Development of organelle single nucleotide polymorphism(SNP) markers and their application for the identification of cytoplasmic inheritance patterns in Pyropia yezoensis(Bangiales, Rhodophyta)*
Lu WANG , Junhao WANG , Yunke ZHU, Zhengcai CUI, Fanna KONG,Xianghai TANG ,**, Yunxiang MAO,2,**
1 Key Laboratory of Marine Genetics and Breeding(Ministry of Education), College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China
2 Key Laboratory of Utilization and Conservation of Tropical Marine Bioresource(Ministry of Education), College of Fisheries and Life Science, Hainan Tropical Ocean University, Sanya 572022, China
Abstract The genus Pyropia contains several important cultivated species. Genetic research in nori species has mainly focused on the cell nucleus, with few studies on organelles (chloroplast and mitochondria).Due to the high copy numbers of organelles in cells, which influence the development and traits of algae,it is necessary to study their genetic mechanism. In this study, the marine red alga Pyropia yezoensis, an important economic macroalga, was selected as the study object. To investigate organelle (chloroplast and mitochondria) inheritance in P. yezoensis, the wild type RZ (maternal strain) was crossed with the red mutant HT (paternal strain) and 30 color-sectors from 11 F1 gametophytic blades were examined. The complete chloroplast and mitochondrial genomes of the red mutant (HT) were assembled for the first time. One reliable and stable single nucleotide polymorphism (SNP) loci filtrated by bioinformatics analysis was used as a molecular marker for chloroplast and mitochondrial DNA, respectively, in subsequent experiments.PCR amplification and sequence analysis showed that the haplotypes of color-sectors detected were consistent with those of the maternal parent, confirming that both chloroplast and mitochondrial genomes were inherited maternally in P. yezoensis. The inheritance pattern of organelles in P. yezoensis can be used to guide the hybridization and breeding of nori. Additionally, the organelle SNP markers developed in this study can be applied in subsequent genetic research.
Keyword: Pyropia yezoensis; organelle single nucleotide polymorphism (SNP) markers; chloroplast;mitochondrial; organelle inheritance; maternal inheritance
1 INTRODUCTION
The commercially important red algal genus,Pyropia/Porphyra(Bangiales, Rhodophyta),popularly known as nori and laver, has a high nutritional value, including up to 25%-30% protein by dry weight, vitamins (in particular, B12), and oligosaccharides (Sutherland et al., 2011; Wu et al.,2017). Due to its high nutritional and economic value,nori is widely cultivated in countries such as South Korea, Japan, and China (Sahoo et al., 2002; Blouin et al., 2011). In recent years, crossbreeding and mutation breeding have been commonly applied in nori germplasm screening, which increase the source of genetic variation. Because of the large number of organelles in each cell, they play an important role in the growth and development of algae. Organelle heredity is an important component of algae genetic research. Moreover, new molecular markers that discriminate closely related species and unreported new genetic features can also be isolated from the organelle genome (Hwang et al., 2013). Up to May 2020, 212 records of Rhodophyta organelle genomes had been submitted to the Organelle Genome Resources of GenBank (NCBI; National Center for Biotechnology Information), including 10 species of the generaPorphyraandPyropia. Understanding the organelle inheritance of nori is of great significance for interspecific or intraspecific hybridization. An understanding of how chloroplast and mitochondrial DNA is transferred between parents and off spring enables their use as key molecular markers in phylogenetic and population genetics studies (Coyer et al., 2002).
In most animals, higher plants, and algae, organelle genomes (chloroplast and mitochondrial DNAs) are usually inherited uniparentally (Bendich, 2013). The eukaryotic genome is distributed among diff erent genetic compartments, which follow contrasting modes ofinheritance (Birky, 2001). Two genetic mechanisms exist for eukaryotic genomes: Mendelian inheritance patterns for nuclear genes and non-Mendelian inheritance patterns for DNA-containing cell organelles. Non-Mendelian inheritance of organelles in diff erent species is diverse (Rebound and Zeyl, 1994), and is mainly divided into three categories: paternal, maternal, and biparental inheritance. The predominant pattern is uniparental and usually maternal (Wilson and Vaughn, 1979;Zhang et al., 2003; Johannessen et al., 2005; Zhong et al., 2011; Greiner et al., 2015). In plants, mitochondrial inheritance is usually maternal, whereas chloroplast inheritance can be maternal (most common),biparental, or paternal (Avise, 1994; Morgensen,1996; Isoda et al., 2000; Coyer et al., 2002).Furthermore, chloroplast and mitochondrial DNA are non-recombinant: variation in DNA bases is caused by mutations, which can be used to trace species evolution and population dynamics (Avise et al.,1987; Moritz et al., 1987).
Research on organelle inheritance patterns in red algae is crucial for population evolution, because ofits unique evolutionary status. Compared to higher plants, studies on organelle inheritance patterns in algae are somewhat lacking, and the few examples reported so far for green and brown algae argue against the regular co-transmission of plastids and mitochondria in isogamous algal species (Miyamura,2010; Motomura et al., 2010). Peters et al. (2004)analyzed organelle inheritance by PCR-sequencing and speculated that the biparental chloroplast inheritance observed inEctocarpussiliculosusmight be the rule in isogamous brown algae. Additionally, inScytosiphonlomentaria(Phaeophyceae), the biparental inheritance of chloroplasts and maternal inheritance of mitochondria have also been reported in isogamous brown algae (Kato et al., 2006). In Ectocarpales species, the mitochondria are usually inherited maternally, both in isogamous and oogamous species and the plastids of oogamous species are also maternally inherited; however, the plastids ofisogamous species are inherited biparentally and distinctive structural genomic rearrangements have been identified (Choi et al., 2020). InSaccharina japonica(Laminariales, Phaeophyta), the maternal inheritance of organellar DNA has been demonstrated using simple sequence repeats (SSR) markers (Li et al., 2016). Earlier studies on the behavior of organelles have been performed in red algae, mainly by microscopic observations of spermatogenesis and fertilization (Hawkes, 1978). Zuccarello et al. (1999a,b) analyzed crosses between diff erent mitochondrial haplotypes ofBostrychiamoritzianaand between isolates ofBostrychiaradicansbased on PCRsequencing and single-stranded conformation polymorphism (SSCP). The results revealed that the mitochondria and plastids are inherited maternally in this species. Furthermore, that was the first study to conclusively demonstrate the maternal inheritance of organelles in marine red algae (Zuccarello and West,2011). Choi et al. (2008) used diff erent genes as molecular markers to distinguish chloroplast and mitochondrial DNA in the cross between diff erent strains ofPorphyrayezoensisbased on cleaved amplified polymorphic sequence (CAPS) analysis.The results indicated that organelle DNA was inherited uniparentally from the maternal parent.Niwa et al. (2010) performed interspecific hybridization in the genusPorphyraand used PCRRFLP analysis to show that organelle genomes were maternally inherited.
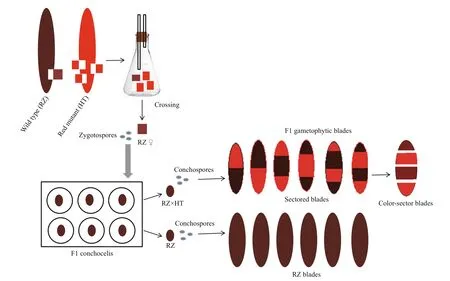
Fig.1 Schematic diagram showing the cross between the maternal strain (RZ) and the paternal strain (HT) of P. yezoensis
In this study, the wild type RZ and the red mutant HT were selected as the maternal and paternal strain,respectively in a cross experiment. Red mutants have been used widely in the genetic research ofPyropiayezoensis(Yan and Aruga, 2000; Niwa, 2010; Niwa et al., 2011; Huang and Yan, 2019; Yu et al., 2020);however, information on their organelle genomes is lacking. Therefore, in this study, the complete mitochondrial and chloroplast genomes of the strain HT were assembled for the first time. Additionally,organelle molecular markers were developed based on alignment to the RZ organelle genomes. Following the cross between wild type RZ and the red mutant HT, color-sectors from F1 gametophytic blades were examined by PCR amplification and sequence analysis of the above organelle molecular markers to determine the pattern of organelle inheritance inP.yezoensis. The study provides a means and strategy for easily and accurately determining the cytoplasmic inheritance pattern ofPyropia. These organelle molecular markers can be applied for the crossbreeding of nori and subsequent genetic research.
2 MATERIAL AND METHOD
2.1 Cross experiment, material culture, and treatment
Two pure strains ofP.yezoensiswere used in our cross experiment: the wild type strain RZ was used as the female parent and the red mutant HT was used as the male parent. Crosses were performed according as described by Niwa et al. (2010). An example of the cross between the maternal strain RZ and the paternal strain HT is shown in Fig.1. Briefly, the ratio of the female parent to the male parent was 1:3, and the blades were cut from the respective blades just before formation of stripy patches with spermatangium and carpogonium. Then, the blade pieces were co-cultured for about 10 days in a 500-mL flask containing PES medium (UTEX Culture Collection of Algae) and agitated by aeration at 15 °C under 80 μmol/(m2·s)photons(10 h/14 h light/dark). After observing the formation of carposporangia in the maternal blade piece, it was transferred into a Petri dish and washed in sterilized seawater while being gently rubbed with a writing brush. To obtain carpospores from the maternal blade piece, the washed piece was chopped with a blade in 20-μL sterilized seawater. Then, a glass micropipette (inside diameter 0.5 mm) was used to suction a carpospore under the light microscope, and F1 conchocelis colonies were cultured in 96-well plates at 20 °C under 30 μmol/(m2·s) photons(12 h/12 h light/dark). Each colony was separated, and filaments were inoculated shells and cultured at 24 °C under 30 μmol/(m2·s) photons (10 h/14 h light/dark).After the formation of conchosporangia were confirmed, cotton monofilaments (approximately 5-cm long) were placed in the beakers to allow the attachment of conchospores, and sampled were stirred by aeration at 20 °C under 135 μmol/(m2·s) photons(12 h/12 h light/dark). After the attachment of the conchospores, the cotton monofilaments were cultured and agitated by ventilation alone at 20 °C under 135 μmol/(m2·s) photons (12 h/12 h light/dark). After two weeks, the beakers were moved to 10 °C under 160 μmol/(m2·s) photons (12 h/12 h light/dark). F1 gametophytic blades were cultured and stirred by aeration at 10 °C under 160 μmol/(m2·s) photons(12 h/12 h light/dark) in flasks containing PES medium. The culture medium was renewed every three days.

Fig.2 Color sectors in F1 gametophytic blades were separated according to the color boundary
Among the F1 obtained gametophytic blades, only the chimeras of color-sectors with two-four colorsectors (Fig.1) showed that the crossing was successful. Consequently, the F1 gametophytic blades with two-four color-sectors were selected as the experimental materials (Fig.2), and each color-sector blade was separated according to the color boundary.DNA was extracted from the parents and the colorsectors using a Plant Genomic DNA kit (Tiangen,China) following the manufacturer’s instructions.DNA was quantified with a Nanophotometer N60(Implen, Germany), and 3-μL DNA was loaded on 1.0% agarose gel to evaluate the quality of DNA.
2.2 Sequencing and assembly of the red mutant(HT) organelle genomes
Following detection, DNA samples were used to construct the Illumina library using the VAHTS Universal DNA Library Prep Kit for Illumina®V3 ND607 (Vazyme Biotech Co., Ltd.; Nanjing, China),according to the manufacturer’s protocol. The eligible libraries were diluted for pair-end sequencing on the Illumina Hiseq™ Xten sequencing platform (Illumina,Inc.; San Diego, CA, USA) at Novogene Bioinformatics Technology Co. Ltd in Tianjin, China. Before assembly,raw reads were trimmed for quality (q>20) and length(n>20) using Trim Galore v 0.5.0 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/),and adapters were removed using Cutadapt (Martin,2011), as run from within Trim Galore. Using the organelle genomes of RZ as the reference, sequencing data for the red mutant were mapped to the reference by Bowtie2 v2.2.1 (Langmead et al., 2009), and all reads that could be mapped to the reference were assembled using SPAdes v3.6.2 (Bankevich et al., 2012) software to obtain contigs. According to the reference data,MUMmer3 (Kurtz et al., 2004) software was used to select contigs and adjust their relative position and direction in the genome in order to obtain a draft genome. Next, the sequencing data of the red mutant HT were mapped to the draft genome for initial gap filling. Finally, the gaps were filled and base correction was performed by Pilon v1.22 (Walker et al., 2014).
2.3 SNP analysis of organelle genomes
The assembled de novo chloroplast genome sequence of HT was then used as a reference to map the clean reads of RZ with Bowtie2 v2.2.6. Similarly,the assembled de novo mitochondria genome sequence of HT was then used as a reference to map the clean reads of RZ. The mapping results were visualized using the Tablet software. After alignment,SNP calling was performed using the SAMtools program (Li et al., 2009) with a Bayesian algorithm.The SNP loci were selected for PCR amplification,and the reliability of the SNP loci was validated by sequencing the PCR products of HT and RZ blades.
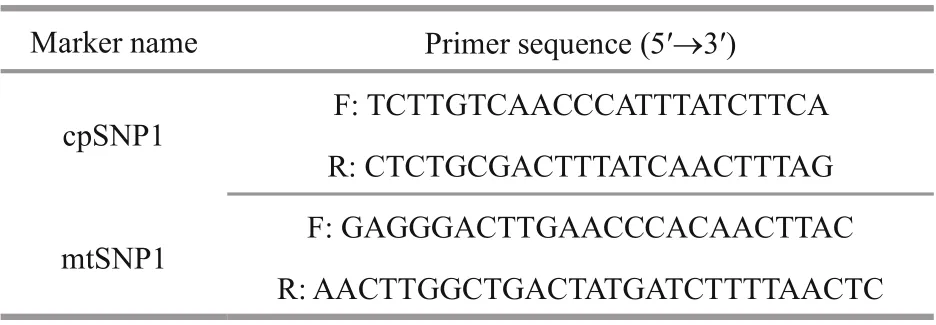
Table 1 Primer sequence of markers used in PCR
SNP primers were designed using the program Primer Premier 5.0 and synthesized by TsingKe Biological Technology (Qingdao) Co., Ltd. (Qingdao,China). PCR for SNP genotyping was performed in a 20-μL volume containing 2 μL of template DNA,6-μL sterilized ddH2O, 10-μL Mix (TaKaRa, China),and 1 μL of each primer pair. Amplification reactions were performed in a thermal cycler A200 (LongGene Scientific Instruments Co., Ltd., Hangzhou, China),with initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 40 s,annealing at 54 °C for 40 s and elongation at 72 °C for 1 min, with a final incubation at 72 °C for 10 min.For analysis of cpSNPs and mtSNPs, PCR products were separated on 1.0% agarose gels and sequenced in both directions (TsingKe, China). Sequence alignment was analyzed using ClustalW embedded in MEGA 7.021.
2.4 SNP markers developed for determining organelle inheritance
By mapping the organelle genomes between RZ and HT, one reliable and variable SNP locus was separately identified and selected in the chloroplast and mitochondrial genome ofP.yezoensis. A 500-bp sequence was identified for the PCR amplification,which contained these two stable SNP loci in the chloroplast and mitochondria, respectively. Flanking primers for the two regions were designed (Table 1).Polymorphism in these two regions for color sectors of F1 gametophytic blades was verified by PCR amplification and sequence analysis, as described above.
3 RESULT AND DISCUSSION
3.1 Cross experiment and the DNA extraction of color-sectors

Fig.3 Results of 1.0% agarose gel electrophoresis for two parents and part of color-sectors
The red algaP.yezoensisis considered a model organism of the intertidal zone because ofits special genetic characteristics (Yan and Aruga, 2000). In our study, the pure strains RZ and HT were crossed successfully. A number of pure conchocelis colonies were obtained, some of which were crossbred and some of which were self-crossbred. The conchocelis colonies were fragmented to inoculate the shells, and after formation of the conchosporangia, the conchospores were released. The ordered tetrad derived from the conchospore continued to divide and develop into two-four pigment-containing chimeras.The generation of pigment chimeras indicated that the intraspecific cross was implemented successfully, and the color sector embedded in the F1 heterozygous blade is a suitable marker to distinguish the types of the ordered tetrads. In genetic analyses of the ordered tetrads, color mutants were frequently selected as cross-experimental material. This can help to determine successful hybridization and identify hybrid off spring with diff erent genotype (Yan and Aruga, 2000; Huang and Yan, 2019; Yu et al., 2020).
Seven types of blades, including one unsectored blade and six sectored blades with two-four sectors were observed in the F1 gametophytic blades. Only the two parental color types were observed in the off spring, including unsectored and sectored blades(Fig.1). The frequency of sectored F1 blades was higher than the frequency of unsectored F1 blades.Among the sectored blades, chimeras with two and three color-sectors accounted for the majority, while the number of four color-sectors was very low.Finally, 11 RZ(♀)× HT(♂) F1 gametophytic blades were selected for further analysis. By cutting each individual F1 gametophytic blade, as selected above,30 color-sectors were obtained overall. DNA was extracted from the blades of two parents and 30 colorsectors for subsequent analysis. The size and integrity of DNA were assessed by 1.0% agarose gel electrophoresis using lambda DNA as the standard(Fig.3, part of the results for presentation).
3.2 Sequencing and assembly of HT organelle genomes
In total, 24.24 Gb clean data were generated for the parents, 8.81 Gb for the female parent (RZ), and 15.43 Gb for the male parent (HT). The average sequencing depth was 75.89× in RZ and 144.21× in HT. The complete chloroplast genome size of HT was 191 977 bp and had 33.09% GC content (Fig.4a),including 209 predicted genes (Supplementary Table S1). The complete mitochondrial genome of HT contained 74 predicted genes (Supplementary Table S2), was 41 692 bp in length and had 32.72% GC content (Fig.4b). Whole sequencing data from RZ and HT ofP.yezoensiswere deposited in NCBI under the BioProject PRJNA606921. Final assembly of the HT chloroplast genome was deposited in GenBank under the accession No. MT876197, and the HT mitochondrial genome was deposited in GenBank under the accession No.MT832023.
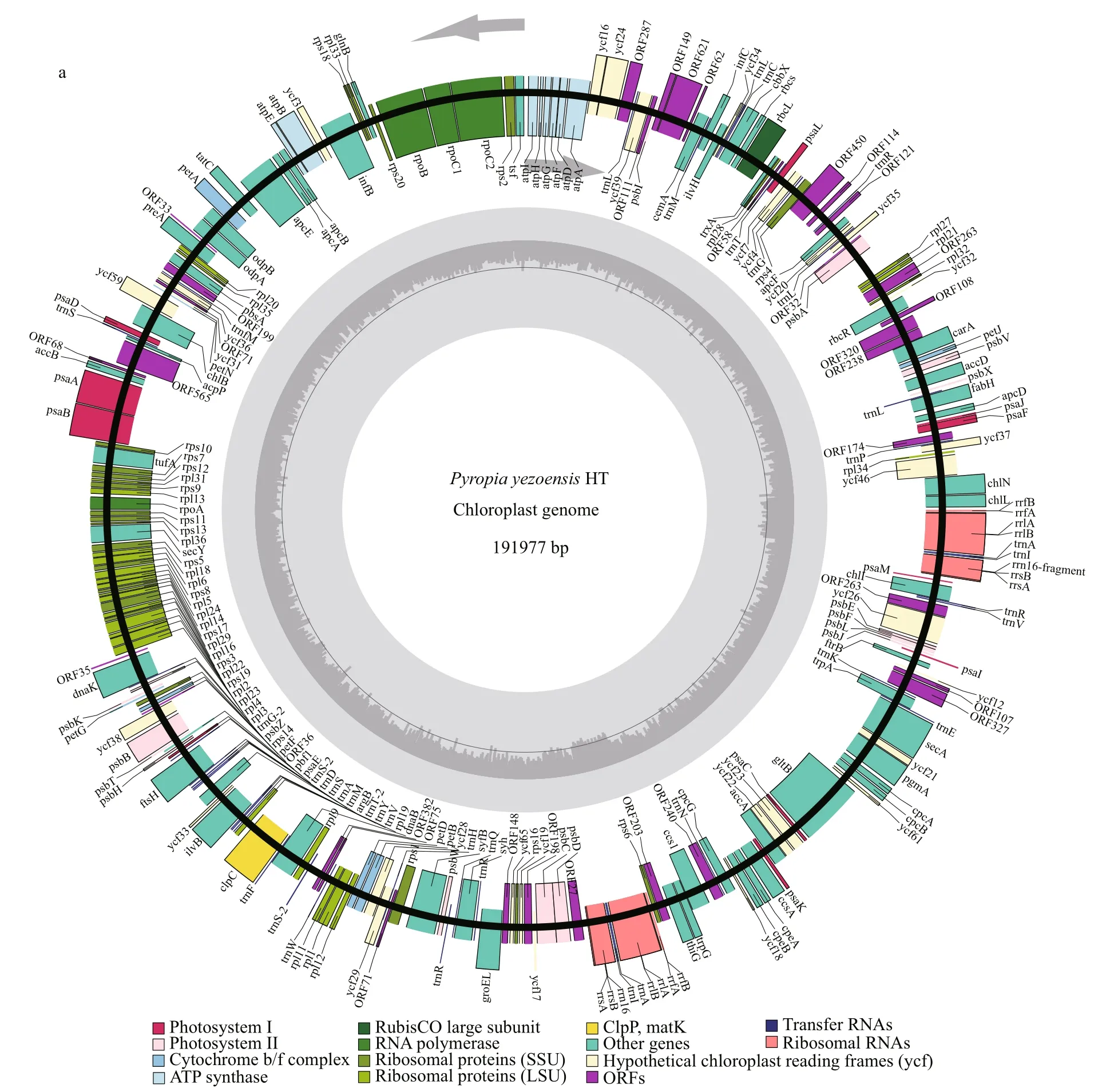
Fig.4 Genome maps of the HT chloroplast (a) and HT mitochondrion (b)
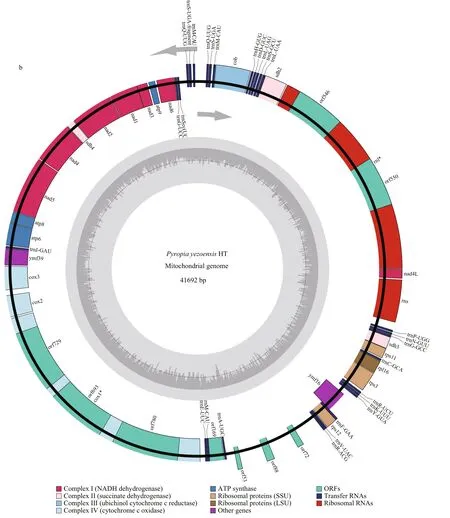
Fig.4 Continued
The plastid (KC517072) and mitochondrial(JQ736809) genomes of RZ have been reported and published (Wang et al., 2013; Kong et al., 2014). By now, the organelle genomes of 10 species in the generaPorphyraandPyropiahave been released in NCBI database. There are eight chloroplast genomes includingPorphyrapurpurea(NC_000925,191 028 bp),Porphyraumbilicalis(NC_035573,190 173 bp),Pyropiahaitanensis(KC464603,195 597 bp),PyropiayezoensisRZ strain (KC517072,191 975 bp),Pyropiafucicola(KJ776837, 187 282 bp),Pyropiakanakaensis(KJ776836, 189 931 bp),Pyropiaperforata(NC_024050, 189 789 bp) andPyropiapulchra(KT266789, 194 175 bp) and nine mitochondrial genomes includingPorphyrapurpurea(NC_002007, 36 753 bp),Porphyraumbilicalis(NC_018544, 29 123 bp),Pyropiahaitanensis(NC_017751, 37 023 bp),PyropiayezoensisRZ strain(JQ736809, 41 688 bp),Pyropiafucicola(NC_024288,35 035 bp),Pyropiakanakaensis(NC_024289,39 300 bp),Pyropiaperforata(KJ708761, 40 042 bp),Pyropianitida(KP890080, 35 313 bp) andPyropiatenera(NC_021475, 42 268 bp). Compared with the organelle genomes mentioned above, the mitochondrial genome size ofPyropiayezoensisHT strain is second only toP.tenerain length, and the chloroplast genome of HT is third toP.haitanensisandP.pulchrain length. The diff erence in length between organelle genomes in the RZ strain and those in the HT stain in this study may result from the use of diff erent sequencing strategies.
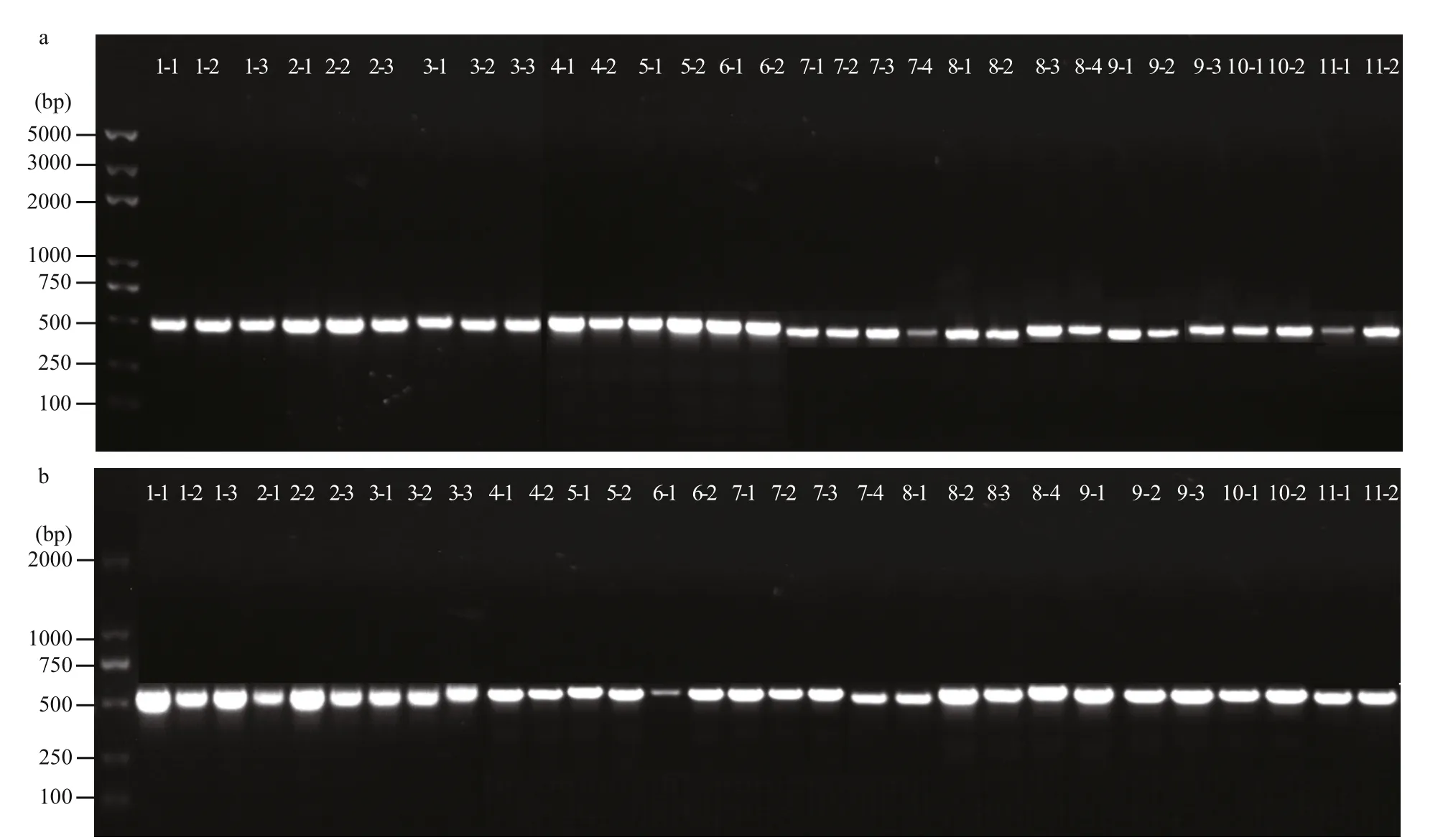
Fig.5 Amplified fragment of PCR products for 30 color-sectors in the chloroplast (a) and mitochondrion (b)
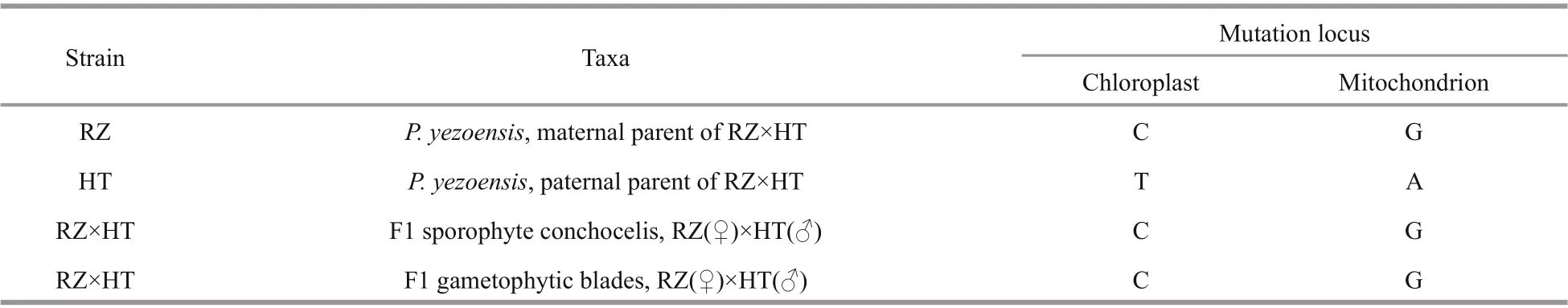
Table 2 Information mutation locus for gametophytes and sporophytes
3.3 Organelle SNPs markers developed in P. yezoensis
By mapping the organelle genomes of RZ and HT,two organelle SNP loci were identified. The SNP loci in the chloroplast genome was located in the 92 270 nt position, where there was a stabile T (HT) to C (RZ)mutation. The SNP loci in the mitochondrial genome were located in the 31 829 nt position, and there was a stabilized mutation of A (HT) to G (RZ). Since the chloroplast and mitochondrial genomes are annular,abnormal alignment of reads can be observed in the sequences of head and tail ends. Thus, indels of the head and tail ends were considered to be false positive loci. In addition to false positive loci, there may also be sequencing errors, heritable variation in algae cells, and other factors. To confirm the reality and stability of the two organelle SNP loci, DNA was extracted from the parent blades, the F1 conchocelis,and color-sectors of F1 gametophytic blades for PCR amplification. Sequences of PCR products showed that the polymorphism of those loci was stable(Table 2) and such SNP loci can be used as an organelle maker in subsequent genetic research. In this study, we identified few diff erences in the organelle genomes between the red mutant HT and the wild type RZ; therefore, this trait of the HT strain must be controlled by nuclear gene (Yu et al., 2020).
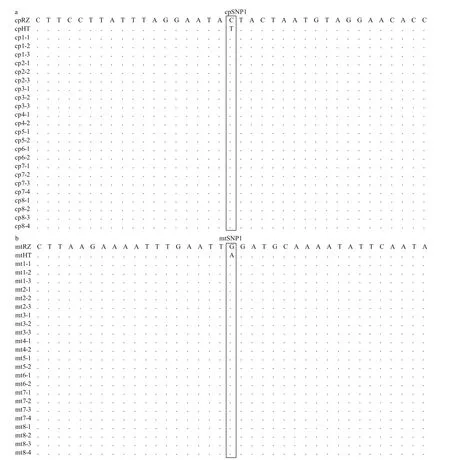
Fig.6 Alignments of sequences amplified from the parent blades and color-sectors of F1 gametophytic blades in chloroplast(a) and mitochondrion (b)
3.4 Chloroplast and mitochondrial DNA inheritance in P. yezoensis
As shown on 1.0% agarose gels, the length of amplified fragments of PCR products were about 500 bp, and these amplified fragments have a high degree of consistency (Fig.5a & b). Alignment analysis showed that all haplotypes of the 30 colorsectors from the 11 F1 gametophytic blades were consistent with the haplotypes of the maternal plant,demonstrating maternal inheritance for both chloroplast and mitochondrial genomes inP.yezoensis(Fig.6a & b). Additionally, in chloroplast, the mutation locus is T (HT) to C (RZ) inerrably, simultaneously in mitochondrion the mutation locus is A (HT) to G (RZ)correctly (Table 2).
Red algae are relatively old eukaryotic algae, most of which are multicellular, and a few of which are monocyte. During sexual reproduction, sperm and spores are produced, and its sexual reproduction is heterogamy (or oogamy). Molecular makers are important in the cytoplasmic inheritance of organelles in red algae, and include the mitochondrialcox2-3spacer,rps11-rnsspacer, andCOI, chloroplastcarA,and RuBisCo spacer (Zuccarello et al., 1999a, b; Choi et al., 2008; Niwa et al., 2010). In this study, organelle SNP makers were developed by mapping the chloroplast and mitochondrial genomes from diff erent strains ofP.yezoensis, and were used successfully to analyze the organelle inheritance patterns and polymorphism in this species. We demonstrated that the organelle genomes inP.yezoensiswere inherited maternally. The methods used in the present study provide an idea and basis for investigating the organelle genetic pattern of other genera of nori.
4 CONCLUSION
In conclusion, SNP markers for the chloroplast and mitochondrial genomes were developed by the assembly of the HT organelle genomes in the present study, and were found to be useful for analyzing the inheritance of organelles inP.yezoensis. Furthermore,the haplotypes of progeny gametophytes were consistent with those of the maternal parents,demonstrating maternal inheritance. Thus, the organelle SNP markers developed in this study has the potential to be applied in germplasm identification and in further genetic studies ofP.yezoensis.
5 DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
 Journal of Oceanology and Limnology2021年4期
Journal of Oceanology and Limnology2021年4期
- Journal of Oceanology and Limnology的其它文章
- Numerical study of the seasonal salinity budget of the upper ocean in the Bay of Bengal in 2014*
- Study on evaluation standard of uncertainty of design wave height calculation model*
- A fast, edge-preserving, distance-regularized model with bilateral filtering for oil spill segmentation of SAR images*
- A Gaussian process regression-based sea surface temperature interpolation algorithm*
- Climatology and seasonal variability of satellite-derived chlorophyll a around the Shandong Peninsula*
- Sources of sediment in tidal flats off Zhejiang coast, southeast China*
