Group Living on Trees Does Not Elevate Inbreeding Risk in Shedao Pit Vipers Gloydius shedaoensis
Xiaoping WANG,Ruifeng WU,Ke JIANG,Yayong WU,Guannan WEN and Yin QI,4*
1 Nature Conservation of Snake Island and Laotieshan Mountain,Dalian 116041,Liaoning,China
2 Chengdu Institute of Biology,Chinese Academy of Sciences,Chengdu 610041,Sichuan,China
3 College of Life Sciences and Food Engineering,Yibin University,Yibin 644007,Sichuan,China
4 Key Laboratory of Southwest China Wildlife Resources Conservation (Ministry of Education),China West Normal University,Nanchong 637009,Sichuan,China
Abstract Group living has increasingly been emphasized due to its potential consequence on inbreeding,nevertheless,the relationship between group living and inbreeding risk is largely unknown.The endangered snake species,Shedao pit viper (Gloydius shedaoensis) inhabiting mostly on Shedao Island (meaning “snake island” in Chinese),provides an excellent model system for research on group living and inbreeding risk.Small island area,high population density and extreme seasonal foraging opportunity prompt many individuals to live on the same tree,which likely increase the potential mating among relatives.To confirm this probability,we used genotypes from 13 microsatellites DNA markers and examined the genetic relationships between pair of individuals lived on the same tree.The results showed that snakes on the same tree did not show closer relationships compared with individuals from different trees.The group constructions based on scenarios of parental-offspring,full-sibling,half-sibling as well as cousin were not consistent with tree-based group living.In addition,we did not find significant correlation between pair of individual genetic distance and geographic distance going beyond trees,suggesting group living on trees might not elevate inbreeding risk.
Keywords group living,inbreeding risk,pairwise relatedness,snakes,Gloydius shedaoensis
1.Introduction
With the fragmentation of critical resources (e.g.habitat or foraging site),a number of threatened individuals choose to live together to ensure their fitness by resource share,parental care or predator avoidance (Støenet al.
,2005;Mckinnonet al.
,2006).Nevertheless,group living likely dramatically increases the likelihood of mating among relatives and aggravate inbreeding risk (Charlesworth and Charlesworth,1987;Vanthournoutet al.
,2016),which contributes a lot in genome-wide homozygosity and expression of deleterious recessive alleles (dominance hypothesis) and/or loss of heterozygous advantage (overdominance hypothesis),thereby has several adverse impacts on population maintaining and individual survival (Charlesworth and Charlesworth,1987;Madsenet al.
,1999;Keller and Waller,2002;Charlesworth and Willis,2009).To alleviate the adverse impacts,we need to manipulate group living and reduce the inbreeding level.The genetic relatedness between individuals should be lower than inbreeding risk level (e.g.0.25) to maintain a sustainable population structure.Nevertheless,the association between group living and inbreeding risk is largely unknown.One recent study around the highly endangered red-cockaded woodpecker (Picoides borealis
) found that inbreeding risk varies inversely with female natal dispersal distance,territory overlap among individuals was found to increase with the proportion of inbreed individuals (Schiegget al.
,2006).Uncovering the relationship between group living and inbreeding risk would contribute a lot to the conservation in endangered species (Caiet al.
,2016;Brzeskiet al.
,2014;Huet al.
,2017),because an increasing number of species are endangered and forced to live in small area with the aggravation of habitat fragmentation.The pit viper (Gloydius shedaoensis
) provides an excellent model system for testing the relationship between group living and inbreeding risk.The snakes are endangered and occur exclusively on the Shedao Island (meaning “snake island” in Chinese),off the coast of Liaodong Peninsula in the Bohai Sea.The snake island is approximately 0.73 kmand 5.3 km away from the nearest continent (Li,1995).The population size of the snake is estimated at 20 281 (Liet al.
,2007).The primary food source for the snakes is migrating birds in April-June and August-October every year (Li,1995),although there are two small mammal species inhabiting on the island (batPipistrellus abramus
and mouseRattus norvegicus
,Li,1995).Small size of the island,high population density of snakes,and strong seasonality in food availability,force many snakes (2-9) to live on the same tree.Individual snakes consistently reused the branch of trees and prey on migrating birds (Shineet al.
,2002),they would also mate on branches or nearby trees during group living (Li,1995),which likely increase the inbreeding risk if they are relatives.The purpose of the present study was to examine whether group living on trees increase the inbreeding risk inG.shedaoensis
.In particular,we asked whether snakes perched on the same tree were kin-related.The pairwise relatedness between snakes in and out of the same tree was estimated using microsatellite DNA phenotypes.We also compared the group construction based on different kin scenarios and trees,and detected the relationship between genetic distance and geographic distances to demonstrate the genetic relationships among individuals both in and out of trees.2.Materials and Methods
2.1.Field sampling
To examine whether group living on trees increases the inbreeding risk in Shedao pit vipers (G.shedaoensis
),a total of 45 snakes (15 males,30 females from 10 trees) were captured using snake clamp from ten trees on Snake Islandin September,2014 (Figure 1).After capturing,the snout-vent length (SVL,to 1 mm) and body mass (to 0.01 g) were measured using plastic ruler and electric scale,respectively,and the sex was determined by checking for the hemi-penis bulge.All snakes were permanently marked using PIT tags (Guangzhou HongTeng Barcode Technology Ltd.,China),and a small drop of blood was collected from caudal vein using FTA card (Whatman International Ltd.,UK) for subsequent genotyping.After carrying out all above procedures,snakes were released back to their dwelling tree.The coordinate of each capture site was recorded using GPS,and detailed sampling information is shown in Table 1.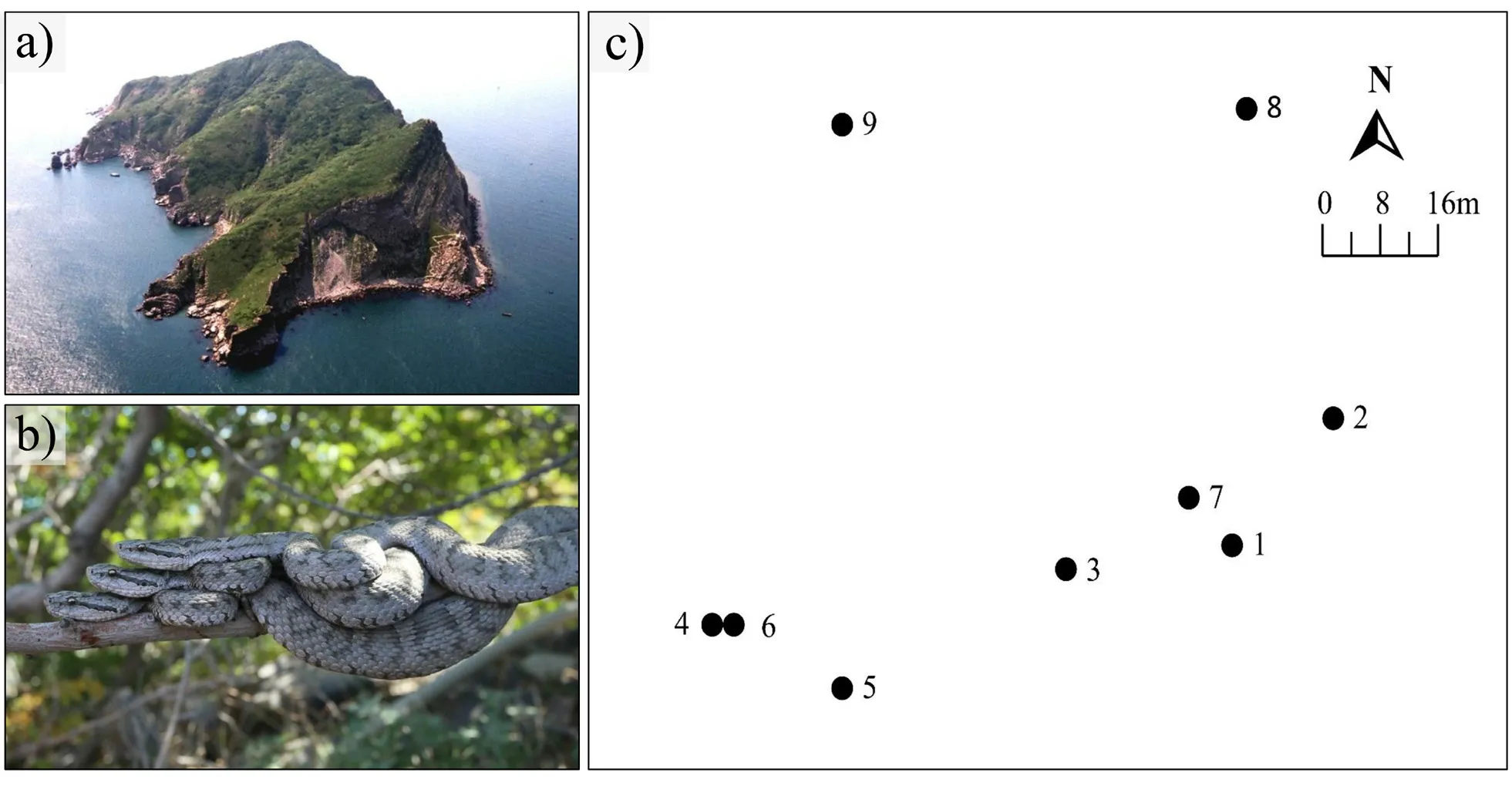
Figure 1 a) an aerial view of Shedao Island;b) three Gloydius shedaoensis lived on the same tree;c) spatial positions of trees No. 1-9 where snake sampling was collected.
2.2 Microsatellite DNA collection
We used microsatellite genotypes to estimate relatedness between pair of snakes.Because of lack in microsatellite markers specifically forG.shedaoensis
,we first obtained eight microsatellite markers development for otherGloydius
species by Goldberget al.
(2003),Holycrosset al.
(2002),Munguia-Vegaet al.
(2009) and Oyler-Mccanceet al.
(2005).The detailed information for those markers is listed in Table S1.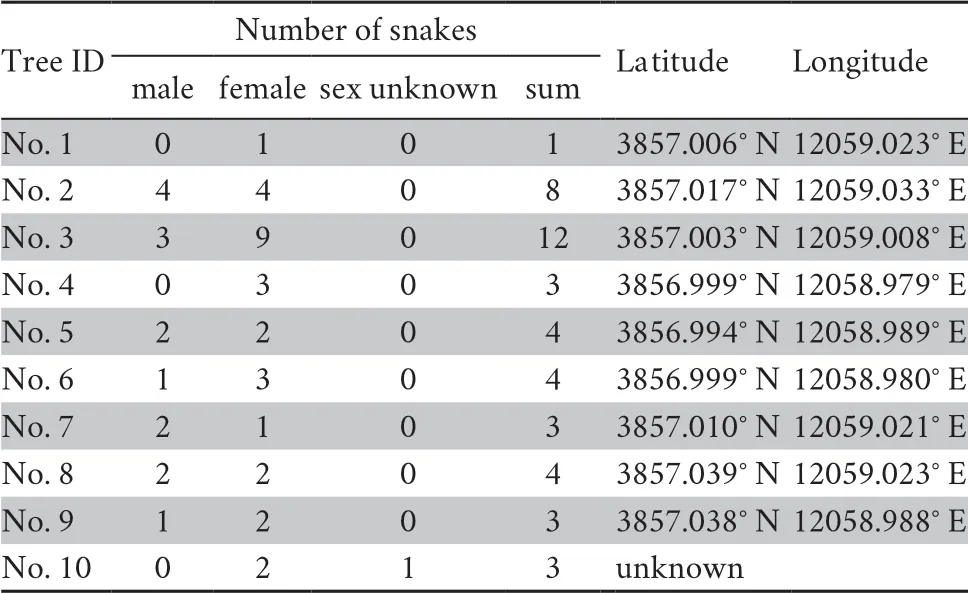
Table 1 Sampling information in Gloydius shedaoensis.
To increase the estimation accuracy,we specifically developed more microsatellite markers forG.shedaoensis
.We acquired the transcriptome sequence of oneG.shedaoensis
from another project,and 40 microsatellite loci were identified and primers were designed to amplify those microsatellite DNA regions.Detailed isolation process has been described in Nieet al.
(2015) and Liet al.
(2015).Briefly,standard PCR amplification with optimized annealing temperature was conducted.PCR products were visualized on 8% denaturing polyacrylamide gels with silver staining,and the polymorphism of each locus was assessed.After multiple tests,nine loci were consistently amplifiable and polymorphic.Cross-species amplification was tested using three relatedGloydius
species,G.intermedius Strauch
,G.strauchi
andG.brevicaudus
Stejneger.The detailed information for those newly developed markers is listed in Table 2.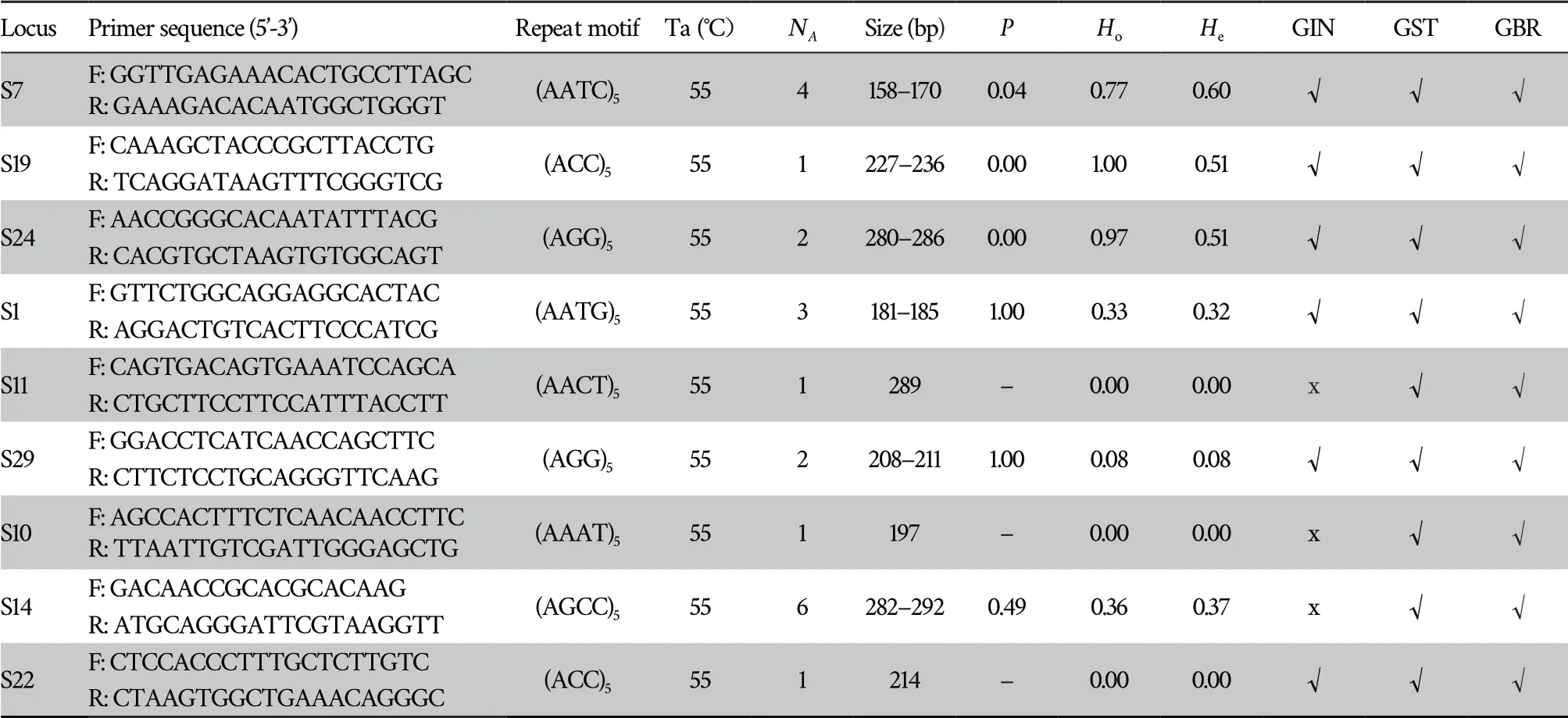
Table 2 Characterization of nine novel microsatellite markers for Gloydius shedaoensis and cross-amplification of those markers in three other Gloydius species.Ta,optimal annealing temperature;NA,number of alleles;P,tests for Hardy-Weinberg equilibrium;Ho,observed heterozygosity;He,expected heterozygosity;F,forward;R,reverse;√,succeeded in amplification;x,failed in amplification;GIN,Gloydius intermedius Strauchi,GST, Gloydius strauchi,GBR,Gloydius brevicaudus Stejneger.
The genotypes of 45 individual snakes were collected using 17 microsatellite DNA loci.In particular,we took a sample disc from the blood card for each snake and incubated it inNaOH
solution (20 mmol/L) for 25min under room temperature.The sample disc was then washed with TE-1 Buffer (10mM Tris-HCl,0.1 mmol/L EDTA,pH 8.0) twice before being used in PCRs.PCRs were conducted with the same parameters as above.The forward primers were labeled with fluorescence dye of FAM,TAMRA,or HEX.PCR products were genotyped on an ABI 3730 genetic analyser (Sangon Biotech Co.Ltd.,Shanghai,China).All eletropherograms were visually inspected and analyzed using GENEMARKER version 1.6 (SoftGenetics,State College,PA).All samples were genotyped at least twice to get consistent results.2.3.Genetic analysis
Before genetic analysis,null alleles were checked for each locus with MICRO-CHECKER Version 2.2.3 (Van Oosterhoutet al.
,2004).Loci of “S19”,“S11”,“S10” and “S22” were excluded due to low polymorphism.Genetic diversity for each locus was assessed using number of alleles (A
),observed heterozygosity (H
),and expected heterozygosity (H
) with GENEPOP version 4.5.1 (Rousset,2008).Deviation from Hardy-Weinberg equilibrium and linkage equilibrium was analyzed with GENEPOP Version 4.5.1.To examine whether snakes perched on the same tree were closely related from genetics,we calculated the mean relatedness between pair of snakes within trees using KINGROUP v2 (Konovalov,2010).Male and female were analyzed as overall or separately to ascertain whether genetic relatedness varied with sex.
We then sorted individuals into subgroups by evaluating alternative partitions according to different likelihood scenario of kin relatedness.The overall likelihood for a given partitionwas calculated from the pairwise likelihood.To create partition,we first arbitrarily assigned one individual to the first subgroup while other individuals were added to the existing subgroup,one snake at a time.The placement of each individual is evaluated by placing it into available subgroups in turn as well as into a new subgroup,and taking the product of likelihood for corresponding pedigree relationships.The addition order was random and decided by the descending ratio search algorithm.We assigned another two different individuals into the first subgroup to examine the possible consequences of arbitrary reallocation,although no difference was found.All genetic group analysis was carried out using KINGROUP v2 (Konovalov,2010).
To go beyond trees,an isolation-by-distance (IBD) pattern was examined to show the correlation between individual genetic relatedness and location distance.We used ‘mantel’ function in R package ‘vegan’ (Oksanenet al.
,2019).The genetic distance was represented byF
/(1-F
) values.All geographic distances were calculated using R package ‘geosphere’ (Hijmans,2019).The analysis was only applied to site 1-9,because the coordinates of site 10 was missing.A Mantel test with 1000 permutations was used to detect significant correlation between the genetic distances and geographic distances.2.4.Ethical approval
All applicable international,national,and/or institutional guidelines for the care and use of animals were strictly followed.All animal sample collection protocols complied with the current laws of China.All animal procedures performed in this research were in accordance with the ethical standards of Experimental Animal Ethics Committee of the Chengdu Institute of Biology,Chinese Academy of Sciences (protocol number 2017005).3.Results
3.1.Newly developed microsatellite loci
Nine microsatellite markers were newly developed forG.shedaoensis
(Table 2).The number of alleles in newly developed loci ranged from 1 to 6,the observed heterozygosity ranged from 0 to 1.00,while the expected heterozygosity ranged from 0 to 0.60.Five of them were significantly deviated from Hardy-Weinberg equilibrium,and six of them were crossly amplifiable in three otherGloydius
species.3.2.Relatedness between snakes
Mean relatedness between pair of snakes for all individuals,males and females perched on the same tree were demonstrated in Figure 2.In high proportion of trees (8/10),snakes did not show close genetic relationships,except the tree No.7 (r
=0.30±0.16,n
=3) and tree No.10 (r
= 0.35±0.26,n
=3).The mean relatedness between males or females perched on the same tree was similar with the overall analysis,except the tree No.3 (r
= 0.40±0.02,n
=3) for males.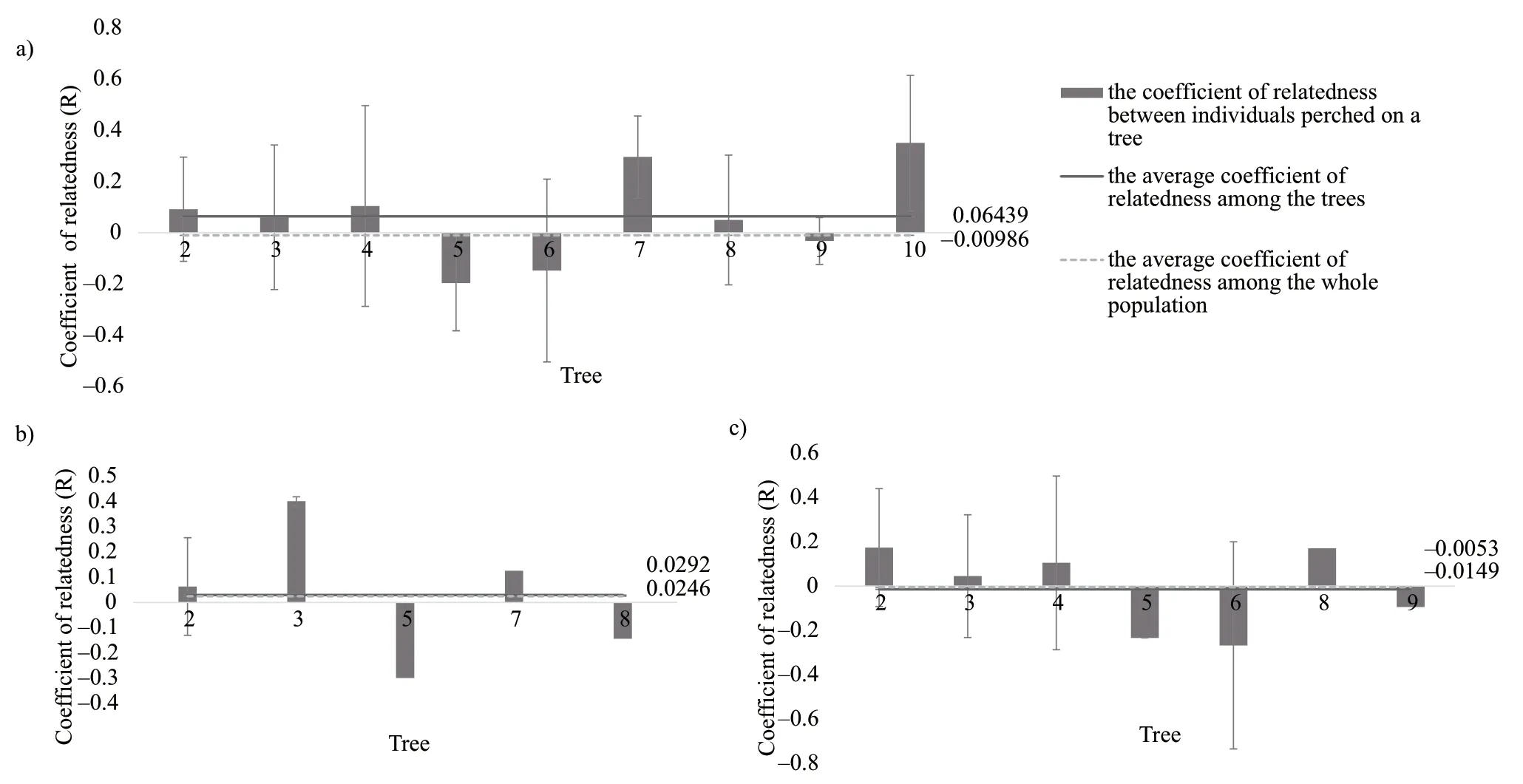
Figure 2 Average coefficient of relatedness (R) between pair of a) all individual Gloydius shedaoensis perched on the same tree;b) males perched on the same tree;c) females perched on the same tree.The solid line represents the average coefficient of relatedness between pair of individuals among the trees,while the dotted line represents the average coefficient of relatedness between pair of individuals from all trees.
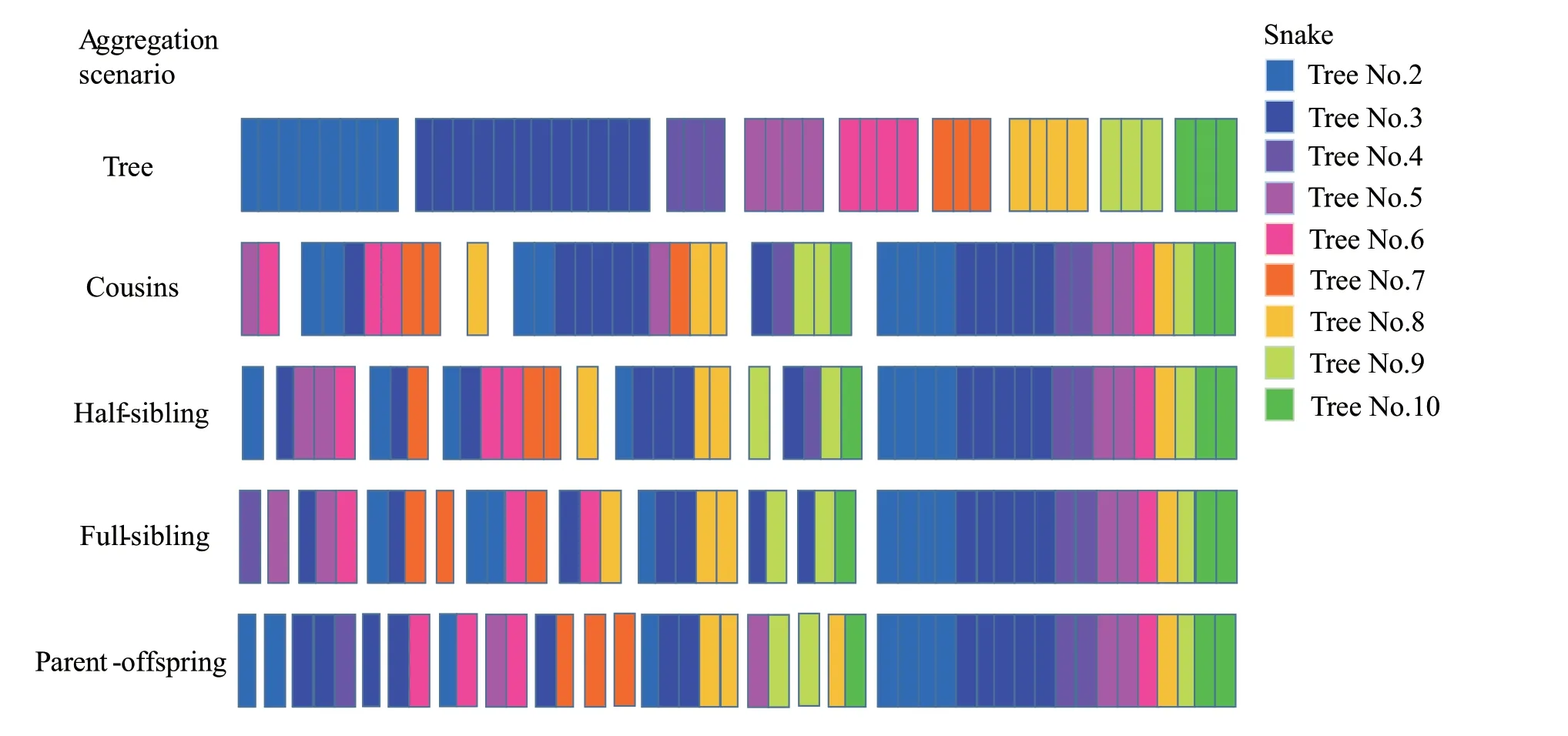
Figure 3 Kin group reconstruction based on scenarios of parental-offspring,full-sibling,half-sibling and cousin.Snake groups based on trees were provided as reference.The same color bar represents Gloydius shedaoensis from the same tree.Snake from tree No.1 was not included because there is only one pit viper from this tree.
3.3.Kin group reconstruction
Kin group reconstructions based on different relatedness likelihood were demonstrated in Figure 3.Under parental-offspring scenario,all snakes were sorted into 15 subgroups.To be conservative,we focused on the first five subgroups.Two snakes from tree No.2 were sorted into first two subgroups respectively.Two snakes from tree No.3 and one snake from tree No.4 were sorted into the third subgroups.One snake from tree No.3 was sorted into the fourth subgroup,while the fifth subgroup consisted of one snake from tree No.3 and one snake from tree No.6.Under full-sibling scenario,eleven subgroups were created.One snake from tree No.4 was sorted into the first subgroup,one snake from tree No.5 was sorted into the second subgroup.The third subgroup consisted of one snake from tree No.3,one snake from tree No.5 and one snake from tree No.6.The fourth subgroup consisted of one snake from tree No.2,one snake from tree No.3 and one snake from tree No.7,while the fifth subgroup consisted of one snake from tree No.7.
Under half-sibling scenario,nine subgroups were created.One snake from tree No.2 was sorted into the first subgroup.The second subgroup consisted of one snake from tree No.3,two snakes from tree No.5 and one snake from tree No.6.The third subgroup composed of one snake from tree No.2,one snake from tree No.3 and one snake from tree No.7.The fourth subgroup consisted of one snake from tree No.2,one snake from tree No.3,two snakes from tree No.5 and two snakes from tree No.7,while the fifth subgroup was made up of one snake from tree No.8.
Under cousin scenario,six subgroups were created.One snake from tree No.5 and one snake from tree No.6 were sorted into the first subgroup.The second subgroup composed of two snakes from tree No.2,one snake from tree No.3,two snakes from tree No.6 and two snakes from tree No.7.One snake from tree No.8 was sorted into the third subgroup.The fourth subgroup consisted of two snakes from tree No.2,five snakes from tree No.3,one snake from tree No.5,one snake from tree No.7 and two snakes from tree No.8,while the fifth subgroup composed of one snake from tree No.2,one snake from tree No.3,two snakes from tree No.9 and one snake from tree No.10.
No matter which scenario,the group construction was different with the tree-based group living,suggesting individuals living on the same tree was not kin-related.
3.4.Isolation-by-distance
The correlation between individual genetic distance and geographic distance was shown in Figure 4.We found no significant correlation between pair of individual genetic distance and geographic distance (r
=0.12,P
=0.34).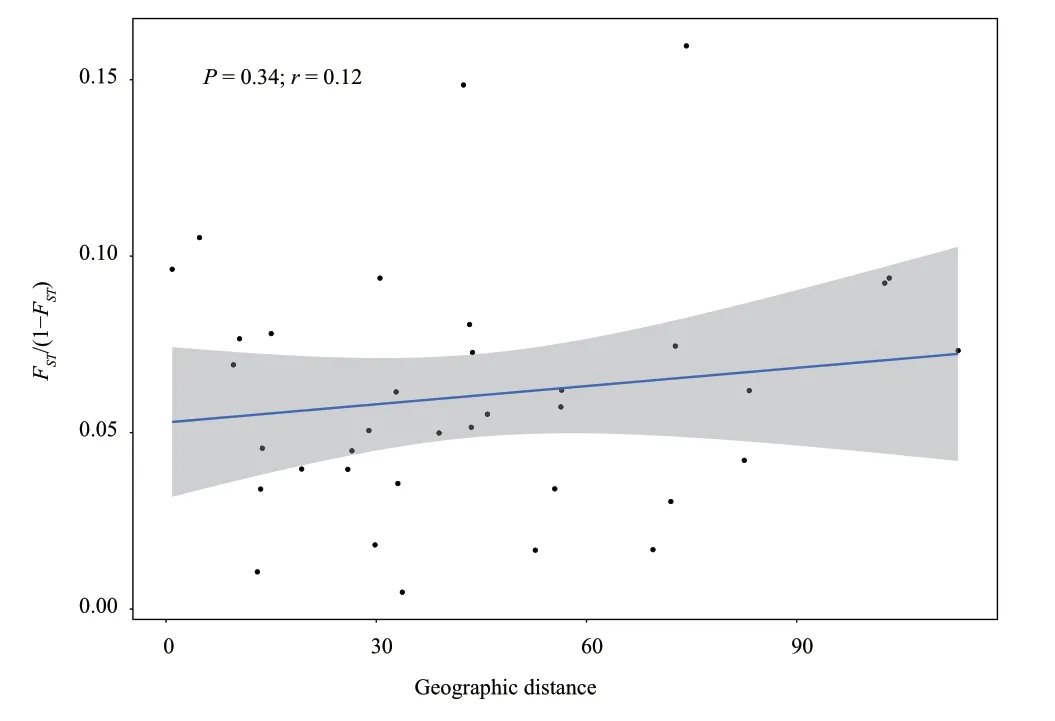
Figure 4 Isolation-by-distance pattern between pair of individual Gloydius shedaoensis genetic distance and geographic distance.
4.Discussion
Using genotypes from 13 microsatellites DNA markers (eight from otherGloydius
species,and five from newly developed microsatellites forG.shedaoensis
),we found snakes living on the same tree were largely not kin-related.We have three evidences for this.First,except the tree No.7 and No.10,snakes on the same tree did not show close pairwise genetic relationships.Second,the group constructions based on parental-offspring scenario,full-sibling scenario,half-sibling scenario as well as cousin scenario,were not consistent with snakes group based on trees.Third,genetic distance and geographic distance was not correlated between pair of individuals.This is very different to social group living lizards of genusEgernia
,among which 23 of 30 reported species are found to aggregate based on kin relatedness (Chapple,2003),andE.striolata
was found to form a tree-dwelling population in group of close kin (Duckettet al.
,2012).None kin-related group living inG.shedaoensis
implies that snakes might have taken crucial ways in avoiding inbreeding risk before tree occupation.Offspring and adult dispersal after hibernation likely play important roles in inbreeding avoidance.As an island-dwelling species,snakesG.shedaoensis
confront many challenges from small living area and high population density.AdultG.shedaoensis
choose to stay in most of time to avoid social conflict.The average daily movement distance of male and female is 1.76 m and 1.99 m,respectively,and a radiotelemetric study shows that adult snakes generally remain within 50 m of their original site,with no tendency to move further (Shineet al.,
2003).Nevertheless,offspringG.shedaoensis
was found to disperse intensively during breeding season,out of 11 marked offspring,six of them have low recapture rate.In addition,adultG.shedaoensis
are very active after hibernation,13 of followed adults have average of 4.59±5.48 m (range from 0 to 39 m) in dispersal distance per day (Yayong Wu,unpublished data).Dispersal behavior has been found to contribute a lot in inbreeding avoidance in many other species (Perrins and Goudet,2001;Olsson and Shine,2003;Le Galliardet al.
,2006;Schiegget al.
,2006).For example,in endangered red-cockaded woodpackerP.borealis
,natal dispersal distance is negatively associated with inbreeding risk,suggesting natal dispersal could alleviate inbreeding pressure (Schiegget al.
,2006).Alternatively,femaleG.shedaoensis
likely avoids inbreeding through strict mate choice.This is inferred from the interesting courtship behavior described by Li (1995).Before copulation,maleG.shedaoensis
always tend to conquer females using forcible ways,such as twining and biting,while females resist males using the same way.As soon as copulation starting,females frequently nod their head (Li,1995).We assumed that head nodding behavior in femaleG.shedaoensis
likely functions as important signal of female acceptance because this behavior only happened during copulation.We still need more direct experiment to verify this,because female mate choice is very tricky in reptiles,although there is one evidence on cryptic mate choice in Swedish sand lizards (Lacerta agilis
) (Olssonet al.,
1996).We admitted that the relationship between group living and inbreed risk we obtained inG.shedaoensis
should be cautious,because in most trees,the number of snakes sampled was low,which likely has consequences on the accuracy of relationship estimation.We need to add more snakes from more trees to verify this in the future.Conclusion
Our study evidenced that tree-based group living in adultsG.shedaoensis
did not aggravate inbreeding risk,because snakes on the same tree are not kin related.This implies that snakes may have taken crucial ways in avoiding mating with relatives,such as offspring natal dispersal,adult dispersal after hibernation and strict mate choice,although much of those are speculated and warrants careful future research.Acknowledgements
We are grateful to Bao YAN for assistance in the field sampling.Thanks to Jinsong SHI in data collection.We thank Jinzhong FU for his constructive comments that improved this manuscript.This project is supported by grants from Liaoning Snake Island Laotie Mountain National Nature Reserve (grant number:Y8Y3041) and Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment (grant number:2019HJ2096001006).Data availability statement
The datasets generated /or analyzed during the current study are available on Figshare (doi number:10.6084/m9.figshare.11369067).Appendix
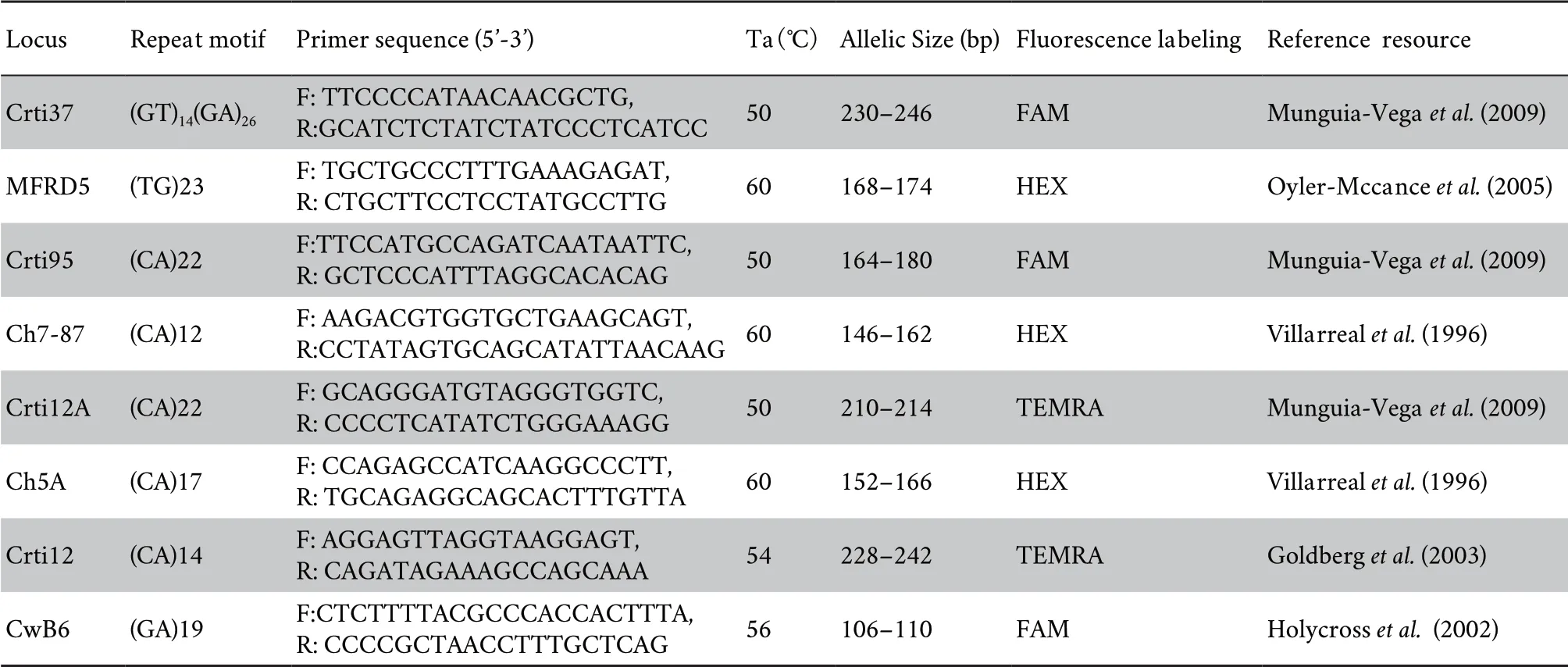
Table S1 Information of eight microsatellite loci from published literature.
Reference
Brzeski K.E.,Rabon D.R.,Chamberlain M.J.,Waits L.P.,Taylor S.S.2014.Inbreeding and inbreeding depression in endangered red wolves (Canis rufus
).Mol Ecol,23:4241-4255Cai B.,Li J.,Chen Y.,Wang Y.2016.Exploring the status and causes of China’s threatened reptiles through the red list assessment.Biodiv Sci,24(5):578-587
Chapple D.2003.Ecology,life-history,and behavior in the Australian Scincid genusEgernia
,with comments on the evolution of complex sociality in lizards.Herpetol Monogr,17:145-180Charlesworth D.,Charlesworth B.1987.Inbreeding depression and its evolutionary consequences.Annu Rev Ecol Syst,18:237-268
Charlesworth D.,Willis J.H.2009.The genetics of inbreeding depression.Nat Rev Genet,10:783-796
Duckett P.,Morgan M.,Stow A.2012.Tree-dwelling populations of the skinkEgernia striolata
aggregate in groups of close kin.Copeia,2012:130-134Goldberg C.S.,Edwards T.,Kaplan E.,Goode M.2003.PCR primers for microsatellite loci in the tiger rattlesnake (Crotalus tigris
,Viperidae).Mol Ecol Not,3:539-541Hu Y.B.,Nie Y.G.,Wei W.,Ma T.X.,Van Horn R.,Zheng X.G.,Swaisgood R.R.,Zhou Z.X.,Zhou W.L.,Yan L.,Zhang Z.J.,Wei F.W.2017.Inbreeding and inbreeding avoidance in wild giant pandas.Mol Ecol,26:5793-5806
Hijmans R.J.2019.geosphere:Spherical Trigonometry.R package version 1.5-10.Retrieved from https://CRAN.R-project.org/package=geosphere.
Holycross A.T.,Douglas M.E.,Higbee J.R.,Bogden R.H.2002.Isolation and characterization of microsatellite loci from a threatened rattlesnake (New Mexico Ridge-nosed Rattlesnake,Crotalus willardi obscurus
).Mol Ecol Not,2:537-539Keller L.F.,Waller D.M.2002.Inbreeding effects in wild populations.Trends in Ecol Evol,17:230-241
Konovalov D.A.,Manning C.,Henshaw M.T.2010.KINGROUP:a program for pedigree relationship reconstruction and kin group assignments using genetic markers.Mol Ecol Resour,4:779-782
Le Galliard J.F.,Gundersen G.,Andreassen H.,Stenseth N.2006.Natal dispersal,interactions among siblings and intrasexual competition.Behav Ecol,17:733-740
Li J.L.1995China Snake Island
.Dalian,China:Liaoning Science and Technology Press.Li J.L.,Sun L.X.,Wang X.P.,Bi H.T.,Wang L.P.,Wu C.,Yu Y.F.,Qu L.X.,Lin X.Z.,Yang C.J.2007.The influence of population distribution pattern ofGloydius shedaoensi
s Zhao on predatory rate.J Snake,19:12-16Li N.,Wen G.,Yang W.,Fu J.2015.Isolation and characterization of fourteen microsatellite loci for Asiatic toad (Bufo gargarizans
) at high altitude through transcriptome sequencing.Conserv Genet Resour,7:407-409Madsen T.,Shine R.,Olsson M.,Wittzell H.1999.Restoration of an inbred adder population.Nature,402:34-35.
Mckinnon L.,Gilchrist H.,Scribner K.2006.Genetic evidence for kin-based female social structure in common eiders (Somateria mollissima
).Behav Ecol,17:614-621Munguia-vega A.,Pelz-Serrano K.,Goode M.,Culver M.2009.Eleven new microsatellite loci for the tiger rattlesnake (Crotalus tigris
).Mol Ecol Resour,9:1267-1270Nie H.,Wu Y.,Qiao L.,Suo L.,Qi Y.2015.Development of novel microsatellite DNA markers for toad-headed agamaPhrynocephalus vlangalii
using next generation sequencing.Conserv Genet Resour,7:385-388Oksanen J.,Blanchet F.G.,Friendly M.,Kindt R.,Legendre P.,McGlinn D.Minchin P.R.,O’Hara R.B.,Simpson G.L.,Solymos P.,Stevens M.H.,Szoecs E.,Wagner H.2019.vegan:Community Ecology Package.R package version 2.5-5.Retrieved from https://CRAN.R-project.org/package=vegan
Olsson M.,Shine R.,Madsen T.,Gullberg A.,Tegelstrom H.1996.Sperm selection by females.Nature,383:585-585
Olsson M.,Shine R.2003.Female-biased natal and breeding dispersal in an alpine lizard,Niveoscincus microlepidotus
.Biol J Linn Soc,79:277-283Oyler-mccance S.J.,John J.S.T.,Parker J.M.,Anderson S.H.2005.Characterization of microsatellite loci isolated in midget faded rattlesnake (Crotalus viridis concolor
).Mol Ecol Not,5:452-453Perrins N.,Goudet J.2001.Inbreeding,kinship,and the evolution of natal dispersal.In:Dispersal (eds.Clobert Jet al
.).Oxford:Oxford University Press,180-190Rousset F.2008.Genepop’007:A complete re-implementation of the GENEPOP software for Windows and Linux.Mol Ecol Resour,8:103-106
Schiegg K.,Daniels S.J.,Walters J.R.2006.Inbreeding in red-cockaded woodpeckers:Effects of natal dispersal distance and territory location.Biol Conserv,131:544-552
Shine R.,Sun L.X.,Fitzgerald M.A.,Kearney M.R.2003.A radiotelemetric study of movements and thermal biology of insular Chinese pit-vipers (Gloydiusshedaoensis
,Viperidae).Oikos,100(2):342-352Shine R.,Sun L.X.,Kearney M.R.,Fitzgerald M.A.2002.Why do junenile Chinese pit-vipers (Gloydius shedaoensis
) select arboreal ambush sites? Ethology,108:897-910Støen O.G.,Bellemain E.,Sæbø S.,Swenson J.E.2005.Kinrelated spatial structure in brown bearsUrsus arctos
.Behav Ecol Sociobiol,59:191-197Van Oosterhout C.,Hutchinson W.F.,Wills D.P.M.,Shipley P.2004.Micro-checker:software for identifying and correcting genotyping errors in microsatellite data.Mol Ecol Not,4:535-538
Vanthournout B.,Greve M.,Bruun A.,Bechsgaard J.,Overgaard J.,Bilde,T.2016.Benefits of group living include increased feeding efficiency and lower mass loss during desiccation in the social and inbreeding spiderstegodyphus dumicola
.Front Physiol,7:12.Wang H.,Wang H.T.,Xiao Y.H.,Wang X.P.,Sun L.X.,Shi J.S.Lin A.Q.,Feng J.,Wu Y.H.2015.Low genetic diversity and moderate inbreeding risk of an insular endemic pit viper (Gloydius shedaoensis
):implication for conservation.J Herpetol,49(2):190-199 Asian Herpetological Research2021年2期
Asian Herpetological Research2021年2期
- Asian Herpetological Research的其它文章
- Trophic Ecology of Ansonia latidisca at Gunung Penrissen,Sarawak,North-Western Borneo
- Trophic Niche Shifts in Mountain Feirana Frogs under Human-mediated Habitat Transformations
- Higher Body Temperatures and Earlier Parturition in Response to Hypoxia Experienced by Pregnant Lizards
- Male Advertisement Call of the Endangered Leptobrachella tengchongensis (Anura:Megophryidae) from Mount Gaoligongshan,Yunnan Province,China
- Evolution of Phenotype and Mitochondrial Genome Reveals Limbless and Body-elongated Squamates may Change Their Energy Basis for Locomotion
- Phylogeny and Biogeography of Common Toad (Bufo bufo) in Xinjiang,China
