)和盛花后3周果实的果皮(FS)、果肉(FF)和果核(FC),每个样品3个重复,共24个样品。取样后液氮速冻并于-80℃保存。
1.2 总RNA提取、文库构建及测序
参照Zhou等[28]的方法提取样品总RNA。得到总RNA后,进行琼脂糖凝胶电泳检测样品RNA的完整性及是否存在DNA污染。使用NEBNext® UltraTM RNA Library Prep Kit for Illumina®试剂盒对总量大于1 μg的总RNA进行文库构建。通过Illumina Hiseq测序平台对制备的文库进行测序,并产生150 bp配对末端读数。测序的原始数据已上传到NCBI,SRA登录号为PRJNA649660。
1.3 基因的功能注释及差异表达分析
以GDDH13 v1.1为苹果参考基因组(×)[29],使用HISAT2 v2.0.5构建参考基因组的索引。使用DESeq2R(1.16.1)软件进行两个基因型之间的差异表达分析,如果基因的adj值<0.05,则被认定为差异表达[30]。校正后的值以及|log2foldchange|作为显著差异表达的阈值。差异表达基因在至少一个文库的FPKM值要大于2以消除低表达的基因。
1.4 苹果ARF和Aux/IAA家族互作关系预测及系统进化分析
根据从转录组数据中鉴定到的和的FPKM值,两两基因之间计算表达量的Pearson相关系数。以|PCC|(pearson correlation ecoefficiency)的值大于0.9为筛选标准,得到候选互作基因。从拟南芥数据库TAIR(http://www.arabiodpsis.org)中下载拟南芥基因组中23个ARFs和34个Aux/IAAs的氨基酸序列,从NCBI上下载番茄基因组中21个ARFs和25个Aux/IAAs,分别与互作候选MdARFs和MdIAAs进行比对并构建系统发育树。使用MEME和TBtools对苹果候选互作对中的MdARFs和MdIAAs蛋白进行motif分析。
1.5 蛋白-蛋白互作网络构建
利用拟南芥STRING数据库进行蛋白-蛋白互作预测,提取其中包含候选MdARFs和MdIAAs的节点,选择互作分数大于700的互作蛋白构建苹果中的互作网络。
2 结果
2.1 RNA-seq结果的统计分析
在前期研究中,获得了miRNA172p过表达的转基因‘皇家嘎啦’。其果实与野生型果实相比,果个显著变小[27](图1)。选取依赖细胞分裂进行果实增大的盛花期后2周的全果和依赖细胞扩张进行果实膨大的盛花期后4周的果皮、果肉和果核进行转录组测序分析。通过Illumina Hiseq 2500 platform,24个库一共生成178.19 Gb数据量。Q20和Q30的值分别大于97.28%和92.37%。Pearson相关分析(2=0.88—0.98)表明,3个生物学重复在所有组织类型上均具有高度一致性。
2.2 苹果ARF和IAA家族成员的表达图谱及差异表达基因
根据基因注释,从转录组数据中一共鉴定到38个和27个在果实中表达(图2-A)。其中分别有17个和14个在WT和miR172OX中的表达量存在差异,为差异表达基因(图2-B)。在差异表达的17个中,有3个为下调基因,14个为上调基因。而14个差异表达的均为下调基因。

A:成熟果实;B:成熟果实果肉组织切片

A:热图显示苹果ARF和IAA基因家族表达量的聚类分析;B:苹果ARF和IAA基因家族中的差异表达基因及差异表达趋势
2.3 生长素信号路径中的蛋白互作预测
ARF与Aux/IAA的特定互作组合对不同组织、发育阶段和生物学过程中生长素响应的方式起着重要的决定作用[31]。通过将上述38个和27个在不同发育时期和不同组织中的表达量逐对进行相关性分析,共鉴定到8对相关系数大于0.9或小于-0.9的ARF-Aux/IAA候选互作组合,其中包括MdARF19(MD02G1096700)和MdIAA29(MD08G1151300)(0.912),MdARF4(MD03G1116000)和MdIAA14(MD16G1206700)(0.960),MdARF4(MD03G1116000)和MdIAA19(MD17G1198100)(0.968),MdARF16(MD04G1096900)和MdIAA22D(MD10G1193000)(-0.912),MdARF6(MD10G1257900)和MdIAA13(MD15G1169100)(-0.901),MdARF6(MD10G1257900)和MdIAA19(MD17G1198100)(-0.943),MdARF9(MD14G1131900)和MdIAA29(MD08G1151300)(0.926),MdARF2(MD14G1148500)和MdIAA22D(MD10G1193000)(-0.930)(图3)。8对候选组合中,共包括6个和5个。候选互作对中涉及的11个基因中,除了外,其余均为差异表达基因(图2-B)。
2.4 候选MdARFs和MdIAAs的系统发育分析
分别将MdARF2、MdARF4、MdARF6、MdARF9、MdARF16、MdARF19和拟南芥()中的23个AtARFs、番茄中的21个SlARFs,将MdIAA13、MdIAA14、MdIAA19、MdIAA22D、MdIAA29和拟南芥中的34个AtIAAs、番茄中的25个SlIAAs进行序列比对后,构建系统进化树(图4-A)。参照与拟南芥ARF家族成员的亲缘关系,MdARF6和MdARF19分别与起转录激活作用的AtARFs聚类到同一分支;而MdARF2、MdARF4和MdARF9则与起转录抑制作用的AtARFs具有较近的亲缘关系。Motif分析结果显示,候选MdARF和MdIAA蛋白中均包含Motif 2和Motif 5。Motif 2和Motif 5分别对应IAA蛋白中的保守结构域Motif IV和Motif III(图4-B)。
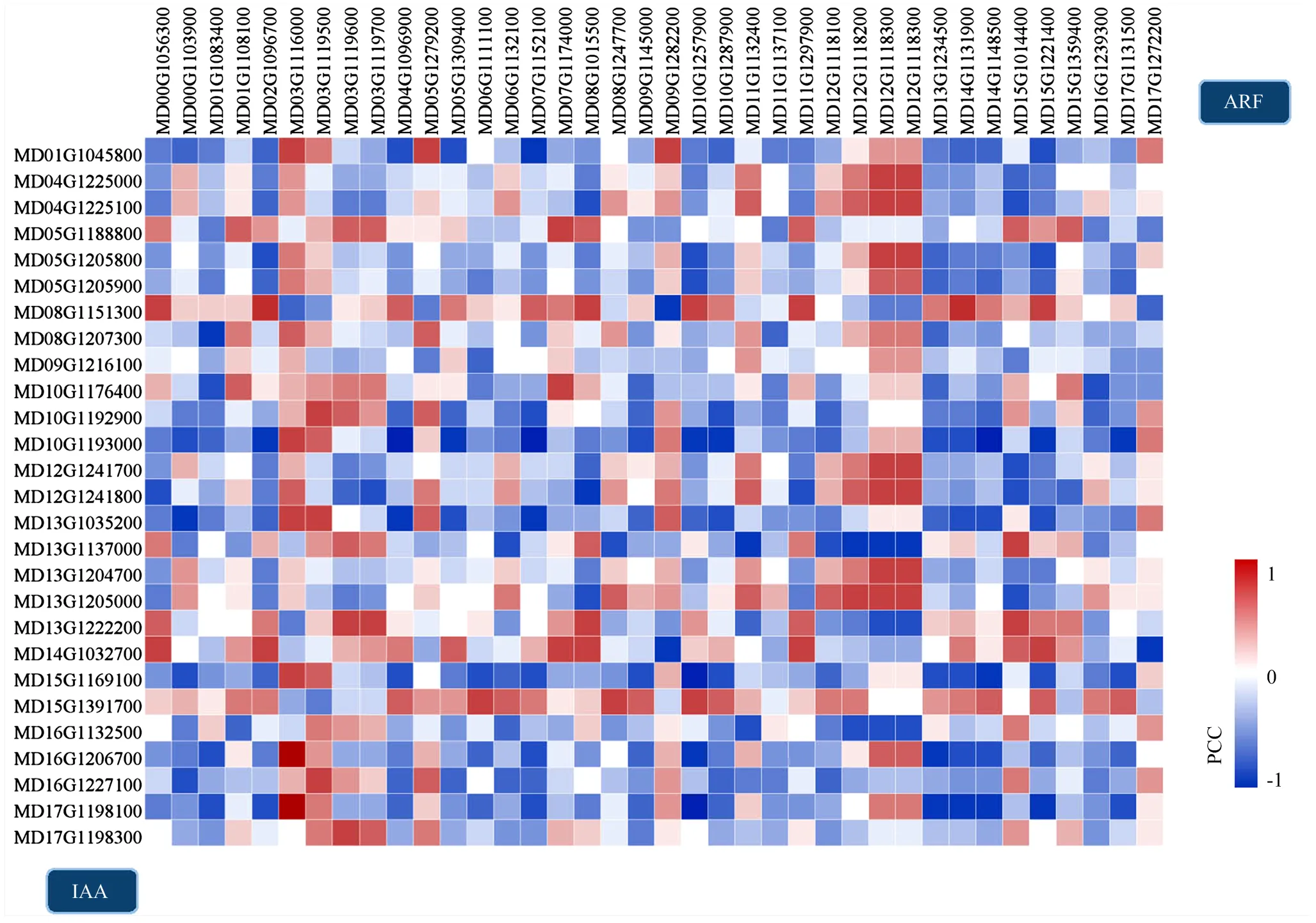
图3 在时空共表达的ARF和Aux/IAA间鉴定蛋白互作

A:苹果候选ARF-Aux/IAA互作对中MdARFs和MdIAAs的系统发育分析;B:苹果候选ARF-Aux/IAA互作对中MdARFs和MdIAAs的保守结构域分析
2.5 蛋白-蛋白互作网络分析
蛋白-蛋白互作网络可以预测两个蛋白间直接的相互作用或间接的功能相关性。在构建的苹果蛋白-蛋白互作网络中搜索上述8对候选ARF-Aux/IAA中的11个基因,共有5个基因出现在互作网络中,包括MdARF4、MdARF19、MdIAA14、MdIAA19和MdIAA29(图5)。其中候选的MdARF4-MdIAA17和MdARF4- MdIAA19互作对在蛋白-蛋白互作网络中也显示互作关系。
3 讨论
许多研究表明,ARF和IAA在植物生长和发育等多个方面起作用[32]。本研究鉴定到的和中接近一半的基因在大、小果两个基因型中表达量存在差异。8对相关性较高的ARF-Aux/IAA 候选组合,其中MdARF19和MdIAA29、MdARF4和MdIAA14、MdARF4和MdIAA19、MdARF9 和MdIAA29呈正相关,而MdARF16和MdIAA22D、MdARF6和MdIAA19、MdARF6和MdIAA13、MdARF2和MdIAA22D呈负相关(图3)。8对组合中,除MdARF19和MdIAA13不满足adj<0.05,为非差异表达基因外,组合中其他基因均在一个或多个时期和/或组织中显示差异表达(图2-B)。对8对候选组合内成员进行保守结构域分析表明,互作组中所有和编码的蛋白,在C端均包含同源的保守结构域(CTD结构域或结构域III、IV)(图4),进一步在结构上证明了8对候选组合互作的可能性。番茄中鉴定到的21个ARF因子中,在果实发育时期呈现高表达的包括、、、、和[33-34]。其中在番茄果实发育初期起负调控作用,而则是番茄果实膨大期重要的负调控因子[35]。可以通过负调控来影响果实中糖分的积累[36]。系统发育结果显示,候选互作对中的MdARF2与SlARF2、MdARF4与SlARF4、MdARF6与SlARF6、MdARF16与SlARF16、MdARF9与SlARF9分别具有较近的亲缘关系(图4-A),因此,苹果中的可能与番茄在果实发育中起相似的调控作用。

节点大小对应节点连接数量,节点颜色对应Log2(Foldchange)值,节点间连线颜色对应PCC值
同源映射是一种比较成熟的预测蛋白质间互作的方法,它主要是基于蛋白之间的互作关系伴随着物种进化而表现出的保守性。本研究最终确定了互作可能性最高的组合为MdARF4-MdIAA17和MdARF4- MdIAA19。除此之外,蛋白-蛋白互作网络还显示MdIAA19与TPR3/4(Topless-related)之间的互作关系。TPR是一类保守的植物转录辅抑制因子家族蛋白,能够与Aux/IAA包含的LxLxL类型的EAR基序结合。在植物缺乏生长素刺激时,Aux/IAA蛋白可与TPR形成蛋白复合物,绑定到植物生长素相关基因的启动子区域,从而抑制ARF转录因子,该机制在植物激素应激反应中发挥重要作用[37]。
到目前为止,关于ARF和IAA调控果实发育的研究多集中在番茄,且多以单独的ARF或Aux/IAA家族成员功能鉴定为主。有关生长素路径中的ARF- Aux/IAA互作单元如何调控果实大小,尤其是多年生果树果实大小的分子机理研究非常有限。通过转录组测序技术,可以获得特定组织在特定时期和特定条件下所有基因的表达情况。借助生物信息学等手段,通过数据量化的方式预测功能基因之间完成生物过程的协作关系,可提高调控网络解析的效率。本研究完成了苹果全基因组范围内果实发育相关ARF- Aux/IAA互作对筛选,为进一步深入解析两个家族基因在生长素路径中扮演的角色及调控果实发育的作用奠定了基础,极大地提高了今后通过分子生物学和基因工程手段进行基因功能验证的效率。
4 结论
本研究共鉴定到8对在表达量上存在显著相关性的果实发育候选MdARF-MdIAA组合,进一步同源映射确认互作后,最终确定了MdARF4-MdIAA17和MdARF4-MdIAA19两对互作组合,可用于进一步的功能验证。
[1] SUGIMOTO-SHIRASU K, ROBERTS K. "Big it up": Endoreduplication and cell-size control in plants. Current Opinion in Plant Biology, 2003, 6: 544-553.
[2] HARADA T, KURAHASHI W, YANAI M, WAKASA Y, SATOH T. Involvement of cell proliferation and cell enlargement in increasing the fruit size ofspecies. Scientia Horticulturae, 2005, 105(4): 447-456.
[3] CHAPMAN E J, ESTELLE M. Mechanism of Auxin-regulated gene expression in plants. Annual Review of Genetics, 2009, 43(1): 265-285.
[4] MARIOTTI L, PICCIARELLI P, LOMBARDI L, CECCARELLI N. Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. Journal of Plant Growth Regulation, 2011, 30: 405-415.
[5] ZHAO Y. Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology, 2010, 61: 49-64.
[6] PEI M S, CAO S H, WU L, WANG G M, XIE Z H, GU C, LING Z S. Comparative transcriptome analyses of fruit development among pears, peaches, and strawberries provide new insights into single sigmoid patterns. BMC Plant Biology, 2020, 20(1): 108.
[7] PIYA S, SHRESTHA S K, BINDER B, NEAL STEWART JR C, HEWEZI T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in. Frontiers in Plant Science, 2014, 5: 744.
[8] AUDRAN-DELALANDE C, BASSA C, MILA I, REGAD F, ZOUINE M, BOUZAYEN M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant and Cell Physiology, 2012, 53(4): 659-672.
[9] HAGEN G, GUILFOYLE T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Molecular Biology, 2002, 49(3/4): 373-385.
[10] BENJAMINS R, SCHERES B. Auxin: The looping star in plant development. Annual Review of Plant Biology, 2008, 59: 443-465.
[11] 胡晓, 侯旭, 袁雪, 管丹, 刘悦萍. ARF和Aux/IAA调控果实发育成熟机制研究进展. 生物技术通报, 2017, 33(12): 37-44.
HU X, HOU X, YUAN X, GUAN D, LIU R P. Research progress on mechanism of ARF and Aux/IAA regulating fruit development and ripening. Biotechnology Bulletin, 2017, 33(12): 37-44. (in Chinese)
[12] LISCUM E, REED J W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology, 2002, 49(3/4): 387-400.
[13] ULMASOV T, HAGEN G, GUILFOYLE T J. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(10): 5844-5849.
[14] GUILFOYLE T J, HAGEN G. Auxin response factors. Current Opinion in Plant Biology, 2007, 10(5): 453-460.
[15] TIWARI S B, HAGEN G, GUILFOYLE T J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell, 2004, 16(2): 533-543.
[16] SZEMENYEI H, HANNON M, LONG J A. TOPLESS mediates auxin-dependent transcriptional repression duringembryogenesis. Science, 2008, 319(5868): 1384-1386.
[17] ULMASOV T, HAGEN G, GUILFOYLE T J. ARF1, a transcription factor that binds to auxin response elements., 1997, 276(5320): 1865-1868.
[18] SHEN C J, YUE R Q, SUN T, ZHANG L, XU L Q, TIE S G, WANG H Z, YANG Y J. Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Frontiers in Plant Science, 2015, 6: 73.
[19] 李俊男, 燕晓杰, 李枢航, 张荣沭. 植物AUX/IAA基因家族研究进展. 中国农学通报, 2018, 34(15): 89-92.
LI J N, YAN X J, LI S H, ZHANG R S. Plants AUX/IAA gene family: Research progress. Chinese Agricultural Science Bulletin, 2018, 34(15): 89-92. (in Chinese)
[20] VERNOUX T, BRUNOUD G, FARCOT E, MORIN V, VAN DEN DAELE H, LEGRAND J, OLIVA M, DAS P, LARRIEU A, WELLS D, GUEDON Y, ARMITAGE L, PICARD F, GUYOMARC'H S, CELLIER C, PARRY G, KOUMPROGLOU R, DOONAN J H, ESTELLE M, GODIN C, KEPINSKI S, BENNETT M, DE VEYLDER L, TRAAS J. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology, 2011, 7: 508.
[21] LLERES D, SWIFT S, LAMOND A I. Detecting protein-protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy (FLIM). Current Protocols in Cytometry, 2007, 42(1).
[22] SHINOZAKI Y, NICOLAS P, FERNANDEZ-POZO N, MA Q, EVANICH D J, SHI Y, XU Y, ZHENG Y, SNYDER S I, MARTIN L B B, RUIZ-MAY E, THANNHAUSER T W, CHEN K, DOMOZYCH D S, CATALA C, FEI Z, MUELLER L A, GIOVANNONI J J, ROSE J K C. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nature Communications, 2018, 9(1): 364.
[23] TATEMATSU K, KUMAGAI S, MUTO H, SATO A, WATAHIKI M K, HARPER R M, LISCUM E, YAMAMOTO K T. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in. Plant Cell, 2004, 16(2): 379-393.
[24] ARASE F, NISHITANI H, EGUSA M, NISHIMOTO N, SAKURAI S, SAKAMOTO N, KAMINAKA H. IAA8 involved in lateral root formation interacts with the TIR1 auxin receptor and ARF transcription factors in. PLoS One, 2012, 7(8): e43414.
[25] SHEN C, WANG S, BAI Y, WU Y, ZHANG S, CHEN M, GUILFOYLE T J, WU P, QI Y. Functional analysis of the structural domain of ARF proteins in rice (L.). Journal of Experimental Botany, 2010, 61(14): 3971-3981.
[26] WANG Y C, WANG N, XU H F, JIANG S H, FANG H C, SU M Y, ZHANG Z Y, ZHANG T L, CHEN X S. Auxin regulates anthocyanin biosynthesis through the Aux/IAA-ARF signaling pathway in apple. Horticulture Research, 2018, 5: 59.
[27] YAO J L, XU J, CORNILLE A, TOMES S, KARUNAIRETNAM S, LUO Z, BASSETT H, WHITWORTH C, REES-GEORGE J, RANATUNGA C, SNIRC A, CROWHURST R, DE SILVA N, WARREN B, DENG C, KUMAR S, CHAGNE D, BUS V G, VOLZ R K, RIKKERINK E H, GARDINER S E, GIRAUD T, MACDIARMID R, GLEAVE A P. A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant Journal, 2015, 84(2): 417-427.
[28] ZHOU Z, CONG P H, TIAN Y, ZHU Y M. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS One, 2017, 12(9): e0185288.
[29] DACCORD N, CELTON J M, LINSMITH G, BECKER C, CHOISNE N, SCHIJLEN E, VAN DE GEEST H, BIANCO L, MICHELETTI D, VELASCO R, DI PIERRO E A, GOUZY J, REES D J G, GUERIF P, MURANTY H, DUREL C E, LAURENS F, LESPINASSE Y, GAILLARD S, AUBOURG S, QUESNEVILLE H, WEIGEL D, VAN DE WEG E, TROGGIO M, BUCHER E. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nature Genetics, 2017, 49(7): 1099-1106.
[30] ANDERS S, HUBER W. Differential expression analysis for sequence count data. Genome Biology, 2010, 11: R106.
[31] LI S B, XIE Z Z, HU C G, ZHANG J Z. A review of auxin response factors (ARFs) in plants. Frontiers in Plant Science, 2016, 7: 47.
[32] CHANDLER J W. Auxin response factors. Plant Cell and Environment, 2016, 39: 1014-1028.
[33] KUMAR R, TYAGI A K, SHARMA A K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics, 2011, 285: 245-260.
[34] WU J, WANG F Y, CHENG L, KONG F L, PENG Z, LIU S S, YU X L, LU G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Reports, 2011, 30(11): 2059-2073.
[35] DE JONG M, WOLTERS-ARTS M, FERON R, MARIANI C, VRIEZEN W H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant Journal, 2009, 57(1): 160-170.
[36] SAGAR M, CHERVIN C, MILA I, HAO Y, ROUSTAN J-P, BENICHOU M, GIBON Y, BIAIS B, MAURY P, LATCHE A, PECH J-C, BOUZAYEN M, ZOUINE M. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiology, 2013, 161(3): 1362-1374.
[37] CAUSIER B, ASHWORTH M, GUO W, DAVIES B. The TOPLESS interactome: A framework for gene repression in. Plant Physiology, 2012, 158(1): 423-438.
Screening of ARF-Aux/IAA Interaction Combinations Involved in Apple Fruit Size
ZHOU Zhe, BIAN ShuXun, ZHANG HengTao, ZHANG RuiPing, GAO QiMing, LIU ZhenZhen, YAN ZhenLi
Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 459000
【】The transcriptomics and bioinformatics methods were used to carry out the genome-wide potential interacting MdARFs and MdIAAs pairs screening, so as to build a foundation for clarifying the function of related genes and elucidating the molecular mechanism underlying auxin-regulated apple fruit size.【】Two apple genotypes, Royal Gala (WT) andtransgenic Royal Gala (miR172OX ) were used as test materials in this study. Fruit materials from different developmental stages and tissue types were collected for both genotypes and were subjected to transcriptome sequencing. Clean reads were aligned to the reference genome and the differential expression analysis was performed. Based on the transcriptome data obtained by sequencing the transgenic small fruit and wild-type large fruit, the pairwise expression analysis was performed across MdARFs and MdIAAs families. The amino acid sequences of 23 ARFs and 34 Aux/IAAs were downloaded from Arabidopsis genome, and 21 ARFs and 25 Aux/IAAs were downloaded from tomato genome, which were further compared with candidate MdARFs and MdIAAs to construction phylogenetic trees. The MEME and TBtools were used to carry out the Motif analysis for candidate MdARFs and MdIAAs. Pairs with high interacting possibilities were further confirmed by a protein-protein interacting network constructed in apple to finalize combinations with the highest probability of involvement in fruit development. 【】The whole fruit at 2 weeks post full bloom and the fruit skin, fruit flesh and fruit core at 4 WPFB were collected from WT and miR172OX, respectively. To achieve research objectives, transcriptome sequencing was carried out. A total of 178.19 Gb paired-end reads of 125 bp/150 bp were generated. All indexes indicated that the three biological replicates had highly consistent transcriptome profiles across all tissue types. FPKM values in at least one library was over 2 were used as a standard to eliminate the low expressed genes, so a total of 38and 27were expressed. In our fruit developmental transcriptome data, eight pairs of MdARF-MdIAA were obtained through Pearson correlation analysis, whose Pearson correlation coefficient was over 0.9 or below -0.9. The systematic phylogenetic analysis showed that MdARF6 and MdARF19 belonged to the same branch with AtARFs, which played a role in transcription activation, while MdARF2, MdARF4, and MdARF9 were closely related to transcriptional inhibitory AtARFs. Motif analysis results showed that both the candidate MdARFs and MdIAAs proteins contained Motif 2 and Motif 5, which were corresponded to the conserved domains Motif IV and Motif III in the IAA protein, respectively. After homolog mapping inspection with Arabidopsis, two potential MdARF-MdIAA interacting pairs were selected for future functional identification. 【】Among apple MdARF and MdIAA family members, eight pairs of MdARF-MdIAA showed significant correlations in terms of their expression patterns during fruit development. Further homology mapping confirmed two pairs of them, including MdARF4-MdIAA17 and MdARF4-MdIAA19, were most likely to participate in the regulation of apple fruit development through mediating auxin signal transduction.
apple; ARF; Aux/IAA; interacting-pair screening

10.3864/j.issn.0578-1752.2021.14.014
2020-08-30;
2020-10-14
国家重点研发计划(2018YFD1000106)
周喆,E-mail:zhouzhe@caas.cn。通信作者阎振立,E-mail:yanzhenli@caas.cn
(责任编辑 赵伶俐)

