OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1
WANG Baoxiang ,LIU Yan ,WANG Yifeng ,LI JingfangSUN ZhiguangCHI MingXING YungaoXU BoYANG BoLI JianLIU JinboCHEN TingmuFANG ZhaoweiLU BaiguanXU DayongBabatunde Kazeem BELLO
(1Lianyungang Institute of Agricultural Sciences,Collaborative Innovation Center for Modern Crop Production,Lianyungang 222006,China;2State Key Laboratory of Rice Biology,China National Rice Research Institute,Hangzhou 311400,China;#These authors contributed equally to this work)
Abstract:We created CRISPR-Cas9 knock-out and overexpressing OsbZIP72 transgenic rice plants to gain a better understanding of the role and molecular mechanism of OsbZIP72 gene in stress tolerance,which has remained largely elusive.OsbZIP72 was expressed and integrated into rice transgenic plant genomes,and the OsbZIP72 transcript in overexpression lines was elicited by salinity,abscisic acid(ABA)and drought stresses.OsbZIP72 overexpressing plants showed higher tolerance to drought and salinity stresses,while knock-out transgenic lines showed higher sensitivity to these stresses.The differentially expressed genes(DEGs)from RNA-sequencing data encompassed several abiotic stress genes,and the functional classification of these DEGs demonstrated the robust transcriptome diversity in OsbZIP72.Yeast one-hybrid,along with luciferase assay,indicated that OsbZIP72 acted as a transcriptional initiator.Remarkably,electrophoresis mobility assay revealed that OsbZIP72 bound directly to the ABAresponsive element in the OsHKT1;1 promoter region and activated its transcription.Overall,our findings revealed that OsbZIP72 can act as a transcriptional modulator with the ability to induce the expression of OsHKT1;1 in response to environmental stress through an ABA-dependent regulatory pathway,indicating that OsbZIP72 can play a crucial role in the ABA-mediated salt and drought tolerance pathway in rice.
Key words:abscisic acid;basic leucine zipper;drought stress;high-affinity potassium transporter;rice;salinity stress;transgenic plant
Environmental abiotic stress,such as high salinity and drought,which constitutes the predominant basis of crop waste worldwide,has posed a serious threat to the existence and development of plants.Plant survival in the environment under various abiotic stresses relies on the stimulation of the molecular mechanism network involved in the transmission of stress signals,the recognition and the manifestation of stress-related genes.Such stress causes specific biochemical and physiological retorts in plants,and understanding the functions of these stress-related genes is essential for unraveling the molecular mechanisms of plant capacity to withstand stress and improving plant adaptability to stress through the genetic scheme.The use of stressinducible genes has improved the ability of plants to tolerate cold,salinity and drought stresses(Roychoudhury et al,2013).
Rice is one of the plants with the largest bZIP family transcription factor.The bZIP family has been discovered in nearly all eukaryotes(Deppmann et al,2006).Currently,56 proteins of the bZIP family have been found in humans(Deppmann et al,2006),and 89 in rice(Nijhawan et al,2008;Ji et al,2009).There are two parts of the bZIP α-helix,namely the N-terminal region,which is positively charged and preferentially binds to ACGT corecis-element containing nucleotides(Izawa et al,1993;Foster et al,1994),and the C-terminal region,which is rich in leucine residue and mediates the bZIPs homo- or hetero-dimerization(Landschulz et al,1988;Ellenberger et al,1992).bZIP transcription factor family members form homo- or hetero-dimers complexes by binding to the target nucleotide sequence.The bZIP family genes consist of a basic region in their domain,which is positively charged and is responsible for the DNA binding associated with the extra sequence of bZIP leucine.The dimer forms of the bZIP family form a structure like chopsticks through the dimerization of their leucinezipper parts,and each basic region fragment connects with a palindromic part in the highly crucial nucleic acid gouge.In plant development,the bZIPs are actively involved in various cellular and physiological activities,and a variety of bZIP families play important roles in the transmission of hormonal stimulation and the signal pathways of plants(Uno et al,2000;Jakoby et al,2002;Rodriguez-Uribe and O’Connell,2006).The bZIP families are effectors of stress activation,such as endoplasmic reticulum,high temperature,oxidative stress,mitogenic activation and cytokine activation.Additionally,bZIPs also play significant roles in different growth processes.Several bZIP family members,for example,OsbZIP52(Liu et al,2012),OsABF2 or OsAREB1(Yang et al,2001;Hossain et al,2010a;Jin et al,2010;Yang et al,2011),OREB1 or OsABI5(Zou et al,2007,2008;Hong et al,2011)and OSBZ8(Nakagawa et al,1996;Mukherjee et al,2006),play crucial roles in the abscisic acid(ABA)signaling pathways by transcriptionally regulating ABA-responsive element(ABRE)-containing genes through binding to ABA elements.OsbZIP52functions in low-temperature response conditions was studied inOsbZIP52overexpressing lines where many abiotic stress-related genes,such asOsTPP1,OsLEA3andRab25,are downregulated,resulting in a greater decrease in the ability to withstand low temperature and drought stresses in rice plants(Liu et al,2012).These results revealed thatOsbZIP52has a negative control over drought and cold stresses.OsABI5transcript,induced by high salt concentration,has a detrimental effect on the response to salinity stress(Zou et al,2007,2008).InArabidopsis,the expression levels ofAtbZIP38(ABF4/AREB2),AtbZIP36(ABF2/AREB1)andAtbZIP37(ABF3)in response to salt stress,ABA signal and dehydration are induced(Uno et al,2000).Unlike OsABI5 and OsbZIP52,OsbZIP46 acts as a promising way to regulate abiotic stress through an ABA-dependent pathway,and is capable of binding to ABRE,which can be induced by numerous environmental stresses such as salinity,drought and low temperature(Hossain et al,2010a;Jin et al,2010;Tang et al,2012).Many other bZIP proteins,including OsABF1(Lu et al,2009;Hossain et al,2010b)OsbZIP23(Xiang et al,2008)and OsbZIP16(Chen et al,2012),are also ABRE-binding factors and function as positive transactivators in response to drought tolerance in rice.A number of studies have shown that OsABF1/OsbZIP72 can withstand high salt stress(Xiang et al,2008;Lu et al,2009;Hossain et al,2010b;Chen et al,2012).Transgenic rice plant overexpressingOsbZIP72exhibits high responsiveness to ABA,increases the expression levels of certain genes such as LEAs in response to ABA,and revamps the tolerance of rice plants to drought(Lu et al,2009).
Despite growing evidence that bZIP72 transcription factor has a biological impact in combating abiotic stress in plants,understanding the molecular mechanism of bZIP72 in rice through integrative and functional studies is still largely limited.In this study,we generated 35S::OsbZIP72overexpressing and CRISPR knock-outOsbZIP72transgenic rice plants,which displayed different phenotypes and higher survival rates under abiotic stress.Our findings indicated that bZIP72 played a crucial role in the transcriptional regulatory pathway of ABA signal transmission via direct activation the ABRE of rice high-affinity potassium transporter geneHKT1;1.
RESULTS
Analysis of OsbZIP72 overexpressing and knock-out transgenic rice plants
To unravel the ability ofOsbZIP72to withstand abiotic stress,transgenic rice plants overexpressingOsbZIP72were generated.PCR using hygromycin specific primers and qRT-PCR were applied to verify thatOsbZIP72has been fused into transgenic plant genomes and expressed.OsbZIP72knock-out lines(crbzip72)were generated using the CRISPR-Cas9 technique,in which different types of deletion were detected by sequencing the target mutation site(Fig.1-A and -B).Thecrbzip72lines harbored two base deletions(GG or TG)in the first exon and shifted the open reading frame ofOsbZIP72,which caused premature termination.The changes of amino acid sequences are shown in Fig.S1.The expression ofOsbZIP72was analyzed in independent transformant lines using qRT-PCR.Higher levels of expression were detected inOsbZIP72transgenic lines compared to the wild type(WT)andcrbzip72(Fig.1-C and -D).Two overexpressed T2lines(OxbZIP72-1andOxbZIP72-2)and knock-out lines(crbzip72-1andcrbzip72-3)were used for further analysis.
OsbZIP72 enhances tolerance to abiotic stress
Plants from the T2generation ofOsbZIP72-overexpressing lines(OxbZIP72)and knock-out lines(crbzip72)were used for salinity and drought screening.WT andcrbzip72lines withered significantly during drought stress compared to theOxbZIP72lines,which remained strong and flowered 10 d after withholding water(Fig.2-A).The survival rates ofOxbZIP72-1andOxbZIP72-2were 66.0% and 66.7%,respectively,compared to 12.0% in WT(Fig.2-B).OsbZIP72-overexpressing plants lost water more slowly and reduced ion leakage compared to the WT andcrbzip72lines(Fig.2-C and -D).The relative water content(RWC)of leaves in the WT andcrbzip72lines were 26.1% and 24.0% respectively,whereas theOxbZIP72transgenic plants retained 75.0% RWC at 10 d after withholding water(Fig.2-E).In the WT andcrbzip72plants,the malondialdehyde(MDA)and H2O2contents were significantly higher than those in the overexpressingOsbZIP72plants,indicating thatOsbZIP72improved resistance to oxidative stress(Fig.2-F and -G).During the 15 d tolerance check on 150 mmol/L NaCl,leaf interveinal chlorosis were noted,OsbZIP72-overexpressing lines were less prone and more salt tolerant than the WT andcrbzip72plants(Fig.3-A).The survival rates ofOxbZIP72-1andOxbZIP72-2under 150 mmol/L NaCl treatment were 70.0% and 72.2%,respectively,compared to a significant reduction noted in WT that was only 14.0%(Fig.3-B).The WT andcrbzip72lines showed a significant decrease in RWC compared with higher values observed inOsbZIP72-overexpressing plants at 10 d after treatment(Fig.3-C).Additionally,a major increase in chlorophyll content was noted during salinity stress inOsbZIP72-overexpressing plants(Fig.3-D).Conversely,the contents of MDA and H2O2were substantially lower in theOxbZIP72plants than in the WT andcrbzip72lines(Fig.3-E and -F).The results suggested that the tolerance ofOxbZIP72transgenic lines to salt and drought stresses was positively correlated with relative expression ofOsbZIP72,indicating thatOsbZIP72can regulate responses to salinity and drought stresses.Taken together,our results revealed that the overexpression ofOsbZIP72in transgenic plants resulted in greater drought and salinity tolerance.
OsbZIP72 regulates transcription of genes involves in abiotic stress

Fig.1.Expression analysis and molecular characterization of OsbZIP72 gene.

Fig.2.Evaluation of drought and oxidative stress tolerance in OsbZIP72 overexpression(OxbZIP72)and knock-out(crbzip72)transgenic plants.
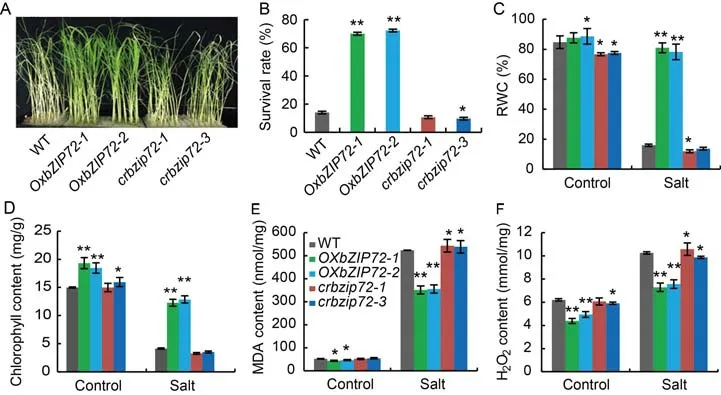
Fig.3.Assessment of salinity stress tolerance in OsbZIP72 overexpression(OxbZIP72)and knock-out(crbzip72)transgenic plants.
To dissect the potential downstream genes inOxbZIP72lines,we conducted RNA-sequencing on theOxbZIP72and WT leaf samples.In total,we found 1 328 differentially expressed genes(DEGs)inOxbZIP72plants,of which 512 genes were up-regulated and 816 were down-regulated(Table S1).The DEGs encompassed many abiotic stress genes.The functional grouping of the DEGs clearly demonstrated the robust transcriptome divergence inOxbZIP72.GO(Gene Ontology)investigation revealed that the DEGs were notably classified into biosynthesis and metabolic functions,regulation of cell organelles and biosynthesis(Fig.S2 and Table S2).In the volcano plot,the degree of differential gene expression was also revealed(Fig.S3).Nevertheless,KEGG(Kyoto Encylopedia of Genes and Genomes)explored that DEGs associated with hormone signal transmission,chlorophyll metabolism,metabolic pathways,porphyrin and biosynthesis of secondary metabolites were prominent(Fig.S4 and Table S3).Our hypothesis that OsbZIP72 may be involved in abiotic stress tolerance was supported by the KEGG results.qRT-PCR investigation of DEGs and certain reported rice abiotic stress regulator genes inOxbZIP72andcrbzip72lines was conducted with multiple biological replicates.Several salinity and drought stress genes,such asOsHKT6(LOC_Os02g07830),OsDST(LOC_Os03g57240),OsHOX24(LOC_Os02g43330),OsCCA1(LOC_Os08g06110),potassium transporter1;1(OsHKT1;1,LOC_Os04g51820),OsNHX1(LOC_Os07g47100.2),rice homeobox geneOsHOX22(LOC_Os04g45810),protein phosphatase 2C(OsPP2C,LOC_Os02g05630.1)and MYB2 transcription factorLOC_Os03g20090were generally up-regulated in the overexpressing plants ofOsbZIP72,demonstrating that the key reported abiotic stress regulator selected and the DEGs are under similar pathways regulated by theOsbZIP72gene(Fig.4-A).To further investigate theOsbZIP72function,we analyzed its expression profile under various abiotic stresses.qRT-PCR analysis revealed thatOsbZIP72transcript levels were strongly induced by drought,salt and ABA treatments(Fig.4-B).These findings showed thatOsbZIP72can participate in modulation of the abiotic stress stimulus.
OsbZIP72 binds to ABRE cis-element of OsHKT1;1 promoter to activate its transcription

Fig.4.Transcript of OsbZIP72 regulated genes in OsbZIP72 overexpression(OxbZIP72)and knock-out(crbzip72)transgenic plants by qRT-PCR.
To uncover the target genes of OsbZIP72,we first investigated the promoter region ofOsHKT1;1due to its higher transcript level inOsbZIP72overexpression lines(Fig.4-A),which may signify thatOsHKT1;1can be the target of OsbZIP72.We discovered that the OsbZIP72 protein can bind to the promoter ofOsHKT1;1through yeast one-hybrid assay(Y1H)(Fig.5-A).Luciferase(LUC)transactivation assay was also performed in rice protoplast to verify that OsbZIP72 mediated activation onOsHKT1;1.Strong activation of LUC was detected in pro35S:bZIP72:tNOS,compared with the negative control,suggesting that OsbZIP72 activated theOsHKT1;1transcriptionin vivo.Besides,the activation ofOsHKT1;1increased significantly with the addition of exogenous ABA,suggesting that ABA may induce the activation ofOsHKT1;1by OsbZIP72(Fig.5-B and -C).In addition,we performed electrophoresis mobility shift assay(EMSA)to further demonstrate the interaction of the OsbZIP72 protein with the promoter fragment ofOsHKT1;1.In the 1 kb upstream transcription start site promoter region ofOsHKT1;1,we found one ABRE element(ACGTG),one MYB regulatory element(TAACTG),one methyl jasmonate(MeJA)responsive element(CGTCA)and onecis-acting element(TGACG)involved in the MeJA-responsiveness by using the online tool Plant Cis-Acting Regulatory Elements(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/PlantCARE)(Lescot et al,2002)(Fig.5-D).Consequently,the shift speed of probe 4(P4),representing the ABRE element(992 bp)to transcription starting site(TSS),was retarded by the OsbZIP72 protein(Fig.5-E),however,the binding was competed off when unlabeled probe was added,indicating the binding is highly specific(Fig.5-F).We then performed site mutation(s)for the 5 bp ABRE element in the 1 kb promoter region and used a number of mutated sequences as probes to further clarify the OsbZIP72-binding sites.When CGT was mutated,the OsbZIP72 binding was prevented(Fig.5-G).This finding showed that no less than three nucleotides are required for the OsbZIP72 binding.However,the OsbZIP72 binding was obviously reduced when performed the mutation of single nucleotide.The mutations of CGT and TG resulted in the inhibition of binding to OsbZIP72,suggesting that these nucleotides are required for OsbZIP72 binding.Taken together,the OsbZIP72 protein binds mainly to the ACGT region of the ABRE element in 992 regions upstream TSS of theOsHKT1;1promoter.
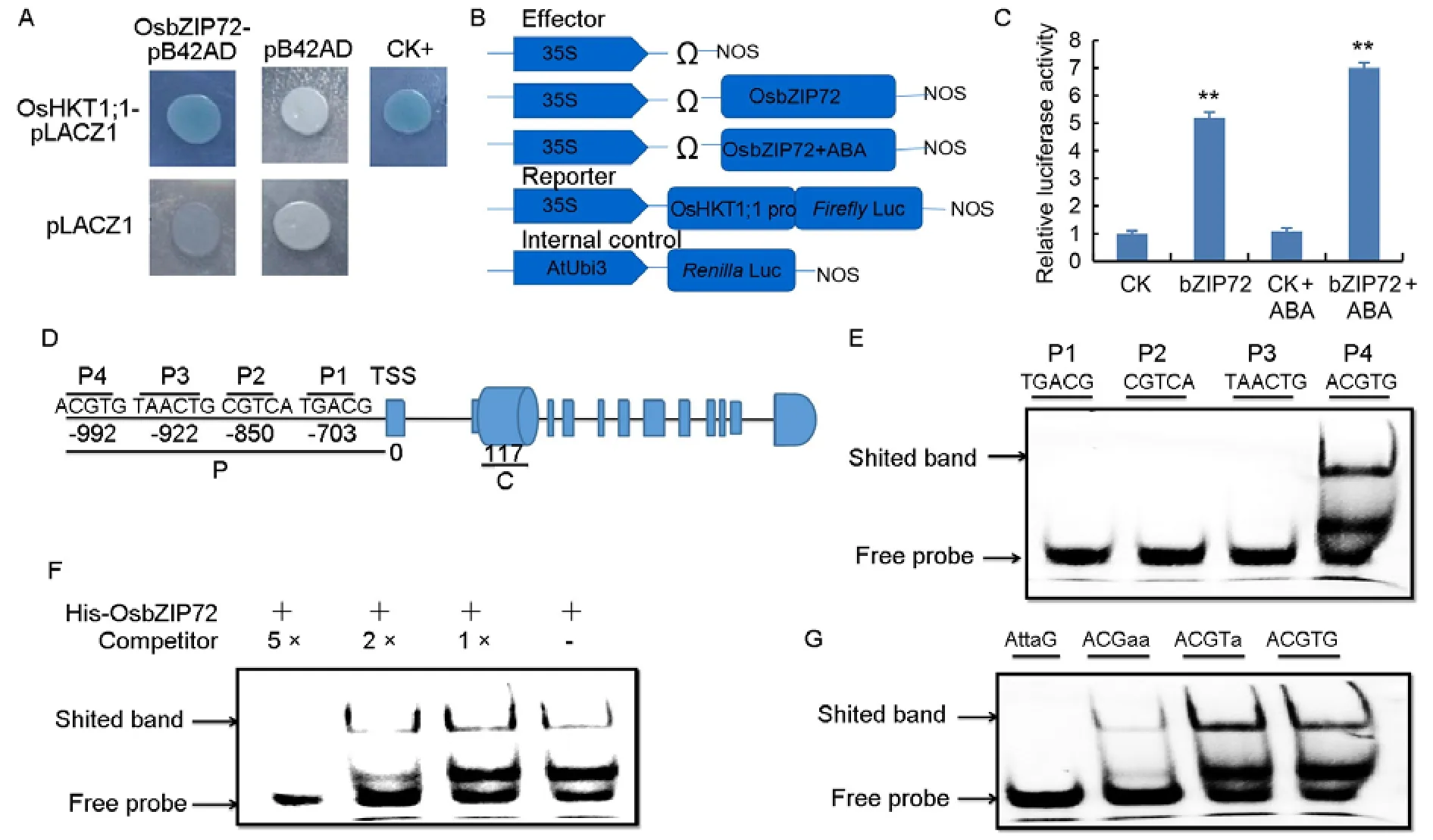
Fig.5.OsbZIP72 directly binds to abscisic acid responsive element(ABRE)in promoter of OsHKT1;1 and activates its transcription.
DISCUSSION
Transgenic approaches offer an efficient tool to enhance crop traits.In the present study,transgenic rice overexpressingOsbZIP72lines were generated and identified.Hygromycin selection was utilized to detect the transgene in the genomes of transgenic lines.The qRT-PCR assay revealed the expression ofOsbZIP72gene in transgenic lines.Transcripts ofOsbZIP72were enhanced by salt,drought and ABA,which demonstrated thatOsbZIPsmight be functionally associated with abiotic stress responses in the ABA signaling pathway.Previous reports have revealed that the expression levels of various members of OsbZIP group,such asOsbZIP10,OsbZIP12,OsbZIP23,OsbZIP46,OsbZIP62,OsbZIP66andOsbZIP72,were significantly enhanced by dehydration,salinity or cold stress treatment(Nijhawan et al,2008).bZIP transcription factors(Groups A,S and G)play an important role in the resistance to various stresses in plants.For example,the Group A bZIP transcription factor,AtAREB1,enhances drought tolerance in vegetative tissues through ABRE-dependent ABA signaling(Fujita et al,2005).Consistent with our result,the expression ofOsbZIP72was induced by salinity and drought stresses.Drought and salt are important stresses in plant habitat,which regularly affect plant development and growth.Improving plant tolerance to these abiotic stresses through biotechnology technique,therefore,holds substantial potential for increasing food production with insufficient availability of freshwater supply.Nevertheless,this requires a deeper insight into the molecular mechanisms of plant tolerance to drought and salt(Xiong et al,2002;Zhu,2002;Wang et al,2003;Yamaguchi-Shinozaki and Shinozaki,2006;Seki et al,2007).Several groups ofAtbZIPsoverexpression plants display enhanced drought-tolerance(Kang et al,2002;Oh et al,2005).Constitutive overexpression ofOsbZIP72drastically increases the resistance of plants to salinity and drought stresses.Conversely,a considerable sensitivity ofcrbzip72lines to drought and salinity stresses was noted.Transgenic overexpressingArabidopsisbZIP geneAtABI5and its rice orthologous geneOsABI5react strongly to exogenous ABA(Lopez-Molina et al,2001;Zou et al,2007).Our study showed that the MDA and H2O2contents of bothOxbZIP72lines were lower than those in the WT andcrbzip72plants in the drought and salt treatments,indicating that less toxic substances were produced in theOxbZIP72lines.
We revealed the genetic evidence thatOsHKT1;1was activated by the OsbZIP72 transcription factor.OsHKT1;1is abundantly located in the leaf blade phloem region and serves to regulate sodium buildup in leaves(Wang et al,2015).Sodium toxicity occurs mainly in plant leaves,and photosynthesis and other metabolic activity also take place in plant leaves(Munns and Tester,2008).In this study,we found that tolerance to salinity stress from overexpressingOsbZIP72plants occurs through transcriptional modulation ofOsHKT1;1.This is corroborated by the previous reports that the expression ofOsHKT1;1is regulated in plants by salinity stress,osmotic stress and potassium deprivation(Wang et al,1998;Horie et al,2005,2007;Ren et al,2005;Shkolnik-Inbar et al,2013).We further demonstrated the interaction between OsbZIP72 andOsHKT1;1promoter using various approaches,indicating that the OsbZIP72 binding was essential for theOsHKT1;1promoter function.bZIP72 transcription factor has been reported to play a substantial role in the ABA signaling pathway by transcriptionally regulating ABRE element in the promoters of the downstream genes through binding to ABRE(Lu et al,2009).Transcriptional modulation induced by stress in plants extensively depends on dehydration responsive element and ABRE that can be found in the promoter segment of stress-triggered genes.Our EMSA results demonstrated that OsbZIP72 precisely and uniquely bound to the ABRE element of theOsHKT1;1promoter,suggesting a novel mechanism of OsbZIP72-mediated gene transcriptional regulation.Previous reports revealed that two OsbZIPtranscription factors,OsABI5 and TRAB1,can bind ABRE element.TRAB1 is cable of binding to Motif I ofOsemgene promoter(Hattori et al,1995),Rab16Apromoter(Skriver et al,1991)andEmpromoter from wheat(Guiltinan et al,1990).ABREs are a group ofcisacting DNA elements that were discovered during promoter exploration of numerous ABA-regulated genes in plants,which are extensively distributed in the upstream region of many plant regulatory genes and need to be notably recognized by appropriate transcription factors for perfect gene expression.Many ABA-mediated physiological responses are modulated by a group of bZIP genes that bind with ABRE element in the promoter of genes(Landschulz et al,1988;Busk and Pagès,1998).In this study,we revealed that the overexpression ofOsbZIP72alleviated various abiotic stresses in rice plants when compared with WT.In line with this finding,our RNA-seq and qRT-PCR results revealed a number of DEGs which are functionally related to abiotic stress.Surprisingly,we observed thatOsHKT1;1,a family of high affinity potassium transporters,was highly up-regulated.Moreover,in ourin vitroY1H andin vivoluciferase transactivation assay,OsbZIP72 transactivatedOsHKT1;1expression,indicatingOsHKT1;1is the exact target of OsbZIP72.OsHKT1;1has been well established to play a crucial role in reducing the accumulation of Na+in shoots to withstand salt stress.HKT1;1recoup sodium ions from the root xylem,thus minimizing the accumulation of sodium salt in the shoots(Mäser et al,2002).Mäser et al(2002)and Møller et al(2009)reported thatArabidopisHKT1;1mutant exhibits hypersensitivity to salinity stress.Consistently,HKT1;1overexpression inArabidopsisshows improved resistance to salinity stress(Møller et al,2009).
Despite the significance of an extensive understanding of the molecular mechanism by which the plant bZIP72 super family recognizes the ABRE element,our work demonstrated the binding of OsbZIP72 to the ABRE element of theOsHKT1;1promoter and to directly regulate its transcription as far as we know.Taken together,we presumed thatOsbZIP72plays crucial roles in various response processes to environmental stresses such as drought and high salinity via binding to ABREs and transcriptionally regulate ABA-induced ABRE-containing gene.
METHODS
Plant growth processes and stress tolerance evaluation
The transgenic rice plant Nipponbare was procured from China National Rice Research Institute,Hangzhou,China.Plants in the T2generation ofOsbZIP72-overexpressing lines,CRISPRCas9 transgenic lines and wild type were used for stress tolerance assay.Seeds were first surface-sterilized in 70%ethanol and drenched in 50% sodium hypochlorite for 30 min,and then rinsed six times in double distilled water and sown on half-strength Murashige and Skoog(MS)medium containing 0.3% plant agar and then moved to a growth chamber(28 °C ±2 °C,12 h light / 12 h dark photoperiod under 60% relative humidity).During salinity stress treatment,14-day-old seedlings were transplanted in solution fortified with 150 mmol/L NaCl for a period of 9 d;then plants were stabilized under ordinary growth conditions for an additional 7 d.The third leaf from the top of the plant was detached after withholding watering of plant for 10 d when plants became evidently droop and used for drought tolerance and RWC determination(Guo et al,2006;Lu et al,2009).For measurement of water loss,the third leaf from the top was detached from plants and immediately weighed,then placed in a hood for a series of periods(up to 140 min).Plant weight was recorded every 20 min,and the percentage of the initial fresh weight at individual time interspace was denoted as the rate of water loss.All data are means of three biological replicates.Rice leaf samples were submerged overnight in 10 mL of distilled water for measurement of ion leakage,and the conductivity denoted as C1 was recorded by heating the leaf in boiling water at 100 °C for 20 min and then allowed to cool down to 25 °C,after which the conductivity denoted as C2 was recorded again.Ion leakage was determined as the C1 to C2 rate.Rice seedlings were incubated in 80%acetone and the extract was obtained for chlorophyll estimation after shaking at room temperature for 24 h in the dark,and then centrifuged at 10 000 ×gfor 10 min.The H2O2and MDA contents were determined using the kits(No.A064-1-1 and A003-1-1,Nanjing Jiancheng Bioengineering Institute,Nanjing,China)according to the manufacturer’s recommendations.
RNA isolation,qRT-PCR and RNA-seq
Total RNA was extracted using plant RNA extraction kit(Takara,Beijing,China).The method of Hou et al(2015)was adopted for reverse transcription to generate cDNA and qRT-PCR analysis.All data generated were reported as mean of three biological replicates.OsActin1was used to normalize the relative expression level of all the analyzed genes.The primers used are listed in Table S4.For the RNA-seq,the qualities of the total RNA extracted from the WT andOsbZIP72overexpression plants were carefully determined using a Thermo Scientific NanoDrop 2000 spectrophotometer(Waltham,MA,USA)and Agilent 2100 Bioanalyzer(Santa Clara,CA,USA).Three independent biological replicates of each sample were used for analyses.The RNA with good quality was then prepared for sequencing library construction.The Illumina HiSeq™ 2000 platform(Illumina,Foster City,CA,USA)was used to perform the high-throughput sequencing to obtain a good quality library,reads with substandard quality,polluted-adaptor,unspecified base and counts lower than 20 reads per million were expunge.The obtained clean reads were aligned with the transcripts of rice genes in RGAP(http://rice.plantbiology.msu.edu)using BOWTIE2(http://bowtie-bio.sourceforge.net/bowtie2/index.shtml),after which the gene expression levels were analyzed using RSEM(RNA-seq by expectation maximization)(Li and Dewey,2011).DEGs between the WT andOxbZIP72were detected using the EBSeq(Leng et al,2013),the cutoff value together with the fragments per kilobase of transcript per million mapped reads(|log 2 ratio| ≥1;Pvalue<0.01)were selected as thresholds to determine the significant differences in gene expression.
Vector construction and plant transformation
The complete open reading fragment(ORF)ofOsbZIP72was ligated into a vector pU1301 driven by theCaMV35Spromoter(Zhang et al,2010).The rice variety Nipponbare was used as the recipient.Agrobacteriumstrain EHA105 was used for transformation.Agrobacterium-mediated transformation was performed as described by Hiei et al(1994).For the CRISPRCas9 construction,the method of Ma et al(2015)was adopted to createcrbzip72plants.Annealed double strand nucleotide sequences of the gDNA were ligated into the pYLgRNA-OsU3 usingBsaI enzyme site.For the recombinant protein expression,the coding sequence ofOsbZIP72was amplified and cloned into pET28a(Merck,Darmstadt,Germany).His-bZIP72,recombinant proteins were induced inE.colistrain Rossetta,and purified by 6× His-Tagged Protein Purification Kit(CWBIO,Beijing,China).
Yeast one-hybrid assay
The ORF ofOsbZIP72was fused with GAL4 AD domain in a pB42AD vector using the Clontech One-Hybrid System(Clontech,Dalian,China),and the promoter fragment ofOsHKT1;1was ligated into a pLacZ2u vector.The confirmed plasmid was transformed to yeast strain EGY48 and plated on SD/-Ura/-Trp plates,then spotted on SD/-Ura/-Trp plates containing 1% raffinose,1% of 10× BU salts(900 mL water,70 g Na2HPO4·7H2O,34.5 g NaH2PO4·H2O),80 mg/L X-Gal and 2% glactose(Clontech,Dalian,China).The transactivation property of OsbZIP72 was confirmed when the blue colonies were visualization on the medium.NF-YB1-pB42AD andWx-pLacZ2u were used as positive controls and the empty vector as a negative control.
Luciferase transient transcriptional activity assay
The coding sequence ofOsbZIP72was cloned into a ‘None’vector as an effector and the promoter region ofOsHKT1;1into a 190fLUC vector as a reporter.The method of Xie and Yang(2013)was adopted for rice protoplast preparation and transformation,after which the transformed protoplasts were re-suspended in 50 μL lysis buffer and the luciferase activity was immediately measured.Then,the firefly luciferase(fLUC)activity was subsequently measured after adding 100 μL firefly luciferase assay buffer into the lysate and finally,the renilla luciferase(rLUC)activity was measured after adding 100 μL stop &renilla luciferase substrate buffer.Luciferase®Reporter Assay System(Promega,Madison,USA)was used to measure all the luciferase activity according to the manufacturer’s instruction.The relative luciferase activity was calculated as the proportion of fLUC and rLUC(fLUC/rLUC).
Electrophoresis mobility shift assay(EMSA)
EMSA probes were commercially synthesized by the Genescript Biotechnology Co.,Ltd.(Nanjing,China).The synthesized probes were labeled by annealing together with equal amount of the complementary single-stranded biotinylated oligonucleotides using the EMSA Biotin Labeling Kit(Cat No.GS008,Beyotime,Shanghai,China).The protein/nucleic acid binding chemical reaction included 5 nmol purified His-tagged OsbZIP72 recombinant protein,gel-shift binding buffer for EMSA and 2 nmol biotin-labeled probes.Non-labeled DNA was used as a competitor.Purified His-bZIP72 recombinant protein was pre-incubated with the binding buffer at 25 °C for 25 min before addition of the biotin-labeled probe and incubated at 25 °C for 25 min.Gel electrophoresis was then performed on 6%polyacrylamide gel which has been pre-run for 30 min at 4 °C using 0.5× Tris-Borate-EDTA buffer,after which the DNA probes were then moved to a charged nylon membrane(Beyotime,Shanghai,China)and subsequently UV crosslinked to the membrane at 120 mJ/cm2for 1 min using a CL-1000 Ultraviolet Crosslinker(Upland,CA,USA).The signal was then visualized using chemiluminescence(Pierce,Waltham,USA).
ACKNOWLEDGEMENTS
This study was supported by the earmarked funds for China Agriculture Research System(Grant No.CARS-01-61),National Science and Technology Support Program of China(Grant No.2015BAD01B01),Science and Technology Support Program of Jiangsu Province,China(Grant Nos.BE2016370-3 and BE2017323),the Natural Science Foundation of Jiangsu Province,China(Grant No.BK20161299),the Financial Grant Support Program of Lianyungang City,Jiangsu Province,China(Grant Nos.QNJJ1704 and QNJJ1912)and National Natural Science Foundation of China(Grant No.31701395).The authors are grateful to Prof.ZHANG Jian for providing the OsbZIP72 seeds and his valuable suggestions during the process of writing the manuscript.We also thank WANG Chunming,YU Jun and GAO Ming for their suggestions and technical supports.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science;http://www.ricescience.org.
Fig.S1.Changes in amino acid sequences ofcrbzip72mutants.
Fig.S2.Gene Ontology(GO)investigation of differentially expressed genes revealing barplot of remarkably enriched GO terms.
Fig.S3.Volcano plot to visualize the extent of differential expression of genes.
Fig.S4.Molecular interaction network of genes and genomes(KEGG pathway).
Table S1.Deferentially expressed genes.
Table S2.Go term investigation for Gene Ontology(GO).
Table S3.Enriched Kyoto Encyclopedia of Genes and Genomes(KEGG)pathway.
Table S4.List of primers used in this study.
- Rice Science的其它文章
- Editing of Rice Endosperm Plastidial Phosphorylase Gene OsPho1 Advances Its Function in Starch Synthesis
- Valuation of Rice Postharvest Losses in Sub-Saharan Africa and Its Mitigation Strategies
- Rice Bran Oil:Emerging Trends in Extraction,Health Benefit,and Its Industrial Application
- Combined Drought and Heat Stress in Rice:Responses,Phenotyping and Strategies to Improve Tolerance
- SB1 Encoding RING-Like Zinc-Finger Protein Regulates Branch Development as a Transcription Repressor
- Genomic Predictio of Arsenic Tolerance and Grain Yield in Rice:Contribution of Trait-Specific Markers and Multi-Environment Models