Circular RNAs: new biomarkers of chemoresistance in cancer
Jiaqi Wang, Yi Zhang, Lianyu Liu, Ting Yang, Jun Song
1Department of General Surgery, The Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, China; 2Institute of Digestive Diseases of Xuzhou Medical University, Xuzhou 221002, China
ABSTRACT Chemotherapeutics are validated conventional treatments for patients with advanced cancer. However, with continual application of chemotherapeutics, chemoresistance, which is often predictive of poor prognosis, has gradually become a concern in recent years.Circular RNAs (circRNAs), a class of endogenous noncoding RNAs (ncRNAs) with a closed-loop structure, have been reported to be notable targets and markers for the prognosis, diagnosis, and treatment of many diseases, particularly cancer. Although dozens of studies have shown that circRNAs play major roles in drug-resistance activity in tumors, the mechanisms by which circRNAs affect chemoresistance have yet to be explored. In this review, we describe the detailed mechanisms of circRNAs and chemotherapeutics in various cancers and summarize potential therapeutic targets for drug-resistant tumors.
KEYWORDS Circular RNA; chemoresistance; drug resistance; cancer
Introduction
Circular RNAs (circRNAs), which were first identified more than 4 decades ago1, are a class of endogenous ncRNAs that have a closed-loop structure, and lack 5′ caps and 3′ poly (A)tails2. Owing to their unique structure, circRNAs are more stable and less easily degraded than mRNAs3. Although circRNAs are abundant in eukaryotes, scientists initially considered them to have no meaningful function4. However, the development of high-throughput deep RNA sequencing and the application of bioinformatics technology has resulted in the confirmation of circRNAs as important molecules that are abnormally expressed and associated with poor prognosis in a variety of diseases, such as cardiovascular diseases5, diabetes6, nervous system diseases7, immune system diseases8, and cancer9.
Chemotherapeutics, which have been validated as conventional treatments for patients with advanced cancer, have widespread application in clinical practice. However, the development of chemoresistance, along with the need for chemotherapy treatment, is often predictive of poor prognosis10. Although the literature regarding drug resistance is extensive, the mechanisms involved require further clarification. In this review, we describe the relationship between circRNAs and chemoresistance, and discuss the mechanisms through which circRNAs contribute to drug resistance in different cancers.
Biogenesis of circRNAs
According to several studies, circRNAs are derived from canonical splice sites, and their biogenesis is dependent on the canonical splicing machinery11,12. However, Liang et al.13have indicated that, when pre-mRNA processing events are slowed down, nascent RNA can be directed to alternative pathways that facilitate backsplicing. The main concept of backsplicing is that looping of the intron sequences flanking the downstream splice-donor (SD) site and the upstream splice-acceptor (SA)site brings these sites close together. Formation of this circular structure is mediated by base pairing between inverted repeat elements located in the upstream and downstream introns,or by the dimerization of RNA-binding proteins (RBPs) that bind particular motifs of lateral introns14. During backsplicing, an upstream branch point attacks a downstream SD site,which subsequently attacks an upstream SA site and results in the formation of exonic circRNAs (EcircRNAs) (Figure 1A) or exon-intron circRNAs (EIciRNAs) (Figure 1B). In addition,during exon skipping, lariat formation, in which alternative exons are spliced out of the final mRNA product and are eventually included in the excised lariat, contribute to circRNA formation when the lariat undergoes internal backsplicing15.Finally, intronic lariats that escape from debranching can lead to the formation of intronic circRNAs16(Figure 1C).
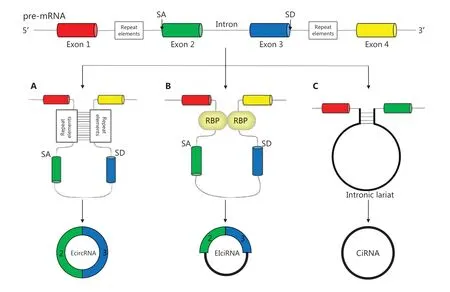
Figure 1 Biogenesis of circRNAs: (A, B) EcircRNAs and EIciRNAs can be generated from an SD site attacking an SA site, with sites with repeat elements close together; (C) Intronic circRNAs can be generated from intronic lariats that escape the debranching step of canonical linear splicing.
Biological functions of circRNAs in cancer
Functions as miRNA sponges
MicroRNAs (miRNAs) are ncRNAs that negatively regulate mRNA translation through specific binding to target sites in the 3′ untranslated regions of mRNA17. Many reported circRNAs contain miRNA response elements, which may play roles as miRNA sponges and serve as competing endogenous RNAs,thus preventing downstream target mRNAs from undergoing miRNA repression. Generally, according to most reports, one circRNA sponges a single miRNA sequence and plays a regulatory role. For example, CDR1as, which is known as a “supersponge,” strongly sponges miR-7, which contains more than 70 selectively conserved binding sites18, thereby affecting the occurrence and development of melanoma19, non-smallcell lung cancer20, and other cancers. However, some circRNAs that act as RNA sponges, such as circHIPK3 and circCCDC66,can interact and regulate multiple miRNAs simultaneously.CircHIPK3, which is derived from exon 2 of theHIPK3gene and consists solely of a large single exon (1,099 bp), may sponge as many as 9 miRNAs, as detected by luciferase reporter assays21. In addition, circCCDC66 has been reported to sponge 4 different miRNAs in various cancers22-25(Figure 2A).
Functions through RNA-binding proteins

Figure 2 Biological functions of circRNAs in cancer: (A) Functions as miRNA sponges: miRNAs can bind miRNA response elements of circRNAs and thereby alleviate repression of downstream mRNAs; (B) Functions through RNA-binding proteins: circRNAs can bind RBPs which have binding sites affecting the expression of associated genes; (C) Functions as transcriptional regulators: circRNAs can regulate transcription by binding polymerase II or U1 snRNP complexes, as well as regulating selective splicing; (D) Functions in protein translation: circRNAs with open reading frames and internal ribosome entry sites can translate protein. Moreover, m6A motif, which is sufficient to initiate translation, is recognized rich in some circRNAs, so as to empower these circRNAs to translate protein.
Some circRNAs, similarly to their roles as miRNA sponges,bind RBPs, which play a crucial role in posttranscriptional modification and mRNA translation, and act as protein sponges affecting the expression of associated genes. A recent report has revealed that circZKSCAN1 competitively binds the RBP fragile X mental retardation protein (FMRP), thus preventing its binding to β-catenin-binding protein-cell cycle and apoptosis regulator 1 (CCAR1) mRNA and consequently leading to downregulation of the transcriptional activity of Wnt signaling targets in hepatocellular carcinoma26(Figure 2B).
Functions as transcriptional regulators
CircRNAs regulate transcription through 2 recognized mechanisms. In one mechanism, circRNAs, particularly EIciRNAs(which are backspliced and retain introns), bind the polymerase II complex and interact with both the U1 small nuclear ribonucleoprotein (snRNP) complex and the promoters of their encoding genes27. Two examples of this subgroup of circRNAs are EIciEIF3J and EIciPAIP2. If the interactions between these EIciRNAs and RNA polymerase II or U1 snRNPs are blocked,the mRNA transcription of their parental genes, eukaryotic translation initiation factor 3 subunit J (EIF3J) and poly(A)-binding protein-interacting protein 2 (PAIP2), respectively, is downregulated28. Furthermore, the circRNA FECR1,which is derived from backsplicing between exon 4 and exon 2 ofFLI1, activates FLI1 expression by interacting with theFLI1promoter and subsequently induces extensive demethylation of CpG islands. Moreover, circFECR1 binds the promoters ofDNMT1andTET1, and regulates the expression of genes associated with DNA methylation and demethylation29.
The other mechanism through which circRNAs (particularly EcircRNAs) regulate transcription is selective splicing. Because some circRNAs have functional binding sites, they can compete with linear mRNAs for canonical splicing and then alter the expression of their parental genes; for instance, circMbl interacts with the splicing complex, such that Mbl promotes the expression of circMbl30. A pioneering study on titin, which was published just before the rediscovery of circRNAs, has shown that a subset of circRNAs that originate from the I-band of the titin gene are regulated by the splicing factor RBM2031. These results demonstrate another aspect of the relationship between circRNAs and selective splicing (Figure 2C).
Functions in protein translation
CircRNAs were previously considered to lack the ability to be translated, because they do not possess 5′ cap structures,poly (A) tails, or internal ribosome entry sites, all of which are essential for translation32. However, more recent studies have debunked this theory. An open reading frame in circZNF609 has been reported to be recognizable by a splicing-dependent and cap-independent mechanism33. The translation mechanism driven by N6-methyladenosine (m6A) modification has become another area of interest. YTHDC1, an m6A reader,has been found to recognize modifications on circNSUN2 and then facilitate circNSUN2 export from the nucleus to the cytoplasm; this activity directly increases the invasion ability of colorectal cancer (CRC)34(Figure 2D).
Mechanisms of chemoresistance in circRNAs
Chemotherapy plays an indispensable role in the treatment of cancer, particularly advanced cancer. However,chemoresistance emerges and then becomes an urgent obstacle to overcome. Many reports have described the mechanisms of drug resistance in cancer, which include (1) promoting drug excretion by expressing ATP-binding cassette (ABC)transporters such as ABCB1, ABCC1, and ABCG2, which are often overexpressed in drug-resistant cancer cells and result in lower drug accumulation in cells35; (2) dysregulating the expression of anti-apoptotic genes (e.g., upregulating the expression of the antiapoptotic genesBcl-2andMDM2, or repressing the expression of tumor suppressors such asp53),thus allowing cancer cells to proliferate indefinitely without constraint36; (3) enhancing the capacity for DNA repair (e.g.,in CD133 positive glioma stem cells, which show increased activation of checkpoint-related proteins and other proteins involved in the DNA damage response)37; and (4) creating a tumor microenvironment (TME) with an altered proportion of stromal fibroblasts, vasculature, and immune cells38(Figure 3). In addition to these mechanisms, many other factors, such as tumor heterogeneity, autophagy, and gene mutations, affect drug resistance39-41.
Although dozens of studies have shown that circRNAs play major roles in drug-resistance activity in tumors, the mechanisms through which circRNAs affect chemoresistance have yet to be explored. In the next section, circRNAs and their mechanisms relating to drug resistance are summarized in detail.
The roles of circRNAs in cancer chemoresistance
The mechanisms through which circRNAs regulate the development of drug resistance in cancer are shown in Tables 1 and 2.
CircRNAs affecting cell apoptosis in chemoresistance
Apoptosis is programmed cell death, which is regulated at the genetic level and removes damaged cells in an orderly and efficient manner42. The elimination of this death process is associated with unchecked cell proliferation, cancer development and progression, and cancer resistance to chemotherapies43.Consequently, dysregulation of molecules involved in apoptotic resistance often leads to treatment failure. According to extensive research, dozens of circRNAs downregulate apoptosis in cancer cells by regulating apoptosis-related genes,thereby leading to drug resistance. In the next paragraph, we briefly introduce the circRNAs that dysregulate apoptosis-related genes in cancers.

Figure 3 Mechanisms of chemoresistance: (A) Promoting drug excretion: ABC transporters enhance drug excretion, thus leading to low drug accumulation in cells; (B) Regulating apoptosis-related genes: upregulation of anti-apoptotic genes or downregulation of apoptotic genes allows cancer cells to proliferate indefinitely without constraint, thus resisting chemotherapy; (C) Enhancing DNA repair: some proteins contribute to repairing damaged DNA, thus resulting in the failure of drugs that destroy the structure of DNA; (D) Creating the TME: a hypoxic,acidic, inflammatory, and immunosuppressive TME with cancer-related fibroblasts and vessels can protect tumor cells from immune cells and killing by drugs.
In breast cancer (BC), dysregulation of circKDM4C and hsa_circ_0006528 is associated with doxorubicin resistance.CircKDM4C, which is downregulated in BC, mitigates doxorubicin resistance by sponging miR-548p and targeting PBLD,a tumor suppressor in BC44. Additionally, hsa_circ_0006528,which is upregulated in doxorubicin-resistant cell lines, functions as an miRNA spongeviathe miR-7-5p/Raf1 axis and contributes to doxorubicin resistance45. CDR1as is thought to promote 5-FU resistance in BC by competitively inhibiting miR-7 and consequently regulating CCNE1 expression46.CircRNF11147and circABCB1048, both of which sponge miRNAs, induce paclitaxel resistance. In cervical cancer, hsa_circ_0023404 and circMTO1 both enhance the resistance to cisplatin and function as miRNA sponges that regulate beclin1 and p6249,50. In CRC, hsa_circ_0007031 and hsa_circ_0000504 promote 5-FU resistance by sponging miR-885-3p and miR-485-5p, and ultimately upregulate AKT3 and BCL2 expression.Moreover, hsa_circ_0048234 represses 5-FU resistanceviathe miR-671-5p/EGFR axis51. In addition, circPRKDC enhances resistance to 5-FUviathe miR-375/FOXM1/Wnt/β-catenin pathway52. A recent study has revealed that circCCDC66 promotes oxaliplatin resistance in CRC by interacting with a set of miRNAs and consequently regulating the expression of multiple genes that facilitate cell apoptosis and survival.In addition, oxaliplatin induces circCCDC66 expression byphosphorylating DHX9, which is involved in multiple pathways associated with the DNA damage response53. In gastric cancer (GC), circPVT154and circFN155are significantly upregulated in paclitaxel-resistant GC tissues and cells, and these circRNAs have been predicted to regulate apoptosisviaan miRNA sponge mechanism. Sun et al.56have found that hsa_circ_0000520 enhances the sensitivity of herceptin in GC cells by targeting at PI3K/AKT. In glioma, circHIPK3 has been found to enhance cell proliferation and temozolomide(TMZ) resistance, not only by sponging miR-421/ZIC557but alsoviathe miR-524-5p/KIF2A/PI3K/AKT axis58. In hepatocellular carcinoma (HCC), both hsa_circ_000341859and hsa_circ_10150560enhance cell apoptosis and the sensitivity to cisplatin through an miRNA sponge mechanism. In lung cancer,3 circRNAs leading to cisplatin resistance by sponging miRNAs have been identified: hsa_circ_0076305, hsa_circ_0007385,and hsa_circ_008513161-63. Moreover, hsa_circ_0004350 and hsa_circ_0092857, both of which decrease cells’ sensitivity to cisplatin, can interact with RBPs and may have a synergistic effect with the parental EIF3a gene64. In addition, hsa_circ_0096157 enhances cisplatin resistanceviaregulating the p21/CCND1/CDK4/BCL2 apoptosis signaling pathway65, and CDR1as has been identified to contribute to both cisplatin and pemetrexed chemoresistance in lung adenocarcinomaviathe EGFR/PI3K signaling pathway66. Other circRNAs functioning as miRNA sponges in lung cancer include hsa_circ_000248367and hsa_circ_001129268against paclitaxel, hsa_circ_0004015 against gefitinib69, hsa_circ_0003998 against docetaxel70,and hsa_circ_0002130 against osimertinib71. In osteosarcoma (OS), hsa_circ_0001258 has been found to decrease the resistance to doxorubicin, cisplatin, and methotrexateviamiR-744-3p/GSTM272. Furthermore, hsa_circ_000467473and hsa_circ_000007374repress cell apoptosis and resist methotrexate by sponging miRNAs, and hsa_circ_001569 represses cisplatin sensitivityviaWnt/β-catenin75. In ovarian cancer,the overexpression of circCELSR176and circTNPO377results in paclitaxel resistance by repressing cell apoptosis and regulating the cell cycle. In addition, CDR1as, in contrast to its role in BC and lung adenocarcinoma, is downregulated in cisplatin-resistant ovarian cancer tissues, and it decreases the resistance of cells to cisplatinviathe miR-1270/SCAI axis78.In prostate cancer (PCa), Gao et al.79have discovered that hsa_circ_0000735 overexpressed in PCa tissue, and downregulated of hsa_circ_0000735 boosts docetaxel sensitivity by promoting cell apoptosis. Hsa_circ_0035483 contributes to gemcitabine-induced autophagy and facilitates resistance of renal cell carcinoma (RCC) to gemcitabineviathe miR-335/Cyclin B1 axis80; similarly, circEIF6 facilitates cisplatinresistanceviamiR-144-3p/TGF-α in thyroid carcinoma81.In chronic myeloid leukemia (CML), hsa_circ_000991082and hsa_circ_008014583repress cell apoptosis and sensitivity to imatinib by acting as miRNA sponges. CircBA9.3, which promotes proliferation and inhibits apoptosis in cancer cells,enhances resistance to tyrosine kinase inhibitors (TKIs) by increasing the expression of the c-ABL1 and BCR-ABL1 oncoproteins, which contribute to the immortality of leukemic cells84. CircPAN3, a circRNA highly expressed in doxorubicin-resistant acute myeloid leukemia (AML) cells, functions
as an miRNA sponge that regulates autophagy and influences the expression of apoptosis-related proteinsviathe AMPK/mTOR signaling pathway, and ultimately contributes to doxorubicin resistance85. In general, circRNAs reverse drug sensitivity by regulating the expression of apoptosis-associated genes, thus repressing cancer cell death.
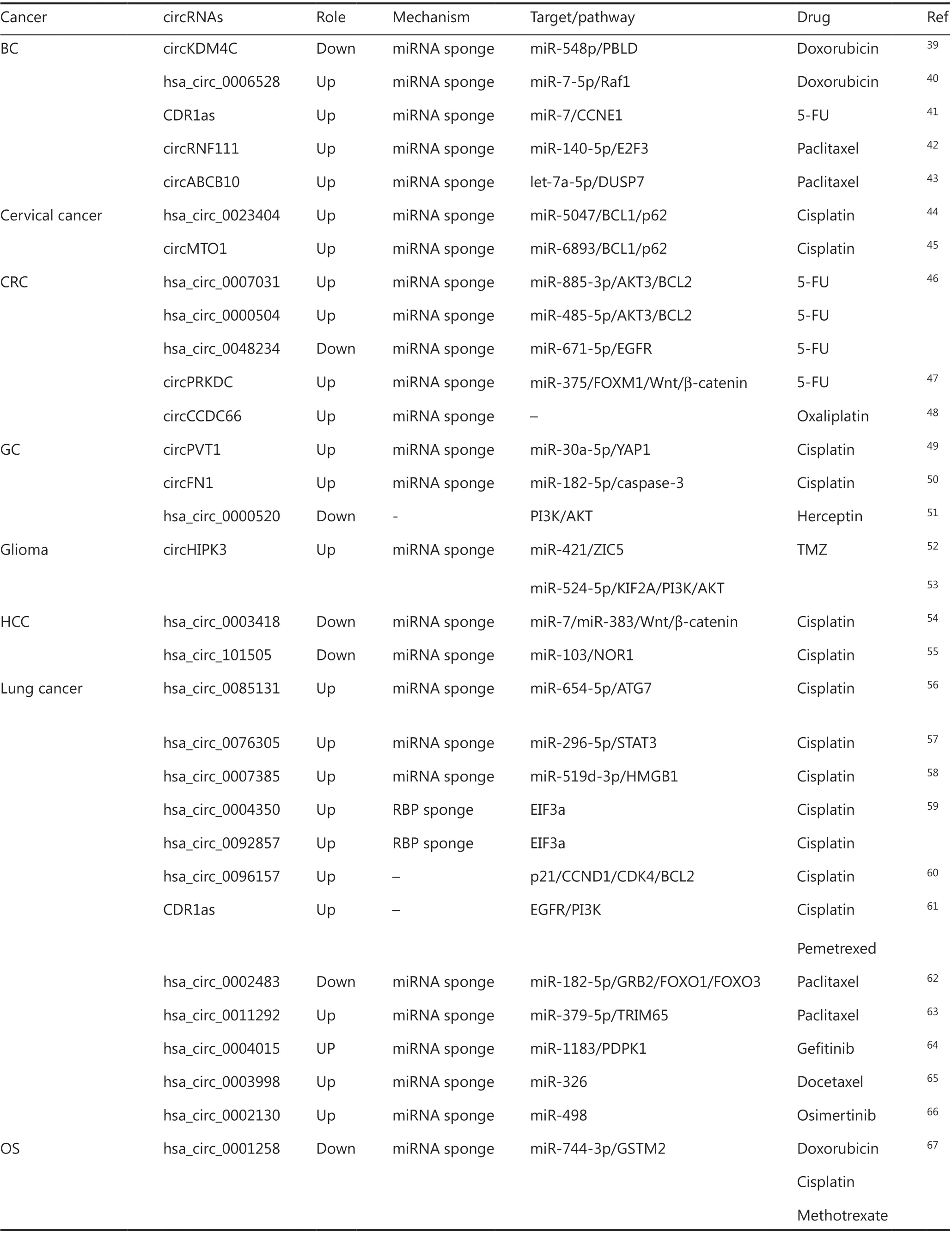
Table 1 CircRNAs affecting cell apoptosis in chemoresistance

Table 1 Continued

Table 2 CircRNAs with other mechanisms in chemoresistance
CircRNAs promoting drug excretion in chemoresistance
The ABC transporter family comprises 48 genes subdivided into 7 subfamilies: ABCA through ABCG86. Most these genes encode large membrane proteins that function in the energy dependent transport of xenobiotics, metabolites, and signaling molecules across cell membranes, often against their concentration gradients87. Two ATP-binding sites in ABC transporters have been reported to participate in the formation of the ATP binding pocket, and their interactions are essential for coupling ATP hydrolysis to drug transport and ultimately cause chemoresistance88. According to recent studies, a group of circRNAs can target at ABC transporters and then promote drug excretion, thereby inducing drug resistance.
On the one hand, some circRNAs indirectly enhance the functionality of ABC transporters. Hsa_circ_0005963 has been found to promote oxaliplatin resistanceviathe miR-122/PKM2 axis89, thus enhancing glycolysis and ATP production and facilitating oxaliplatin excretion by ABC transporters in CRC. On the other hand, circRNAs can upregulate the ABC transporter family directly. Overexpression of hsa_circ_0004674 confers resistance to doxorubicin and cisplatinviathe miR-490-3p/ABCC2 axis in OS73. CircPVT1 not only facilitates resistance to cisplatin and doxorubicin by enhancing the expression of ABCB1 in OS90, but also functions through miR-145-5p/ABCC1 in lung cancer, thus resulting in resistance to cisplatin and pemetrexed91. Hsa_circ_0109291,like circPVT1, targets ABCB1 by sponging miR-188-3p and consequently rendering cisplatin ineffective in oral squamous cell carcinoma (OSCC)92. He et al.93have found that hsa_circ_0007031 represses 5-FU sensitivityviathe miR-133b/ABCC5 axis in CRC. Xu et al.94have demonstrated that circ-MTHFD2 enhances GC resistance to pemetrexed by sponging miR-124 and consequently increases the protein expression of ABCB11. CircCEP128 significantly promotes the expression of ABCG2 and resistance to TMZ by sponging miR-145-5p95in glioma. In addition, in multiple myeloma (MM), overexpression of hsa_circ_0007841 leads to high levels of ABCG2 and consequently decreases doxorubicin’s efficacy96. These results suggest that circRNAs can directly or indirectly regulate the expression of ABC transporters and eventually induce drug resistance by promoting drug excretion.
CircRNAs promoting DNA repair in chemoresistance
DNA repair is a biological event in which cells identify and correct DNA damage induced by chemotherapy and other incidents. The main mechanisms comprise base excision repair, nucleotide excision repair, mismatch repair, homologous recombination repair, nonhomologous end-joining, and interstrand crosslink repair, which can confer resistance to DNA-targeting chemotherapeutics97. Therefore, understanding the activity of different DNA repair pathways in individual tumors and the correlation between DNA repair function and drug response will be key in the selection of targeted drugs for patients. Because ncRNAs are a widespread concern, multiple circRNA-mediated DNA damage repair mechanisms leading to drug resistance have been discovered.
Eg5, a member of the kinesin family, whose members are crucial for maintaining separation of the half-spindles, is a microtubule-based motor protein required for the formation and maintenance of the bipolar spindle98,99. Monastrol,a reversible, cell-permeable small molecule, selectively inhibits Eg5 and consequently disrupts the structure of tumor cells’chromosomes and DNA100. A recent study has identified a circular RNA, circMTO1, that is significantly downregulated in monastrol resistant cells and reverses monastrol resistance through RBP targeting at TRAF4/Eg5101.
Cisplatin, first synthesized by Michele Peyrone and approved by the US FDA for use in testicular and ovarian cancer in 1979102, binds DNA and forms adducts once inside tumor cells. The primary forms of DNA damage are N7-d (GpG) and N7-d (ApG) intrastrand DNA-platinum adducts, which cause substantial kinking of DNA103. In GC, both circAKT3 and hsa_circ_0026359 have been verified to induce cisplatin resistance. Huang et al.104have identified that circAKT3 promotes DNA damage repairviamiR-198/PIK3R1. Hsa_circ_0026359 sponges miR-1200 and upregulates POLD4105, and low expression of hsa_circ_0026359 has been reported to weaken DNA repair systems106. In addition, circPRMT5 overexpression in lung cancer enhances cisplatin resistance by upregulating REV3 L, which encodes the catalytic subunit of DNA polymerase ζ and is responsible for translesional replication107.In summary, circRNAs activate related proteins or pathways,thus maintaining chromosome or DNA stability, and promote DNA damage repair, thereby leading to chemotherapy failure.
CircRNAs influencing the TME in chemoresistance
The TME, the cellular environment in which tumors exist,has been characterized as hypoxic, acidic, inflammatory, and immunosuppressive. Beyond tumor cells, the TME contains the extracellular matrix, surrounding blood vessels, and other non-malignant cells38. The TME is considered a “sanctuary of the devil,” in that immune cells in the TME secrete cytokines,inflammatory factors, and chemokines and drive the epithelial to mesenchymal transition (EMT) process in cancer cellsviavarious pathways. In turn, cancer cells interact with immune cells and induce cellular plasticity and the release of immunosuppressive substances; consequently an immunosuppressive microenvironment that promotes immune escape and drug resistance is created108.
Several studies have shown that circRNAs alter the TME by regulating EMT and immune escape, and ultimately result in drug resistance. Hu et al.109have reported that circUBE2D2 accelerates the progression of EMT and enhances resistance to tamoxifen in BC. CircPVT1, which is expressed at high levels in paclitaxel-resistant GC cells, sponges miR-124-3p and upregulates ZEB1, a crucial transcriptional inhibitor of E-cadherin that expedites EMT110. CircMET overexpression promotes an HCC immunosuppressive tumor microenvironment by inducing EMT through the miR-30-5p/Snail/DPP4/CXCL10 axis and leads to anti-PD1 therapy resistance111. In lung cancer, circFGFR1 has a critical role in immune evasion and induces resistance to anti-PD1 based therapyviathe miR-381-3p/CXCR4 axis112. In addition, circCCDC66 promotes EMT and represses cisplatin sensitivity in lung adenocarcinoma113. Through this mechanism, circRNAs mainly alter the TME by regulating EMT, thus allowing cancer cells to escape drug and immune injury.
CircRNAs regulating CSCs in chemoresistance
CSCs are a group of self-renewing cells that have high tumorigenic capacity and play crucial roles in chemoresistance, accelerating tumor regrowth after therapy114. CSCs share many characteristics with regenerative stem cells, such as self-renewal, multipotency, and the reversibility of their quiescent state115,116. After chemotherapy, residual bodies enriched in CSCs remain, which can promote tumor recurrence, re-grow metastatic tumors, and enhance resistance to drugs117,118.
To date, 3 circRNAs have been reported to influence CSClike phenotypes. CircELP3, which is elevated in hypoxic environments, enhances cisplatin resistance in bladder cancer by displaying a high self-renewal capacity, as evidenced by high levels of sphere formation and stem cell marker expression119.Similarly, both hsa_circ_001680, resisting irinotecan in CRC120,and circCAP4, resisting cisplatin in lung cancer121, strengthen stem cell-like traits, self-renewal and tumor-initiating capacityviaincreasing stemness associated signatures and sphere formation. In conclusion, circRNAs increase cancer cells’ stemness, thus protecting tumors against chemotherapeutics.
CircRNAs mediating endocrine therapy in chemoresistance
Endocrine therapy plays a unique role in the treatment of cancer, particularly in BC and PCa122. The estrogen receptor,which is expressed in approximately 70% of all BC, is considered the main factor inducing BC123. Tamoxifen, an anti-estrogen that decreases estrogen-induced effects by blocking the estrogen receptor in breast tissue, remains a cornerstone treatment that has significantly improved clinical outcomes124.Simultaneously, as an effective therapeutic target in PCa, the androgen receptor, is expressed in nearly all PCa and results in an abnormal gene expression profile, including cell cycle regulators, transcription factors, and genes important for cell survival, lipogenesis, and secretion125. Antiandrogenic drugs such as enzalutamide inhibit the progression of PCa by blocking androgens126. However, the development of drug resistance is inevitable after the first chemotherapy, and some circRNAs have been reported to participate in the progression of endocrine-related resistance.
In BC, 2 circRNAs that are downregulated in BC cells have been found to act synergistically with tamoxifen. Sang et al.127have demonstrated that overexpressed hsa_circ_0025202 sponges miR-182-5p and upregulates FOXO3a, an inhibitor of tamoxifen resistance in BC. CircBMPR2 acts as an miR-553 sponge, thereby preventing it from repressing the tumor suppressor USP4, and subsequently mitigates tamoxifen resistance128. To date, downregulation of 3 circRNA in PCa cells has been reported to lead to increased resistance to enzalutamide.Low hsa_circ_0004870 expression results in the upregulation of its parental gene RBM39 and the downstream target gene AR-V7, whose expression is positively correlated with enzalutamide resistance129. Another circRNA that targets AR-V7 is hsa_circ_0001427. Interestingly, hsa_circ_0001427 is positively correlated with miR-181c-5p, which directly binds and degrades the ARv7 3′ untranslated region and may stabilize miR-181c-5p, thus ultimately decreasing cells’ resistance to enzalutamide130. Finally, CircUCK2, whose expression is downregulated in enzalutamide-resistant cells, may function as an miRNA sponge through the miR-767-5p/TET1 axis131.These results suggest that circRNAs regulate the effects of endocrine drugs by changing relative receptor levels, thereby potentially providing new directions in hormone-dependent cancer therapy.
Discussion
Because of the vast amount of research on cancer, a variety of targeted chemotherapy drugs have been widely used in clinical settings and have achieved good efficacy. The most common mechanism of chemotherapy is inducing cancer cell death by damaging DNA to inhibit tumor growth. In contrast, the mechanisms of tumor drug resistance include promoting drug excretion and DNA repair, inhibiting apoptosis, and creating a TME resistant to the damage caused by chemotherapy drugs.
According to the literature, the multi-drug resistance of circRNAs may manifest different roles in different cancers. For example, CDR1as promotes resistance to cisplatin, 5-FU, and pemetrexed in BC and lung cancer, but it enhances the sensitivity of ovarian cancer to cisplatin. Although CDR1as appears to play a crucial role in the chemoresistance of cancers, whether an opposite effect might occur in different cancers, owing to the heterogeneity and origin of tumors, remains unknown.Moreover, reports on circRNA-related chemoresistance have largely focused on their mechanisms as miRNA sponges,which functionviainteractions between circRNA and miRNAs that regulate downstream genes and ultimately influence drug effects. In contrast, only few studies have reported the mechanisms of circRNAs as they relate to RBPs. Although circRNAs are known to regulate transcription and translation,these 2 mechanisms have not been reported in terms of circRNA-related drug resistance. Instead, dozens of studies have shown that ncRNAs influence chemoresistance by regulating transcription and translation; for example, miR-5787 affects the translation of mitochondrial cytochrome c oxidase subunit 3 (MT-CO3), thus regulating cisplatin resistance132, and miR-182-5p, regulates GLI2, a transcriptional regulator of the hedgehog signaling pathway133. Therefore, a knowledge gap regarding the mechanisms of circRNA-related drug resistance remains to be further explored.
Currently, RNA-seq, owing to its high throughput, accuracy, and sensitivity, is the gold standard for screening circRNAs associated with drug resistance. Most studies use RNA-seq to screen circRNAs with differential expression patterns between resistant and nonresistant cell lines, to explore mechanisms of chemoresistance. Interestingly, some studies have shown not only that circRNAs influence multi-drug resistance but also that drug feedback regulation on circRNAs exists. For example, circCCDC66 is a circRNA that is induced by treatment with oxaliplatin and subsequently promotes resistance to oxaliplatin-induced apoptosis.Future studies may reveal new mechanisms for the occurrence and enhancement of drug resistance. Blocking these mechanisms could substantially support the elimination of chemoresistance. This research direction is especially applicable to the clinical study of drug resistance and may provide new directions for the exploration of the mechanisms of circRNA-related chemoresistance.
With the recent development of liquid biopsy, examination of secretions, and other clinical techniques, circRNAs have broad application prospects as early screening indicators of drug sensitivity. For example, as mentioned above,hsa_circ_0025202 has an anti-oncogenic role in HR-positive breast cancer, and it may be exploited as a novel marker for tamoxifen-resistant breast cancer. Meanwhile, with research advances, many clinical trials are underway that are targeting ncRNAs, including a mimic of the tumor suppressor miRNA miR-34, which has reached phase I clinical trials134. Because circRNAs are emerging research targets, relevant clinical studies remain lacking, although many reports have already shown that circRNAs are associated with cancer progression and drug resistance, possibly because of the high toxicity and low specificity of the inhibitors, which are difficult to put into clinical use. In general, how to accurately target circRNAs to tumor cells and minimize the damage to healthy cells is an urgent problem that must be solved.
Conclusions
In summary, circRNAs have been shown to be involved in tumor progression and to be closely associated with drug resistance. We summarized the mechanisms through which circRNAs interact with RBPs and miRNAs, and consequently influence drug delivery, reverse drug-induced apoptosis, and ultimately lead to drug resistance. However, many unknown circRNAs and mechanisms of drug resistance remain to be discovered and elucidated. Therefore, in-depth exploration in this field may provide a broader space for further research and treatment of chemoresistance mechanisms in tumors. circRNAs may have extensive roles in the treatment of tumors in the future.
Grant support
This research was supported in part by the National Natural Science Foundation of China (Grant Nos. 81702435 and 82073133), the Natural Science Foundation of Jiangsu Province (Grant Nos. BK20170264 and BK20191154), and the Six Talent Peaks Project in Jiangsu Province (Grant No.WSW-050).
Conflict of interest statement
No potential conflicts of interest are disclosed.
 Cancer Biology & Medicine2021年2期
Cancer Biology & Medicine2021年2期
- Cancer Biology & Medicine的其它文章
- Biological roles and potential clinical values of circular RNAs in gastrointestinal malignancies
- Biomaterial-based platforms for cancer stem cell enrichment and study
- sLex expression in invasive micropapillary breast carcinoma is associated with poor prognosis and can be combined with MUC1/EMA as a supplementary diagnostic indicator
- Nanomaterial-based delivery vehicles for therapeutic cancer vaccine development
- Targeting FGFR in non-small cell lung cancer: implications from the landscape of clinically actionable aberrations of FGFR kinases
- Three-dimensional collagen-based scaffold model to study the microenvironment and drug-resistance mechanisms of oropharyngeal squamous cell carcinomas
