MULTI-FLORET SPIKELET 4 (MFS4) Regulates Spikelet Development and Grain Size in Rice
Wang Yan, Zeng Xiaoqin, Lu Lu, Cheng Qinglan, Yang Fayu, Huang Mingjiang, Xiong Mao, Li Yunfeng
Research Paper
() Regulates Spikelet Development and Grain Size in Rice
Wang Yan#, Zeng Xiaoqin#, Lu Lu, Cheng Qinglan, Yang Fayu, Huang Mingjiang, Xiong Mao, Li Yunfeng
()
In rice, the spikelet is the basic unit of inflorescence, and its development is important for determining the grain yield and quality. We reported a rice spikelet mutant() which resulted in the production of extra floral organs or a whole extra floret, and elongated sterile lemmas. The results suggested that the mutation of thegene interfered with spikelet meristem determinacy and floral organ identity. In addition, the plant height and the grain length and width in themutant were all less than those in the wild type. Using the bulked segregant analysis method, thegene was localized in a 557-kb region on the long arm of chromosome 1. Sequence analysis showed that there was a C-base deletion at the open reading frame of. Further tests indicated that a wild type copy ofwas able to reverse thedefects, which indicated thatwas thegene. Thegene encodes a lipase located in the mitochondria and isexpressed strongly in the young inflorescence. qRT-PCR results showed that the expression of some genes that were known to regulate spikelet meristem determinacy and grain size were decreased in themutant, which indicated that thegene regulates spikelet meristem determinacy and grain size by modulating the expression of these genes.
rice; sterile lemma; multi-floret spikelet; meristem; grain size
Rice (L.) is one of the world’s most important food crops. Its yield is determined by three key factors: grain number per panicle, seed-setting rate and 1000-grain weight. To optimize these parameters, it is crucial to understand the genetic and molecular mechanisms of the development of the panicle, spikelet and grain. It is well known that the number and length of the branches determine the grain number per panicle to a large degree. Many genes, such as(),/(),() and(), are involved in the regulation of initiation and growth of the branch meristem and affect the number of spikelets per grain, and have been identified as yield-related factors (Ashikari et al, 2005; Xue et al, 2008; Huang et al, 2009; Zhang et al, 2017). The spikelet is a basic and unique inflorescence structure in grass, which consists of a pair of bracts and 1‒40 florets per grain (Malcomber et al, 2006). However, few researchers have considered if and how the number of florets per grain within the spikelet affects the grain number per panicle or the yield.
As with the inflorescence, the spikelet can be classified as determinate or indeterminate according to whether the apical meristem transforms into a terminal flower meristem (FM). In determinate spikelet species such as rice and maize, the spikelet meristem (SM) is terminated by the formation of the terminal FM after developing a fixed number of lateral FMs. In contrast, in indeterminate spikelet species such as wheat, there are no terminal FMs, thus the SM cannot be terminated and always maintains the ability to form an indeterminate number of lateral FMs (Malcomber et al, 2006). Several genes that encode the AP2/ERF domain have been reported to regulate SM fate determinacy, for example,() in maize (Chuck et al, 2008), and() andin rice (Lee et al, 2007; Lee and An, 2012; Ren et al, 2013). Themutant of maize produces an extra floret within the spikelet, which suggests that thegene is involved in maintaining SM determinacy (Chuck et al, 2008). Bothandare orthologs of thegene in rice. A mutation ofresults in the production of extra rudimentary glumes or florets (Lee et al, 2007). Although there are no obvious defects insingle mutants, and minimal defects insingle mutants, thedouble mutant shows a more severe phenotype than the single mutants (Lee and An, 2012). In addition, the() gene, another AP2/ERF transcription factor, can also promote SM-to-FM transformation by activating theandgenes in rice (Ren et al, 2013). These findings suggest that these-like AP2/ERF factors may initiate SM-to-FM transformation before terminating the SM. These-like genes are targets of(). The suppression of their expression byis necessary for SM determinacy(Zhu et al, 2009; Lee and An, 2012). In addition,()/(),()/() and()/() encode lipase, AT-hook protein and MYB protein, respectively, as well as regulating spikelet determinacy in rice. Loss of function of these genes results in the formation of two florets or extra glumes in spikelets, which indicates that the mutation of these genes may enable the transformation of the meristems from determinate to indeterminate (Li et al, 2009; Jin et al, 2011; Ren et al, 2018b, 2020; Zheng et al, 2019; Li et al, 2020).
In the present study, we identified a rice-() gene, which encodes a lipase and is located in the mitochondria. Themutant generated extra floral organs or a whole extra floret, which suggested delayed SM-to-FM transformation. In addition, decreased grain length and width in themutant indicated thatalso regulated the development of the grain size. Our results showed that thegene regulated the determinacy of SM by affecting the expression of-like genes, and regulated grain size by modulating the expression of related genes.
RESULTS
Phenotypes of mfs4 spikelets
The wild type (WT) spikelet was composed of a pair of rudimentary glumes, a pair of sterile lemmas and a fertile terminal floret. The WT floret was composed of four whorls of floral organs from the outside to the inside. The lemma and the palea (whorl 1) were closely coupled, two lodicules (whorl 2) were located on the lemma side, and six stamens (whorl 3) surrounded a pistil (whorl 4) with two stigmas (Fig. 1-A1 and -A2). The lemma and the palea, collectively termed the hulls, hooked together, and enclosed the inner floral organs. The sterile lemma and rudimentary glumes were formed in a 1/2 alternate arrangement with the hulls (Fig. 1-A3).The sterile lemma had a smooth abaxial surface with a few trichomes (Fig. 1-A4). The palea can be considered a combinational organ consisting of two parts: the body of the palea (bop) and the marginal region of palea (mrp). The bop was anatomically similar to the lemma, and the four cell layers (silicified abaxial epidermis, fibrous sclerenchyma, spongy parenchymatous cells and non- silicified adaxial epidermis) were discernible. The mrp consisted of a smooth non-silicified epidermis, a large amount of spongy parenchymatous tissues and a small number of sclerenchyma cells (Fig. 1-A5 to -A7; Fig. 2-A and -B). The lemma consisted of a silicified abaxial epidermis and bore trichomes and protrusions (Fig. 1-A6). The lemma contained five vascular bundles, the palea contained three vascular bundles and the sterile lemma contained one vascular bundle (Fig. 1-A7 and Fig. 2-C).
Most significantly, themutant spikelets produced extra floral organs or occasionally a whole additional floret. Three types of phenotypes with indeterminate spikelets were found in themutant from the statistical analysis. In the Type I spikelets (62% ofspikelets), an additional lemma-like organ and a whole fertile terminal floret were formed on the spikelet rachilla (Fig. 1-B1 to -B6, -F and Fig. 2-E). In the Type II mutant spikelets (11% ofspikelets), instead of the terminal floret, two incomplete florets were formed. In one Type II mutant spikelet, in addition to rudimentary glumes and sterile lemmas, the spikelet consisted of two lemma-like organs, two degraded paleas (each one consisted of one residual bop-like structure and two normal mrps, or two independent mrp-like structures without a bop), two pairs of lodicules, 9‒12 stamens and 1‒2 pistils (Fig. 1-C1 to -D7, -F and Fig. 2-D and -G). In the Type III mutant spikelets (2% ofspikelets), two almost complete and independent florets developed, although the identity and the number of stamens in some lower florets showed differences from the WT (Fig. 1-E1 to -E5 and -F).

Fig. 1. Phenotypes of spikelet in wild type (WT) andmutants.
A, Spikelet of the WT. A1, WT spikelet; A2, The lemma and palea were removed in A1; A3 to A6, The WT spikelet surface characters of sl, pa and le; A7, Transverse section of WT spikelet.
B‒E, Spikelets of themutants. B1 and B2, The Type Imutant spikelet; B3‒B5, The Type Imutant spikelet surface characters of spikelet, le and ele; B6, Transverse section of the Type Imutant. C1 and C2, The Type IImutant spikelet; C2, The lemma and extra lemma were removed; C3‒C6, The Type IImutant spikelet surface characters of sl, pa, le and ele; C7, Transverse section of the Type IImutant. D1 and D2, The Type IImutant spikelet; D2, The lemma, palea and elongate sterile lemma were removed; D3‒D6, The Type IImutant spikelet surface characters of sl, pa, le and mrp; D7, Transverse section of the Type IImutant. E1, The Type IIImutant spikelet; E2‒E4, The Type IIImutant spikelet surface characters of sl, pa and le; E5, Transverse section of the Type IIImutant.
F, Percentage of mutant organs inspikelets.
bop, Body of palea; ele, Extra lemma; le, Lemma; lo, Lodicule; mrp, Marginal region of palea; lsl, Lemma-like sterile lemma; ov, Ovule; pa, Palea; pi, Pistil; rbs, Residual bop-like structure; rg, Rudimentary glume; sl, Sterile lemma; st, Stamen.
Scale bars are 1 000 μm in A1, A2, B1, B2, C1, C2, D1, D2 and E1; Scale bars are 500 μm in A3‒A6, B3‒B5, C3‒C6, D3‒D6 and E2‒E4; Scale bars are 100 μm in A7, B6, C7, D7 and E5.
Fig. 2. Histological and qRT-PCR analysis of lateral organs in spikelets of wild type (WT) andmutants.
A‒C, Transverse sections of lateral organs in the WT spikelet. A and B, The lemma and palea of WT. C, The sterile lemma of WT. Red box indicates the vascular bundle. Scale bars are 500 μm.
D‒H, Transverse sections of lateral organs in themutant spikelets. D‒G, The lemma, palea and extra lemma of themutant spikelets. H, The elongate sterile lemma of themutant spikelets. Red boxes indicate the vascular bundles. Scale bars are 500 μm.
I‒P, qRT-PCR analysis of,,,,andexpression.was used as a control. RNA was isolated from the flower organs of the WT andmutant spikelets. Data are Mean ± SE (= 3). *,≤ 0.05 and **,≤ 0.01 by the Student’s-test.
bop, Body of palea; ele, Extra lemma; le, Lemma; lo, Lodicule; lsl, Lemma- like sterile lemma; mrp, Marginal region of palea; Pa, Palea; bs, Residual bop-like structure; sl, Sterile lemma.
To further clarify the identities of the extra floral organs in themutant, the expression of several genes for the lemma and/or the palea identity (for the lemma and the palea,for the palea andfor the lemma; Jeon et al, 2000; Nagasawa et al, 2003; Li et al, 2010) was investigated. In WT, whilewas expressed in both the lemmas and the paleas,was highly expressed in the paleas and lowly in the lemmas. Conversely,was expressed highly in the lemmas and weakly in the paleas. In the extra lemma-like organs of thespikelets, the expression levels of these genes were similar to those in the WT lemmas and the upper floret-lemmas of thespikelets (Fig. 2-I to -K). This indicated that these additional lemma-like organs were just lemmas. In the degraded paleas of themutant spikelets, the transcription levels ofandwere similar to those in the WT paleas, whereas the transcriptionlevel ofwas down- regulated significantly in themutants (Fig. 2-I to -K). This result suggested that these palea-like organs were indeed paleas and consistent with the phenotype of themutants.
The sterile lemma was elongated or transformed into a lemma-like organ in somemutantspikelets. The WT sterile lemma had a relatively smooth upper epidermis bearing a few trichomes (Fig. 1-A4). However, the lemma-like organ was similar to the sterile lemma in size and shape. Scanning electron microscope (SEM) images showed that the upper epidermis of the elongatedmutant sterile lemma developed many trichomes and silicified cells similar to those in the upper epidermis of the WT lemma (Fig. 1-D3). Histological analysis also showed that the WT sterile lemma only contained one vascular bundle (Fig. 2-C), but themutant sterile lemma contained three vascular bundles (Fig. 2-H). We then detected the expression levels of(a sterile lemma identity gene), and,,and(four lemma/palea identity genes) in themutant elongated sterile lemmas (Nagasawa et al, 2003;Arora et al, 2007; Yoshida et al, 2009; Wang et al, 2010). The qRT-PCR results indicated thatthree lemma/palea identity genes (,and) were up-regulated significantly in the elongated sterile lemmas of themutants, while the sterile lemma identity genewas down- regulated sharply (Fig. 2-L to -O). These results indicated that the elongated sterile lemmas in themutant spikelets had gained lemma-like identities to a large extent.
Early morphological analysis of mfs4 mutant
The early development of rice spikelets can be divided into stages Sp1‒Sp8 (Ikeda et al, 2004). SEM was used to observe the difference between the WT andmutantspikelets at the early stage. In the WT spikelets, the palea primordia were initiated at Sp4 (Fig. 3-A). Then, two pairs of lodicule primordia formed at Sp5 were covered by the lemma and palea. At Sp6, six stamen primordia formed, and a carpel primordium formed at Sp6‒Sp7. During the Sp8 stage, the primordia of lemma palea were hooked together and enclosed the inner floral organs (Fig. 3-B to -D). In themutant, the development of floral organ primordia differed from the WT. From the Sp5 stage, extra lemma primordia between the lemma and the sterile lemma primordia were observed and the extra lemma primordia progressed in a manner/sharp size similar to that of the lemma primordia (Fig. 3-E to -H). These results showed that the defects in themutant spikelets were occurred during the early developmental stages.
Phenotypic analysis of mfs4 mutant
In addition to the spikelet defects, we noted that themutants showed some changes in the plant and grain. Firstly, the plant height of themutant was significantly shorter than that of the WT (Fig. 4-A and -G). In the WT plants, five nodes were elongated, while only four elongated nodes were observed in theplants (Fig. 4-B). Furthermore, the first three internodes and the panicle length of themutant were significantly shorter than those of the WT (Fig. 4-E and -F), which indicated thataffected stem development in rice. Secondly, the grains of themutants were significantly shorter and narrower than those of the WT, which resulted in a decrease in 1000-grain weight (Fig. 4-I to -K). Finally, there was significantly fewer grain number per panicle in themutants than that in the WT (Fig. 4-H). These results showed thataffected the number and size of the grains, and hence the yield in rice.

Fig. 3. Scanning electron micrographs of spikelets at early developmental stages in wild type (WT) andmutant.
A–D, WT. E–H,with ele. Asterisks indicate the stamens. Scale bars are 100 μm.
ele, Extra lemma; fm, Flower meristem; le, Lemma; pa, Palea; rg, Rudimentary glume; sl, Sterile lemma.
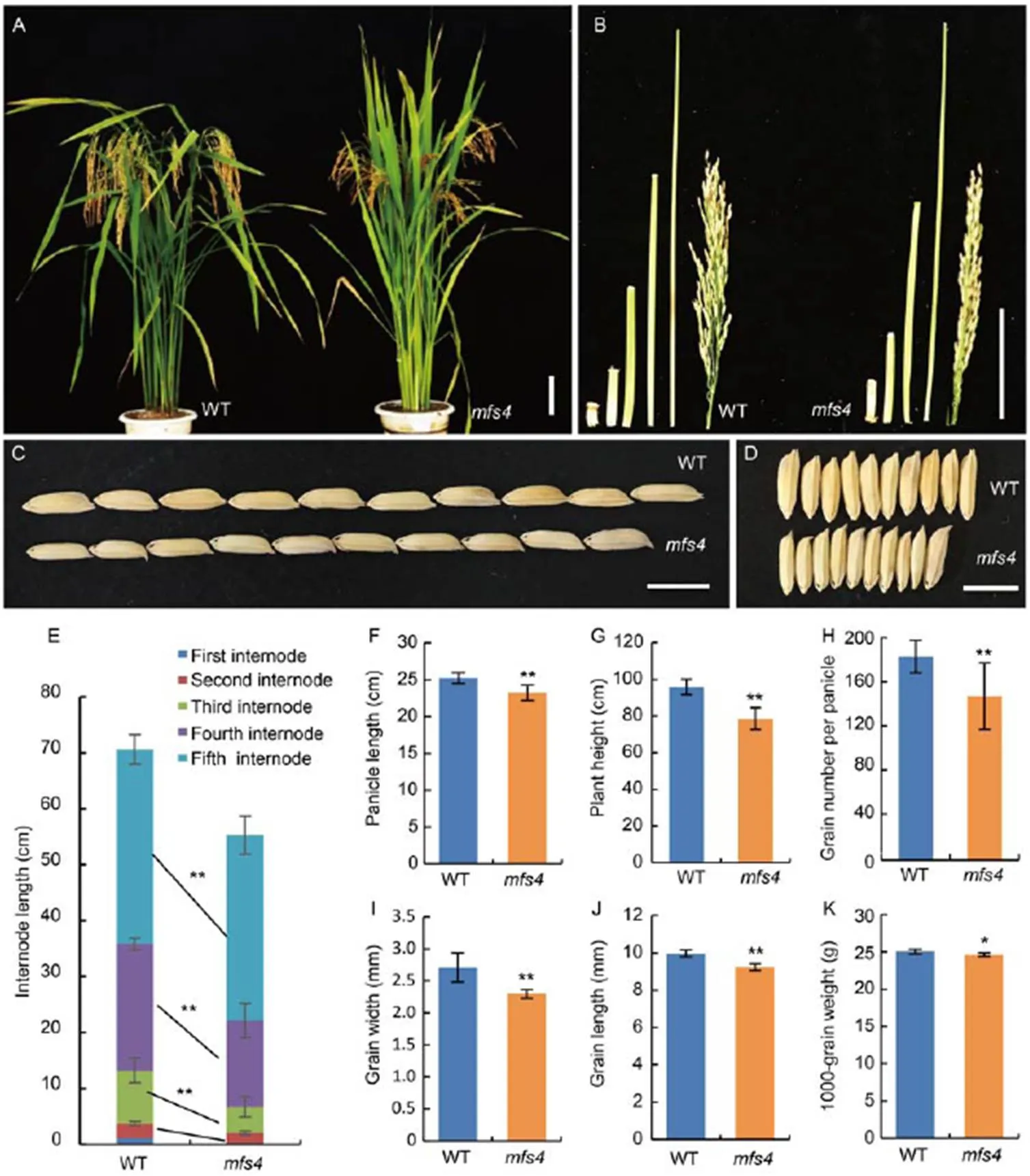
Fig. 4. Comparison of morphological characters between wild type (WT) andmutant.
A, Plant morphology of WT andmutant at the heading stage.
B, Internodes and panicles of WT andmutant.
C and D, Comparisons of grain length (C) and width (D) in the WT andmutant.
E‒K, Comparisons of internode length (E), panicle length (F), plant height (G), grain number per panicle (H), grain width (I), grain length (J) and 1000-grain weight (K) between the WT andmutant.
Scale bars are 10 cm in A and B, and1 cm in C and D. *,≤ 0.05 and **,≤ 0.01 by the Student’s-test.
Mapping-based clone of MFS4 gene
To isolate thegene, themutant was crossed with a sterile line 56S (). All F1plants manifested the WT phenotype. F2population resulted in 1 080 plants in total, among them, 831 plants were normal and 249 plants manifested themutant phenotype. The segregation ratio of WT to mutant plants was conformed to 3:1 (χ2= 3.11 < χ20.05= 3.84), which indicated that the multi-floret spikelet trait is controlled by a single recessive gene.
Recessive individuals in the F2population were used as a mapping population to localize thegene. Within the F2segregating population, 15 WT plants and 15 mutant plants were selected randomly to construct WT and mutant DNA pools, respectively. Linkage analysis showed that thegene was linked to the polymorphic SSR markers CHR1-WY-4, CHR1-TJ-4, CHR1-WY-5, CHR1-WY-6, CHR1-WY-7 and CHR1-WY-8 on chromosome 1. We used these markers to survey 100 mutant individuals, and thegene was further localized in the 557 kb region between CHR1-WY-5 and CHR1-TJ-4 on the long arm of chromosome 1. Through genome re-sequencing and general PCR sequence analysis, we found a C-base deletion in the open reading frame ofat the 1102th base. This deletion caused a frame shift and the premature termination of translation (Fig. 5-A)Next, we constructed a complementary vector, which contained 2000-bp upstream sequence and the full genomic sequence of, and then introduced the recombinant plasmid into themutant. In total, 15 positive transgenic lines were obtained. In eight of these, themutated traits were rescued by designedendogenous and exogenous primers via PCR verification (Fig. 5-B to -D). The results verified thatis thegene

Fig. 5. Fine mapping and sequencing analysis ofgene.
A,Gene mapping ofon rice chromosome 1. The red triangles indicate the target gene. The red line in the exon indicates the mutational site of candidate gene. WT, Wild type.
B, Schematic structure of the complementary vector pCAMBIA1300-MFS4-GFP andgenome. MFS4-COM-F/MFS4-COM-R are complementary vector primers, F1/R2 are primers for amplifying endogenous, and F1/R1 are primersfor amplifyingexogenous.
C, Defect inflowers were completely rescued by introduction of pCAMBIA1300-.-com,complementary flower. Scale bars are 1 mm.
D, Identification of transgenic plants. M, DNA marker; Lanes 1‒3, Detection of positive transgenic plants by F1/R2 primers; Lanes 4‒6, Detection of positive transgenic plants by F1/R1 primers.
Spatiotemporal expression pattern and subcellular localization of MFS4
To further understand the functions of thegene, we examined its spatial expression patternby qRT- PCR. In the vegetative tissues,had an obvious expression in the stem, while its expression levels were significantly lower in other vegetative tissues (root, leaf, sheath and bud) (Fig. 6-A). In the reproductive tissues,was expressed highly in young panicles and decreased gradually in inflorescences with different lengths. At the reproductive growth stage,was expressed mainly in the young panicle and spikelet organs, and especially highly in the sterile lemmas and paleas (Fig. 6-B). Further, we detected the expression pattern ofusinghybridization and found strong signals in the rice panicles (Fig. 6-C to -H). Thetranscripts were firstly detected in the branch meristem, spikelet meristem and floral meristem (Fig. 6-C). At the Sp3 stage when the primordia of the rudimentary glume and the sterile lemma had been initiated, thegene was highly expressed in these organs, but not in the spikelet meristem (Fig. 6-D and -E). After the Sp4 stage, when the primordia of the floral organs were initiated, a strongsignal was detected in the primordia of floral organs, including the lemma, palea, lodicule, stamen and floral meristem. Simultaneously,was still expressed obviously in the rudimentaryglume and sterile lemma (Fig. 6-F to -H). Theseresultsindicated thatwas mainly expressed in the spikelets and florets, consistent with its role in influencing spikelet meristem development and grain size.
To explore the subcellular localization of the MFS4 protein, we expressed an MFS4::GFP fusion protein in rice protoplasts. We also coexpressed a mitochondrial marker MTS::Cherry fusion protein in rice protoplasts (Zhang et al, 2016). While the GFP protein alone was uniformly expressed in the cytoplasm and nucleus (Fig. 7-A to -C), it was found that the MFS4::GFP protein was co-located with the mitochondrial marker MTS::Cherry protein (Fig. 7-D to -F). These results indicated that theprotein was predominantly located in the mitochondria.
MFS4 regulates spikelet meristem determinacy genes and grain-size-related genes
To confirm how SM development was affected in themutant, the expression levels of genes that regulate SM development, including,,,,,,/and/(Zhu et al, 2009; Huang et al, 2009;Zheng et al, 2019; Li et al, 2020; Ren et al, 2020) were detected by qRT-PCR. Compared with the WT, the expression levels of all these genes were clearly down-regulated in themutant (Fig. 8-A). This result indicated thatwas a key upstream regulator in SM determinacy by activating-like genes,/and/.

Fig. 6. Spatiotemporal expression pattern ofgene.
A and B, qRT-PCR of.was used as a control. RNA was isolated from young panicles < 0.5 cm, 0.5‒1.0 cm, 1.0‒2.0 cm, 2.0‒5.0 cm and ≥ 5.0 cm, as well as vegetative organ and floral organ of wild type plants. Data are Mean ± SD (= 3).
C‒H,hybridization expression in panicles of wild type (C and D), and spikelets at Sp2‒Sp3 (E), Sp4‒Sp5 (F), Sp5‒Sp6 (G) and Sp7‒Sp8 (H).
bm, Branch meristem; fm, Floral meristem; fo, Floral organ; le, Lemma; lo, Lodicule; pa, Palea; rg, Rudimentary glume; sl, Sterile lemma; sm, Spikelet meristem; st, Stamen.Scale bars are 500 μm in C and D, and 100 μm in E‒H.

Fig. 7. Subcellular localization of MFS4 protein.
A and D, GFP illuminant. B and E, mCherry illuminant which indicates the mitochondria marker. C and F, Merge illuminant.
Scale bars are 50 μm.
In themutant panicles, around 75% of the spikelets showed defects surrounding the meristem and organ identities, but the remaining 25% showed a ‘normal’ phenotype (Fig. 1-F). Wefurther investigated the grain traits of these ‘normal’ grains in themutants, and found that these ‘normal’grains were shorter and narrower than the WT grains(Fig. 4-I and -J). To determine howregulates grain size, we firstly investigated the expression of several genes that regulate both the cell cycle and cell expansion. Five cell cycle-related genes,,,,and,showedsignificantly lower expression levels in themutant than in the WT (Fig. 8-B).Theexpression ofwas slightly up-regulated in themutant compared with WT (Fig. 8-B). We also investigated the expression of several genes that regulate grain size by hull cell proliferation and expansion.,,,,,,,,,andinfluence cell expansion, and,,,,,,,,,,,,andare involved in regulating cell proliferation (Kitagawa et al, 2010; Heang and Sassa, 2012a; Hu et al, 2015; Ren et al, 2018a; Xiong et al, 2018; Dong et al, 2020; Guo et al, 2020). Compared with WT, the expression levels of,,,,,,,,,,,,,,,andwere reduced, whileandwere increased in themutant panicles. In addition, there were no obvious differences in the expression levels of,,,andbetween theand WT panicles (Fig. 8-C and -D).Together, these results supported thataffected grain size by regulating cell proliferation and expansion in the lemmas and paleas.

Fig. 8. Expression levels of related genes in wild type (WT) andmutant.
A, Expression analysis of related genes influencing spikelet meristem determination in young panicles.
B, Expression analysis of related genes influencing grain size by cell cycle-related genes in panicles.
C, Expression analysis of related genes influencing grain size by hull cell expansion-related genes in panicles.
D, Expression analysis of related genes influencing grain size by hull cell proliferation-related genes in panicles.
was used as a control. RNA was isolated from panicles of WT andmutant. At least three replicates were performed and the mean value was used. Error bars mean standard error. *,≤ 0.05 and **,≤ 0.01 by the Student’s-test.
DISCUSSION
MFS4 is a new allele of EG1/DF1
In this study, a spikelet-defectivemutant was identified. Map-based cloning and genome DNA sequencing showed thatis a new allele of/.was identified definitively as a deletion mutant allele at the/locus. The genetic background ofisrice ZF802, in which the abnormal phenotype includes the occurrence of glume-like organs, and in some extreme cases, alteration of the number of floral organs and losses of flower determinacy (Li et al, 2009). The genetic background ofisrice ZH11, in which only the empty glumes, lodicules and stamens are affected in the abnormal phenotype (Zhang et al, 2016).The genetic background ofisrice 9522, which manifests extra glume-like structures between the sterile lemma and the lemma, together with altered floral organ numbers and identities (Cai et al, 2014).,andare derived from Nipponbare CRISPR-Cas mutations (Li et al, 2009; Zhang et al, 2016). Themutant produces a ‘two-flower’ phenotype and shows increased yield (Ren et al, 2018b). However, themutant possesses the/allele for all phenotypes, as found in previous studies (Li et al, 2009; Cai et al, 2014; Ren et al, 2018). This suggests that thegene might manifest different phenotypes in different genetic background materials and mutation sites. More detailed analyses of the functional alteration of the novel mutant allelecan yield new insights into the function of thelocus in the development of spikelet determinacy in rice and other grasses.
MFS4 regulates spikelet meristem determinacy
SM determinacy is associated with the number of lateral organs and florets produced in the spikelet. In the WT rice, because one terminal FM is formed on the top of the SM, the determinate spikelet ultimately produces one pair of rudimentary glumes, one pair of sterile lemmas and one terminal floret. Our findings revealed that themutant spikelets produced an extra lemma and formed two florets instead of one terminal floret. Similarly, in the,/andmutants, some spikelets produce an additional lemma-like organ or two florets (Ren et al, 2013, 2020; Zheng et al, 2019; Li et al, 2020). Moreover, in themutant, some spikelets develop supernumerary rudimentary glumes, additional lemma/palea-like structures or extra florets.mutants manifest a more severe phenotype thanandsingle mutant (Lee et al, 2007; Lee and An, 2012). These findings suggested that, together with,,,and, affect the SM to FM transition, and their mutations induce SM indeterminacy.
Loss of SM determinacy in these mutants tends to produce ‘two-floret’ spikelets (Fig. 1-E) (Lee and An, 2012; Ren et al, 2013; Li et al, 2020), which suggested the possibility of a rice spikelet transforming into an indeterminate spikelet and producing two or more florets in one spikelet, similar to other indeterminate spikelet species such as wheat. If confirmed, the number of grains per panicle, and hence the rice yield, can increase significantly.
MFS4 regulates grain size
Grain size is determined by grain length, width, thickness and fullness. The development of the hulls (the lemma and the palea) is very important for grain length and width in rice and other grass crops. Many grain size regulating genes related to hull development have been identified and are widely used in rice breeding. For example,encodes a RING-type E3 ubiquitin ligase and negatively regulates cell division by anchoring its substrate to the proteasomes responsible for degradation. The loss of function ofactivates the division of the hull cells, thus increasing the hull width, grain width, grain weight and yield in rice (Song et al, 2007; Yan et al, 2011).regulates grain size and grain weight through a ubiquitin-proteasome pathway that is similar to. The loss of function ofactivates the division of the hull cells, thus increasing the grain size (Wan et al, 2008).encodes a DUF640 domain protein. In themutant, the number of hull cells decreases, leading to reduced grain length, grain thickness and grain weight (Li et al, 2012; Ma et al, 2013). Besides these genes,(Xia et al, 2018),(Li et al, 2011),(Su’udi et al, 2012),(Wang et al, 2012),(Ruan et al, 2020),(Ying et al,2018),(Ishimaru et al, 2013),(Ren et al, 2016),(Li et al, 2012),(Xiong et al, 2018) and(Dong et al, 2020) are also involved in regulating cell proliferation, and their mutants produce small and light grains. The grain length geneis grouped as a member of the kinesin-13 subfamily. In themutant, a reduction in the longitudinal cell length of the grains results in small and round seeds (Kitagawa et al, 2010; Wu et al, 2014; Deng et al, 2015). PGL1 and PGL2 are helix- loop-helix proteins that interact with APG to determine rice grain length by controlling the length of lemma/ palea cells (Heang and Sassa, 2012a, b). Genes,,,,,andalso influence cell expansion-regulated hull size, and their mutants display decreased hull size (Segami et al, 2012; Aya et al, 2014; Duan et al, 2016; Feng et al, 2016; Huang et al, 2017). In addition, cell cycle- related-like genes are involved in hull development by affecting cell proliferation and expansion (La et al, 2006; Ren et al, 2018a).
In the present study, we found that the grain length, grain width and 1000-grain weight of themutant were significantly decreased compared to the WT, and thegene was strongly expressed in the lemma and palea. Further, in themutant, the expression of several genes related to hull cell proliferation and expansion (,,,,,,,,,,and) was significantly down-regulated compared to the WTIn light of allthese results, we suggested that thegene plays a crucial role in regulating the development of grain size via genes related to hull cell proliferation and expansion, and thus affecting the yield of in rice. Further understanding of the molecular mechanisms ofregulating hull development could be of significance in the design of grain morphology in molecular breeding.
METHODS
Rice materials
Themutant was derived from the ethyl methane sulfonate (EMS) mutagenesis population of a maintainer line XD1B (). Themutant was crossed with 56S (a temperature- sensitive sterile line) to generate an F1population. In the second year, the F1population was cultivated and self-crossed to generate an F2population. All plants with the mutant phenotype in the F2population were used to locate thegene
Morphological and histological analysis
During the flowering stage, the spikelets of the WT andmutants were observed and photographed using a stereoscope (SMZ1500, Nikon, Japan) and a scanning electron microscope (SU3500, Hitachi, Japan) under a -20 ºC vacuum environment.
Histological analysis of the spikelets at the flowering stage was performed by paraffin section. The WT and mutant spikelets at the heading stage were immersed in FAA solution (50% anhydrous ethanol, 0.9 mol/L glacial acetic acid and 3.7% formaldehyde) and fixed at 4 ºC for more than 16 h after vacuumed. Then, the spikelets were dehydrated with ethanol series and infiltrated with xylene before being embedded in paraffin (Sigma-Aldrich Inc., Shanghai, China). Next, the paraffin block that contained individual spikelets was cut into 8‒10 µm-thick slices, which were then pasted onto microscope slides (RM2245, Leica, Hamburg, Germany). These slices were dyed sequentially with 1% safranin (Amresco Inc., Framingham, MA, USA) and 1% Fast Green (Amresco Inc., Framingham, MA, USA), then dehydrated through an ethanol series, infiltrated with xylene, and finally mounted beneath a coverslip. Light microscopy was performed using a microscope (E600, Nikon, Japan).
Molecular mapping
The mutant individuals in the F2population were used as the mapping population to locate the target gene by the bulked segregant analysis (BSA) method. The DNA of the parents, F2population, WT and mutant gene pools were extracted from fresh leaves following the cetyl trimethyl ammonium bromide (CTAB) method. SSR markers distributed evenly on the 12 rice chromosomes were used for gene mapping. All the primers were synthesized by the Tsingke Company (Beijing, China). The total volume of the PCR amplifications was 15 µL, which comprised 1.5 µL of 10× PCR buffer, 1 µL of 50 ng/μL DNA, 0.75 µL of 2.5 mmol/L dNTPs, 9.5 µL of ddH2O, 1 µL of 10 mmol/L forward and reverse primers and 0.25 µL of 5 U/µLDNA polymerase. Amplification was done under the following conditions: 5 min at 94 ºC for DNA strand separation, followed by 35 cycles of denaturing at 94 ºC for 30 s, annealing at 56 ºC for 30 s, extension at 72ºC for 30 s, and finally extension at 72 ºC for 7 min. The amplified products were observed and analyzed after separated by electrophoresis on 10% polyacrylamide gels and the silver staining. The relative distances between thelocus and the linkage SSR or the insertion/deletion (InDel) markers were marked by the number of recombinants. The primers are shown in Table S1.
Expression analysis of related genes
RNAs from different tissue samples of the WT and mutant were extracted using Eastep Super Total RNA Extraction Kit (Promega, China). The first-strand cDNA was synthesized from 1 µg total RNA with oligo (dT)18primers in a 20 µL reaction volume using the PrimeScript® RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). The qRT-PCR analysis was performed with three replicates using SYBR Premix Ex Taq II Kit (Takara, Dalian, China) with an ABI 7500 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA).(,) was used as an endogenous control. Allprimers are shown in Table S2.
Complementation test
To construct the complementary vector of, a fragment containing a 2 000 bp upstream sequence and complete open reading frame sequence was amplified from rice genomic DNA using the primers MFS4-com-GFP-F-EcoRI and MFS4-com- GFP-R-BamHI. The fragment was cloned into the binary vector pCAMBIA1300 at theRI/HI site. After verification by sequence analysis, the vector was transformed into themutant by the-mediated method. The primer sequences used are shown in Table S3.
Subcellular localization
To carry out the subcellular localization of MFS4 protein, thecoding sequence was amplified from rice genomic DNA using the primers 580-MFS4-XbalI-F and 580-MFS4-BamHI-R. The fragment was cloned into the vector pAN580 at theI/HI site. After verification by sequence analysis, the pAN580-GFP, MTS-GFP and MFS4-GFP plasmids were transformed into rice protoplasts. After incubation at 28 ºC for 12–16 h, GFP fluorescence was detected using a confocal laser scanning microscope (LSM710, Zeiss, Jena, Germany). The primers are shown in Table S3.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31971919 and 31730063), the Natural Science Foundation Project of Chongqing Science and Technology Commission, China (Grant No. cstc2020jcyj-jqX0020), National Key Program for Research and Development of China (Grant No. 2017YFD0100202), and Chongqing Graduate Research and Innovation Project Funding, China (Grant No. CYS20123).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
Table S1. Sequences of primers for gene mapping.
Table S2. Sequences of primers for qRT-PCR.
Table S3. Primers used in this study.
Arora R, Agarwal P, Ray S, Singh A K, Singh V P, Tyagi A K, Kapoor S. 2007. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress., 8(1): 242.
Ashikari M, Sakakibara H, Lin S Y, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production., 309: 741‒745.
Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. 2014. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway., 55(5): 897‒912.
Chuck G, Meeley R, Hake S. 2008. Floral meristem initiation and meristem cell fate are regulated by the maizegenesand.,135(18): 3013‒3019.
Deng Z Y, Liu L T, Li T, Yan S, Kuang B J, Huang S J, Yan C J, Wang T. 2015. OsKinesin-13A is an active microtubule depolymerase involved in glume length regulation via affecting cell elongation., 5: 9457.
Dong N Q, Sun Y W, Guo T, Shi C L, Zhang Y M, Kan Y, Xiang Y H, Zhang H, Yang Y B, Li Y C, Zhao H Y, Yu H X, Lu Z Q, Wang Y, Ye W W, Shan J X, Lin H X. 2020. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice., 11(1): 2629.
Duan P G, Ni S, Wang J M, Zhang B L, Xu R, Wang Y X, Chen H Q, Zhu X D, Li Y H. 2016. Regulation ofby OsmiR396 controls grain size and yield in rice., 2(1): 1‒5.
Feng Z M, Wu C Y, Wang C M, Roh J, Zhang L, Chen J, Zhang S Z, Zhang H, Yang C Y, Hu J L, You Xi M, Liu X, Yang X M, Guo X P, Zhang X, Wu F Q, Terzaghi W, Kim S K, Jiang L, Wan J M. 2016.controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice., 67(14): 4241‒4253.
Guo T, Lu Z Q, Shan J X, Ye W W, Dong N Q, Lin H X. 2020.acts upstream of the OsMKKK10-OsMKK4- OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice., 32(9): 2763‒2779.
Heang D, Sassa H. 2012a. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice., 7(2): e31325.
自2015年李克强总理提出了“新旧动能转换”以来,开始出现在国家领导人的讲话和文件中,并在2016年开始频繁出现在互联网中。进入2017年以来,“新旧动能”的内涵才逐渐丰富和完善起来。所以“新旧动能”作为官方用语来说,没有严格的概念界定,但我们从一系列政府文件中和领导讲话中可以理解为:新旧动能转换的实质就是经济发展方式转变的过程,是转型升级的过程。在这个过程中,壮大新动能、提升传统动能,推动经济保持中高速增长、产业迈向中高端水平。在实施新旧动能转换的经济驱动下,我们职业及教育的教学模式也应该进行相应的转换才能够适应社会经济的发展。
Heang D, Sassa H. 2012b. An atypical bHLH protein encoded byis involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG., 62(2): 133‒141.
Hu J, Wang Y X, Fang Y X, Zeng L J, Xu J, Yu H P, Shi Z Y, Pan J J, Zhang D, Kang S J, Zhu L, Dong G J, Guo L B, Zeng D L, Zhang G H, Xie L H, Xiong G S, Li J Y, Qian Q. 2015. A rare allele ofenhances grain size and grain yield in rice., 8(10): 1455‒1465.
Huang K, Wang D K, Duan P G, Zhang B L, Xu R, Li N, Li Y H. 2017., which encodes an otubain- like protease with deubiquitination activity, influences grain size and shape in rice., 91(5): 849‒860.
Huang X Z, Qian Q, Liu Z B, Sun H Y, He S Y, Luo D, Xia G M, Chu C C, Li J Y, Fu X D. 2009. Natural variation at thelocus enhances grain yield in rice., 41(4): 494‒497.
Ikeda K, Sunohara H, Nagato Y. 2004. Developmental course of inflorescence and spikelet in rice., 54(2): 147‒156.
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E. 2013. Loss of function of the IAA-glucose hydrolase geneenhances rice grain weight and increases yield., 45(6): 707‒713.
Jeon J S, Jang S, Lee S, Nam J, Kim C, Lee S H, Chung Y Y, Kim S R, Lee Y H, Cho Y G, An G. 2000.is a homeotic mutation in a rice MADS box gene affecting rice flower development., 12(6): 871‒884.
Jin Y, Luo Q, Tong H N, Wang A J, Cheng Z J, Tang J F, Li D Y, Zhao X F, Li X B, Wan J M, Jiao Y L, Chu C C, Zhu L H. 2011. An AT-hook gene is required for palea formation and floral organ number control in rice., 359(2): 277‒288.
Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y. 2010. A novel kinesin 13 protein regulating rice seed length., 51(8): 1315‒1329.
La H G, Li J, Ji Z D, Cheng Y J, Li X L, Jiang S Y, Venkatesh P N, Ramachandran S. 2006. Genome-wide analysis of cyclin family in rice (L.)., 275(4): 374‒386.
Lee D Y, An G. 2012. Two AP2 family genes,() and(), synergistically control inflorescence architecture and floral meristem establishment in rice., 69(3): 445‒461.
Lee D Y, Lee J, Moon S, Park S Y, An G. 2007. The rice heterochronic generegulates the transition from spikelet meristem to floral meristem., 49(1): 64‒78.
Li H F, Liang W Q, Jia R D, Yin C S, Zong J, Kong H Z, Zhang D B. 2010. The AGL6-like generegulates floral organ and meristem identities in rice., 20(3): 299‒313.
Li H G, Xue D W, Gao Z Y, Yan M X, Xu W Y, Xing Z, Huang D N, Qian Q, Xue Y B. 2009. A putative lipase generegulates both empty-glume fate and spikelet development in rice., 57(4): 593‒605.
Li X J, Sun L J, Tan L B, Liu F, Zhu Z F, Fu Y C, Sun X Y, Sun X W, Xie D X, Sun C Q. 2012., a DUF640 domain-like gene controls lemma and palea development in rice., 78(4/5): 351‒359.
Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F. 2011. Natural variation inplays an important role in regulating grain size and yield in rice., 43(12): 1266‒1270.
Li Y F, Zeng X Q, Li Y, Wang L, Zhuang H, Wang Y, Tang J, Wang H L, Xiong M, Yang F Y, Yuan X Z, He G H. 2020. MULTI-FLORET SPIKELET 2, a MYB transcription factor, determines spikelet meristem fate and floral organ identity in rice., 184: 988‒1003.
Ma X D, Cheng Z J, Wu F Q, Jin M G, Zhang L G, Zhou F, Wang J L, Zhou K N, Ma J, Lin Q B, Lei C L, Wan J M. 2013.is required for lateral development of lemma and palea in rice., 31(1): 98‒108.
Malcomber S T, Preston J C, Reinheimer R, Kossuth J, Kellogg E A. 2006. Developmental gene evolution and the origin of grass inflorescence diversity., 44: 425‒481.
Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. 2003.andgenes control floral organ identity in rice., 130(4): 705‒718.
Ren D Y, Li Y F, Zhao F M, Sang X C, Shi J Q, Wang N, Guo S, Ling Y H, Zhang C W, Yang Z L, He G H. 2013., which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice., 162(2): 872‒884.
Ren D Y, Rao Y C, Wu L W, Xu Q K, Li Z Z, Yu H P, Zhang Y, Leng Y J, Hu J, Zhu L, Gao Z Y, Dong G J, Zhang G H, Guo L B, Zeng D L, Qian Q. 2016. The pleiotropicaffects plant height, floral development and grain yield in rice., 58(6): 529‒539.
Ren D Y, Hu J, Xu Q K, Cui Y J, Zhang Y, Zhou T T, Rao Y C, Xue D W, Zeng D L, Zhang G H, Gao Z Y, Zhu L, Shen L, Chen G, Guo L B, Qian Q. 2018a.determines grain size and sterile lemma fate in rice., 69(20): 4853‒4866.
Ren D Y, Yu H P, Rao Y C, Xu Q K, Zhou T T, Hu J, Zhang Y, Zhang G H, Zhu L, Gao Z Y, Chen G, Guo L B, Zeng D L, Qian Q. 2018b. ‘Two-floret spikelet’ as a novel resource has the potential to increase rice yield., 16(2): 351‒353.
Ren D Y, Rao Y C, Yu H P, Xu Q K, Cui Y J, Xia S S, Yu X Q, Liu H, Hu H T, Xue D W, Zeng D L, Hu J, Zhang G H, Gao Z Y, Zhu L, Zhang Q, Shen L, Guo L B, Qian Q. 2020.encodes a MYB transcription factor that regulates spikelet development in rice., 184(1): 251‒265.
Ruan B P, Shang L G, Zhang B, Hu J, Wang Y X, Lin H, Zhang A P, Liu C L, Peng Y L, Zhu L, Ren D Y, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. 2020. Natural variation in the promoter ofdetermines grain width and weight in rice., 227(2): 629‒640.
Segami S, Kono I, Ando T, Yano M, Kitano H, Miura K, Iwasaki Y. 2012.gene encodes alpha-tubulin regulating seed cell elongation in rice., 5: 4.
Song X J, Huang W, Shi M, Zhu M Z, Lin H X. 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase., 39(5): 623‒630.
Su’udi M, Cha J Y, Ahn I P, Kwak Y S, Woo Y M, Son D Y. 2012. Functional characterization of a B-type cell cycle switch 52 in rice ()., 111(1): 101‒111.
Wan X Y, Weng J F, Zhai H Q, Wang J K, Lei C L, Liu X L, Guo T, Jiang L, Su N, Wan J M. 2008. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allelein a recombination hotspot region on chromosome 5., 179(4): 2239‒2252.
Wang K J, Tang D, Hong L L, Xu W Y, Huang J, Li M, Gu M H, Xue Y B, Cheng Z K. 2010.andregulate reproductive habit in rice., 6(1): e1000818.
Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. 2012. Control of grain size, shape and quality byin rice., 44(8): 1‒6.
Wu T, Shen Y Y, Zheng M, Yang C Y, Chen Y L, Feng Z M, Liu X, Liu S J, Chen Z J, Lei C L, Wang J L, Jiang L, Wan J M. 2014. Gene, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice., 33(2): 235‒244.
Xia D, Zhou H, Liu R J, Dan W H, Li P B, Wu B, Chen J X, Wang L Q, Gao G J, Zhang Q L, He Y Q. 2018., a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts withto produce extra-long grains in rice., 11(5): 754‒756.
Xiong H Y, Yu J P, Miao J L, Li J J, Zhang H L, Wang X, Liu P L, Zhao Y, Jiang C H, Yin Z G, Li Y, Guo Y, Fu B Y, Wang W S, Li Z K, Ali J, Li Z C. 2018. Natural variation inincreases drought tolerance in rice by inducing ROS scavenging., 178(1): 451‒467.
Xue W Y, Xing Y Z, Weng X Y, Zhao Y, Tang W J, Wang L, Zhou H J, Yu S B, Xu C G, Li X H, Zhang Q F. 2008. Natural variation inis an important regulator of heading date and yield potential in rice., 40: 761.
Yan S, Zou G H, Li S J, Wang H, Liu H Q, Zhai G W, Guo P, Song H M, Yan C J, Tao Y Z. 2011. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice., 123(7): 1173‒1181.
Ying J Z, Ma M, Bai C, Huang X H, Liu J L, Fan Y Y, Song X J. 2018., a major QTL that negatively modulates grain length and weight in rice., 11(5): 750‒753.
Yoshida A, Suzaki T, Tanaka W, Hirano H Y. 2009. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet., 106(47): 20103‒20108.
Zhang B Y, Wu S H, Zhang Y E, Xu T, Guo F F, Tang H S, Li X, Wang P F, Qian W F, Xue Y B. 2016. A high temperature- dependent mitochondrial lipase EXTRA GLUME1 promotes floral phenotypic robustness against temperature fluctuation in rice (L.)., 12(7): e1006152.
Zhang L, Yu H, Ma B, Liu G F, Wang J J, Wang J M, Gao R C, Li J J, Liu J Y, Xu J, Zhang Y Y, Li Q, Huang X H, Xu J L, Li J M, Qian Q, Han B, He Z H, Li J Y. 2017. A natural tandem array alleviates epigenetic repression ofand leads to superior yielding rice., 8(1): 14789.
Zheng H, Zhang J, Zhuang H, Zeng X Q, Tang J, Wang H L, Chen H, Li Y, Ling Y H, He G H, Li Y F. 2019. Gene mapping and candidate gene analysis of() in rice (L.)., 18(12): 2673‒2681.
Zhu Q H, Upadhyaya N M, Gubler F, Helliwell C A. 2009. Over- expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice ()., 9: 149.
27 August 2020;
17 November 2020
Copyright © 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2021.05.005
Li Yunfeng (liyf1980@swu.edu.cn)
(Managing Editor: Wu Yawen)
- Rice Science的其它文章
- SDF5 Encoding P450 Protein Is Required for Internode Elongation in Rice
- RGB1 Regulates Rice Panicle Architecture and Grain Filling Through Monitoring Cytokinin Level in Inflorescence Meristem and Grain Abscisic Acid Level During Filling Stage
- Application of Rice Husk Biochar for Achieving Sustainable Agriculture and Environment
- OsbZIP09, a Unique OsbZIP Transcription Factor of Rice, Promotes Rather Than Suppresses Seed Germination by Attenuating Abscisic Acid Pathway
- A New Approach to Select Doubled Haploid Rice Lines under Salinity Stress Using Indirect Selection Index
- iTRAQ-Based Proteomics Investigation of Critical Response Proteins in Embryo and Coleoptile During Rice Anaerobic Germination

