SB1 Encoding RING-Like Zinc-Finger Protein Regulates Branch Development as a Transcription Repressor
ZENG Xiaoqin,ZHUANG Hui,CHENG Qinglan,TANG Jun,YANG Fayu,HUANG Mingjiang,WANG Ziyi,LI Zhongcheng,ZHU Honghui,CHEN Rui,HE Guanghua,LI Yunfeng
(Rice Research Institute,Key Laboratory of Application and Safety Control of Genetically Modified Crops,Academy of Agricultural Sciences,Southwest University,Chongqing 400715,China;#These authors contributed equally to this work)
Abstract:Inflorescence structure of rice,including the number and length of branches,and the density of the spikelet,can greatly affect the number of grains per panicle,which is one of the key factors in yield compositions.Here we identified five allelic mutants sb1-1/2/3/4/5 that related to branch development of rice.In these mutants,the branch meristem fate was prolonged sharply,resulting in delay of transition from branches to spikelets,and then increased the numbers of branches and spikelets per panicle.SB1 encodes a nuclear RING-like domain protein of SHI/LRP/SRS family and strongly expressed in branch meristems.The results of protein interaction and chromatin immunoprecipitation further suggested that SB1 directly repressed the expression of DEP1,TAW1,MOC1 and IP A1 by interacting with a co-repressor complex to affect acetylation level of histone H3 on target regions.Thus,we proposed that SB1 is a transcription repressor of branch meristem activity by widely and negatively regulating a series of genes that maintain branch meristem fate.
Key w ords:branch development;grain number per panicle;rice;RING-like zinc-finger;transcription repressor;panicle arthitecture
In grass crops,grain number per panicle(GNP)is one of the most important yield components(Wang and Li,2011).Panicle architecture consisting of the number of branches,the length of branches,and grain density on branches,determines the GNP(Li et al,1998;Teng et al,2002).In the past two decades,dozens of genes related to panicle architecture have been characterized in rice,some of which play crucial roles in determining GNP and then yield.
In rice,primary branches(PBs),secondary branches(SBs)and lateral spikelets(LSs)are all originated from axillary meristem,which compose the panicle with main axis and topic spikelet(TS)originating from apical meristem.Therefore,the genes participating in the initiation,formation and maintaining of axillary meristem in panicle are first regulators for panicle architecture and GNP,such asLAX PANICLE1(LAX1),LAX2andMONOCULM1(MOC1)in rice.They all encode transcription factor,and the loss-of-function mutants of them all display sharp decreasing of branch and spikelet number(Li et al,2003;Skirpan et al,2008;Tabuchi et al,2011).It is proved thatLAX1can interact withLAX2andMOC1respectively to regulate the development of axillary meristem(Tabuchi et al,2011),but the targets of them are still unclear up to date.InArabidopsis,LATERAL SUPPRESSOR(LAS)(an ortholog ofMOC1)can directly regulate REVOLUTA(REV,a HD-ZIPIII protein),which can activate meristem maintaining geneSHOOT MERISTEMLESS(STM),to promote the initiation and formation of axillary meristem(Shi et al,2016).LATERAL FLORET 1(LF1,an ortholog ofREVin rice)regulates lateral floret formation in rice spikelets by activatingOSH1(an ortholog ofSTMin rice)(Zhang et al,2017).However,a similar pathway ofMOC1-LF1-OSH1has not been proved in branch formation in rice panicle development.
The transition from branch meristem(BM)to spikelet meristem(SM)is also considered as a key process for panicle architecture.The delay of the transition can elongate the BM fate and then results in more and longer branches,and then more spikelets,whereas the earlier transition results in less spikelets.InArabidopsis,the mutual restriction ofTERMINAL FLOWER 1(TFL1,an inflorescence meristem identity gene)andLEAFY(LFY,a floral organ identity gene)determines the inflorescence architecture(Bradley et al,1997;Ratcliffe et al,1999;Mimida et al,2001).RICE CENTRORADIALIS1(RCN1)andRCN2,two orthologs ofTFL1,maintain the BM fate in rice,overexpression of which leads to the delayed transition from BM to SM,and then more branches and spikelets(Nakagawa et al,2002).However,ABERRANT PANICLE ORGANIZATION 2(APO2)/RFL,functions on BM but not SM in rice,an opposite effect with its orthologLFY.It is also proved thatAPO2/RFLcan be activated by a DOF-domain transcription activatorSHORT PANICLE 3(SP3),and interact withAPO1,to delay the transition from BM to SM.In mutants ofapo1,apo2andsp3,the numbers of branches and spikelets are all decreased significantly(Rao et al,2008;Ikeda-Kawakatsu et al,2012).In addition,TAWAWA1(TAW1)encodes a nuclear protein and also participates in determining the fate of BM.In gain-of-function mutanttaw1-D,the BM activity is increased and the differentiation of SM is delayed,which improves the formation of more SBs and spikelets(Yoshida et al,2013).Two E-class MADS-box factors,OsMADS1andOsMADS34/PAP2,might accelerate the differentiation of SM.Overexpression ofOsMADS1leads to the less numbers of branches and spikelets,whereas thepap2mutant(one of the loss-of-function ofOsMADS34)displays the more numbers of branches and spikelets(Kobayashi et al,2010;Wang et al,2017).
Extensive research has demonstrated that plant hormones play crucial roles in panicle architecture,such as cytokinin.Grainnumber1a(Gn1a),a major QTL of GNP,encodes cytokinin oxidase 2(OsCKX2)which is responsible for cytokinin degradation.In the natural allelic variant with low-level expression ofGn1a,the accumulation of cytokinin leads to largely increased branch and spikelet numbers and thus grain yield(Ashikari et al,2005).It has been confirmed that multiple genes participate in panicle development by regulating the expression ofGn1a.The zinc finger transcription factor DROUGHT AND SALT TOLERANCE(DST)activatesGn1aexpression by directly binding to its promoter(Li et al,2013),while the chromatin interacting factor VIN3-LIKE 2(VIL2)directly restrictsGn1aexpression by binding with its promoter region(Yang et al,2019).When the loss-of-function ofDSTresults in low-levelGn1aexpression and then increases branches,overexpression ofVIL2leads to increased branches as low expression ofGn1a.ThemicroRNA156-IPA1-DEP1pathway regulates panicle development through cytokinin pathway,in whichIDEAL PLANT ARCHITECTUTRE1(IPA1)(OsSPL14/WEP)can directly activateDENSE AND ERECT PANICLE1(DEP1)expression by binding with its promoter,and then high expression ofDEP1repressesGn1a(Huang et al,2009;Lu et al,2013).The mutation ofLARGERPANICLE(LP)increased panicle branching by down-regulating theGn1atranscript(Li et al,2011).In addition,LONELYGUY(LOG)(a cytokininactivating enzyme),Grain Number per Panicle1(GNP1,the gibberellin oxidaseGA20ox1)and transcript activatorSP3regulate branch development by affecting cytokinin synthesis(Kurakawa et al,2007;Wu et al,2016).
Some genes which are involved in heading date and transporting nutrients can also control panicle architecture,maybe because of the delayed heading date or increased nitrate in panicles prolonging the BM activity.Grains.Height.Date-7(Ghd7)encodes a CCT-domain protein,high expression of which greatly increases GNP and the overall yield per plant under long-day conditions,whileGhd8andOsCOL13have similar effects(Xue et al,2008;Wei et al,2010;Yan et al,2011;Sheng et al,2016).Nitrate transporter 1/peptide transporter(NPF)family members(NPF2.2,NPF4.1/sp1,NPF6.3,NPF7.3andNPF7.7)have been proved to regulate panicle branching by transporting nitrate or other unknown nutrients.Overexpression of most of them can increase panicle branch number by enhancing nitrogen utilization(Li et al,2009;Hu et al,2015;Huang et al,2018;Wang et al,2018).
To sum up,the genes related to GNP are involved in multiple networks and they have many members,some of which have been used to improve GNP and thus yield in the long period of productive practice,such asSP1,DEP1,IPA1,Gn1aandTWA1(Ashikari et al,2005;Xue et al,2008;Li et al,2009;Jiao et al,2010;Li F et al,2010;Li W Q et al,2010;Lu et al,2013).However,as the exhausted potential of old genes,there is an urgent need to characterize new genes related to panicle architecture and GNP in rice.In the present study,we identified five allelic mutants,which were characterized by significant increase of the numbers of branches and spikelets,and namedsuper branch 1(sb1).SB1gene encodes a new RINGlike zinc finger protein,locates in nuclear and is expressed highly in BM.By analysis of qPCR and chromatin immunoprecipitation(ChIP)assays,we foundSB1participates in direct repressing multiple known genes that related to panicle development,includingIPA1,DEP1,TAW1andMOC1.Therefore,our data implied thatSB1acts as a transcription repressor and functions on accelerating the transition of BM to SM in panicle architecture of rice.
RESULTS
Characterization of sb1 mutant phenotype
We identified five allelic recessive mutants namedsb1-1/2/3/4/5.Compared with the wild type plants,the mutants showed no significant difference in plant height or tiller number but the panicles ofsb1generated more branches.A mature wild type rice panicle was composed of a main axis,branches [PBs,SBs,barely tertiary branches(TBs)] and spikelets(TS and LS)produced on the branches(Fig.1-A and -B).In contrast,although there was no significant difference in the numbers of PBs betweensb1-1/2/3/4/5and the wild type(Fig.1-C),the numbers of SBs,TBs and spikelets per panicle of the five allelic mutants were significantly increased(Fig.1-D to -F).Among them,the numbers of SBs ofsb1-1/2/3/4/5reached 92(+104.4%),59(+31.1%),79(+75.5%),64(+42.2%)and 68(+51.1%),respectively,whereas it was only 45 in the wild type(Fig.1-D).Particularly,the wild type panicle usually produced few TBs,while thesb1-1/3/5produced 84,34 and 13 TBs,respectively(Fig.1-E).The numbers of spikelets per panicle were increased remarkably in thesb1mutants.There were the highest number of spikelets per panicle insb1-1with an average of 473(+125.2%),and the lowest number insb1-2with an average of 291(+38.6%),whereas the average number of spikelets in the wild type was only 210(Fig.1-F).However,the increase of spikelet numbers in thesb1mutants did not increase grain number,and thesb1-1/2/3/4/5mutants generated only about 82,175,130,190 and 176 grains per panicle,respectively.Some of them were significantly lower than the average number of grains in the wild type(190 grains per panicle)(Fig.1-G).It was because the seed-setting rates ofsb1-1/2/3/4/5were only 17.7%,64.8%,33.6%,70.0% and 56.4%,which were significantly lower than 90.2% in the wild type(Fig.1-H).

Fig.1.Phenotypes of panicles in wild type(WT)and sb1-1/2/3/4/5 mutants.
In addition,we observed the development of branches and spikelets in young panicles using a scanning electron microscopy.As expected,more branch primordia were observed in thesb1-1panicles(about 0.5 cm in length),while the branch primordia rarely appeared at the similar stage of the wild type panicles(Fig.S1-A to -D).Next,we detected the expression of genes that involving in the BM activity.The results showed that the expression levels ofLAX1,RCN1,OSH1andMOC3were significantly upregulated in thesb1-1panicles(Fig.S1-E).Therefore,our data suggested thatSB1might be responsible for accelerating the transition from BM to SM in panicles,and the mutations ofSB1prolonged the BM fate and then resulted in more secondary and tertiary branches instead of producing spikelets.
Map-based cloning of SB1
All F1plants of the cross 56S ×sb1showed normal phenotype.In the F2population,2 874 plants were obtained in total,among which 2 198 plants showed normal phenotype and 693 plants with mutant phenotype.Genetic analysis indicated that the numbers of wild type and mutant individuals fit the expected 3:1 ratio(χ2= 1.73<χ20.05= 3.84),which meant thesb1phenotype was controlled by a single recessive gene.
To identify theSB1gene,the 693 mutated individuals were used as mapping population.Linkage analysis revealed that theSB1locus was associated with the simple sequence repeat(SSR)markers SSR1 and SSR2 on the long arm of chromosome 9.Subsequently,the candidate region was further delimited to a 122-kb interval between two single nucleotide polymorphism(SNP)primers SNP-1 and SNP-2,where 13 annotated genes were predicted(Fig.2-A).Sequencing analysis revealed that a G-T substitution in the last base of the first exon ofLOC_Os09g36160occurred in thesb1-1mutant.The mutation caused the 183th codon changed into termination codon and led to premature translation termination.Sequencing analysis also revealed that other four mutants had mutational sites in the candidate geneLOC_Os09g36160.A 29-bp fragment deletion since the 166th base and a 4-bp deletion since the 298th base occurred insb1-2andsb1-3,respectively,which both led to frameshift mutation.sb1-4had a T-A substitution on the 661th base of the second exon and led to Tyr-Asn substitution of the 221th amino acid.sb1-5had a 51-bp fragment deletion since the 664th base and led to 17 amino acids deletion in the SB1 protein(Fig.2-B).To verify whetherLOC_Os09g36160was responsible for thesb1phenotype,a 4091-bpSB1-GFP(green fluorescent protein)fusion expression vector which contained 3070-bp upstream sequence and 1021-bpSB1open reading frame(ORF)sequence without termination codon was transformed intosb1-1(Fig.2-C).Totally,17 transgenic plants were obtained,and we then analyzed the coding sequence ofLOC_Os09g36160and examined the GFP signal to identify the positive plants.Among them,11 plants showing double peaks at the mutant site also showed obvious GFP fluorescent signal,which revealed that the combination plasmid was successfully transformed into these 11 plants.All these 11 transgenic plants displayed normal panicles with similar numbers of branches and spikelets per panicle with those in the wild type(Fig.2-D).These results finally confirmed thatLOC_Os09g36160gene was indeed theSB1gene.
SB1 gene encodes a nuclear-located SHI/LRP/SRS family protein
SB1encodes a RING-like zinc finger transcription factor that belongs to SHI/LRP/SRS family.InArabidopsis,the SHI family contains 10 members:SHORT INTERNODES(SHI),STYLISH1 and 2(STY1 and STY2),SHI-RELATED SEQUENCE3(SRS3)to SRS8,and LATERAL ROOT PRIMORDIUM(LRP)(Fridborg et al,2001).Phylogenetic analysis revealed that there were five SHI/LRP/SRS protein family members in rice.SB1 had a poor homology with all SHI/LRP/SRS proteins inArabidopsisand had a close evolutionary relationship with ZmLRL4 and ZmLRL5 in maize.

Fig.2.Map-based cloning of SB1.

Fig.3.SB1 encodes a RING-like zinc finger protein.
TheLOC_Os01g724901andLOC_Os05g320701had high homology and close branching relationship with LRP,which was widely studied inArabidopsisand maize(Fig.3-A).Previous studies have shown that the sequences of the SHI/LRP/SRS family proteins were highly diverse,with highly conserved regions:a C3HC3H RING-like domain and an IGGH domain(Fridborg et al,2001;Kuusk et al,2002,2010).Then,the protein motif prediction program MEME and protein sequence alignment program DNAMAN were used to analyze the full-length amino acid sequences of SHI/LRP/SRS family proteins in rice,maize andArabidopsis.The result showed that there were three highly conservative domain/motifs:a RING-like structure domain,an IGGH domain and an FRCVR domain in the SHI/LRP/SRS family proteins.In previous results,the FRCVR domain was not identified separately(Figs.3-B,S2 and S3).
To confirm the subcellular localization of SB1 protein,the fluorescence signal of the SB1-GFP fusion protein was observed directly in the SB1P:SB1:GFP complementary transgenic plants using a confocal laser scanning microscope.The GFP signal of SB1-GFP fusion protein was localized in the nucleus(Fig.S4).These results together demonstrated that SB1 was a nuclear-located SHI/LRP/SRS family transcription factor.
Spatiotemporal expression pattern of SB1
Firstly,the expression ofSB1gene in different rice tissues was detected by qPCR.The results revealed thatSB1gene was strongly expressed in the young panicle,while it was weakly expressed in the shoot,tiller bud,leaf and sheath,and almost not expressed in the root(Fig.4-A).Furthermore,we analyzed the signal of GFP in detail using young panicles of the SB1P:SB1:GFP complementary transgenic lines(Fig.4-B to -M).Strong GFP signals were first observed in the primary branch primordia at the inflorescence stage In2(Fig.4-B and -C),and then the GFP signal was enhanced as the number of primary branch primordia increased during the stage In3(Fig.4-D and-E).During the stages In4-5,when the PBs continuedly elongated and generated secondary branch primordia,strong GFP signal was observed in the second branch primordia as expected(Fig.4-F to -I).With the growth of primary and secondary branches,the GFP expression was still observed in the elongated SBs and the potential ancestral cells of SM(Fig.4-J to -M).These results indicated thatduring panicle development,SB1expression was always strongly detected in the branch primordia,which was well corresponded with the mutant phenotype of increased branch number.
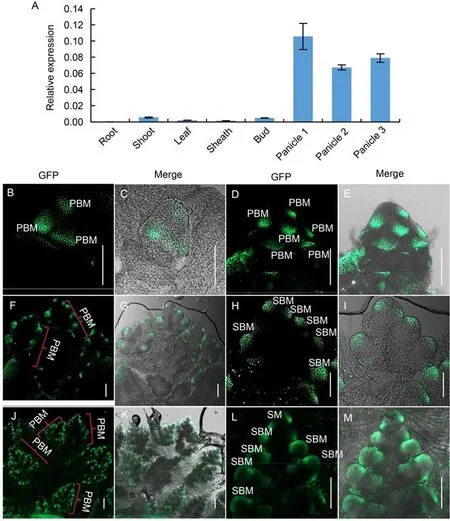
Fig.4.Expression pattern of SB1.
SB1 interacts with transcriptional corepressor complex
To illuminate the function of SB1,we carried out a yeast twohybrid(Y2H)assay to find the interacting proteins of SB1.Firstly,the full-length SB1-BD plasmid was transformed into Y2HGold yeast cells.The yeast cells that harboured pGBKT7-FZP(positive control)can grow on both SD/-Trp and SD/-Trp-His-Ade media,whereas the yeast cells that harboured pGBKT7-SB1 and the empty pGBKT7 can only grow on the SD/-Trp medium but not on the SD/-Trp-His-Ade medium,indicating that SB1 protein showed no auto-activation ability(Fig.5-A).Then,pGBKT7-SB1 was used as a bait to screen its partners in a cDNA library from rice young panicles.According to the screening results,we found two proteins:Os06g0126000(OsSEUSS-like 1,OsSEU1)and Os04g0510200(OsLEUNIG-like 1,OsLUG1)(Table S1),which are annotated as transcriptional corepressors and are homologous of SEU and LUG inArabidopsis(http://rice.plantbiology.msu.edu/).Previous research had shown that the corepressor complex SEU-LUG plays an important role in plant development(Conner and Liu,2000;Franks et al,2002;Sridhar et al,2004;Liu and Karmarkar,2008;Sitaraman et al,2008).Then,a Y2H assay was performed to test the interaction of SB1 with OsSEU1 and OsLUG1.As expected,after about 3 d,the yeast that co-expressed SB1 and OsLUG1 grew on the SD/-Trp-Leu-His-Ade selective medium,and the yeast that co-expressed SB1 and OsLUG1 grew after about 5 d.The results revealed that SB1 can interact with OsLUG1 and OsSEU1,and the interaction between SB1 and OsSEU1 was weaker than that of SB1 and OsLUG1(Fig.5-B).Next,a bimolecular fluorescent complimentary(BiFC)assay was conducted to further detect whether the SB1 can interact with SEU and LEU in leaf epidermal cells ofNicotiana benthamiana.Yellow fluorescent protein(YFP)signal was found in the nuclear of epidermal cells of tobacco leaves when co-expressing the SB1-YFPCfused protein with OsSEU1-YFPNand OsLUG1-YFPNfused proteins,respectively(Fig.5-C).The results revealed that SB1 was capable of interaction with OsSEU1 and OsLUG1 in plant cells,and then acted as a transcriptional repressor in regulating the downstream genes.
SB1 directly and negatively regulates expression of DEP1,TAW1,MOC1 and IPA1
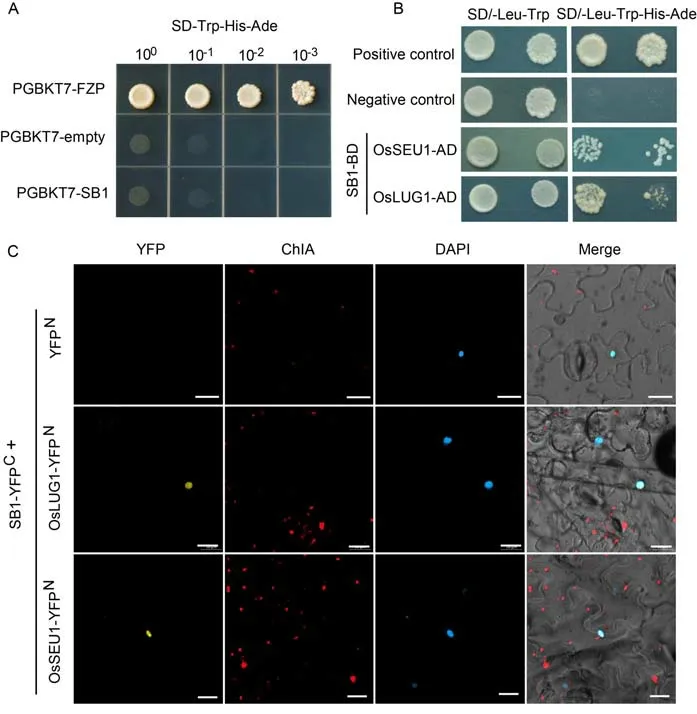
Fig.5.Interaction of SB1 with corepressors of OsSEU1 and OsLUG1.
To clarify the mechanism ofSB1regulating branch development,chromatin immunoprecipitation-sequence(ChIP-seq)was performed to screen the direct down- stream target genes ofSB1using the young panicles of SB1P:SB1:GFP complementarytransgenic lines with a GFP-antibody.A total of 3 535 significant binding peaks which covering 2 323 genes were screened by data analysis(fold enrichment > 2 andq-value<0.001).Among these 2 323 genes,several genes such asDEP1,TAW1,MOC1andIPA1that were known to regulate rice branch development showed obvious potential binding peaks withSB1.There were three binding peaks in the promoter ofDEP1and one peak in the coding sequence ofDEP1,one peak in the 3′-untranslated regions ofTAW1,two peaks in the coding sequence ofMOC1and one peak in the promoter ofIPA1(Fig.6-A).Next,we detected the expression levels of these four genes in the young panicles of the wild type andsb1mutants.qPCR results revealed that the expression levels ofDEP1,TAW1andMOC1were significantly increased among different lengths of panicles in thesb1-1andsb1-2mutants.The expression level ofIPA1was significantly increased in panicles(<0.5 cm)while slightly increased in other panicles(Fig.6-B).To further confirm whetherSB1directly regulated the expression of these genes,ChIP-qPCR was used to examine the binding between SB1 with these four genes.Chromatin isolated from young panicles of SB1P:SB1:GFPtransgenic lines was immunoprecipitated with a GFP antibody and then subjected to qPCR analysis.The results verified that SB1 can directly bind withDEP1(P2 site),TAW1(P2 site),MOC1(P2 site)andIPA1(P1 site)(Fig.6-C).Therefore,our data indicated thatSB1directly and negatively regulated the expression ofDEP1,TAW1,MOC1andIPA1genes.

Fig.6.SB1 directly repressed expression of DEP1,TAW1,MOC1 and IPA1.
Furthermore,it is reported that the LUG and SEU complex can interact with histone deacetylases to mediate the acetylation level of histone protein in the targeted region(Gonzalez et al,2007;Liu and Karmarkar,2008).We further detected the acetylation levels of histone protein H3 in the promoter and coding sequence region ofDEP1,TAW1,MOC1andIPA1by ChIP-qPCR with an anti-H3K9Ac antibody.The result indicated that the acetylation levels of H3K9 inDEP1(P1,P2 and P3 sites),TAW1(P2 and P3 sites),MOC1(P1,P4 and P5 sites)andIPA1(P1 and P2 sites)were significantly increased in thesb1mutant compared with those in the wild type(Fig.6-D).These results suggested thatSB1regulated the transcription ofDEP1,TAW1,MOC1andIPA1genes by the modification of histone acetylation.
DISCUSSION
SB1 is an important factor regulating GNP in rice
In rice,GNP as one of three key factors of grain yield,is heavily influenced by the inflorescence architecture,especially the number of branches.The previous research showed that a large number of genes can affect GNP to increase/decrease the yield by regulating the number of branches.Mutations ofGn1a,TAW1,DEP1,LPandIPA1all lead to increased branches and GNP to finally increase the yield.Gn1agene acts as a negative regulator of the yield by degrading the cytokinin.Mutation ofGn1aincreases the number of spikelets by 20% and significantly increases the yield of rice(Ashikari et al,2005).In the gain-functional mutanttawawa1-D,the spikelet specialization is delayed,resulting in prolonged branch formation time,increased the numbers of SBs and spikelets per panicle and finally the yield(Yoshida et al,2013).The functional acquired mutation ofDEP1makes rice panicle dense and increases the numbers of branches and grains per panicle,thus promoting rice yield of 15%-20%(Huang et al,2009).LPgene encodes an F-box protein,and its mutation leads to a significant increase in the numbers of branches and spikelets,with a yield increase of about 11% per plant(Li et al,2011).QTLIPA1is an allele ofOsSPL14.Mutation ofOsSPL14disturbsOsmiR156’s regulation ofOsSPL14,resulting in reduced tillering and significantly increasing branch number,and then panicle grain number and the yield by 10%(Jiao et al,2010;Miura et al,2010).
In this study,all the five mutantssb1-1/2/3/4/5were manifested as extremely significant increase in the numbers of SBs and TBs(which barely appeared in the wild type plants),and finally showed an increase in the number of spikelets per panicle.Among them,the average ‘number of spikelets per panicle’ ofsb1-1reached nearly twice as much as the wild type.However,it was regretful that the number of grains per panicle in thesb1mutant was obviously decreased because of the sharply decreased seed-setting rate.That meant the increasing numbers of spikelets in thesb1mutant was meaningless unless handling the defect of seed-setting rate.In fact,it was reported that the advantageous traits(such as the increased number of branches)and disadvantageous factors(such as abnormal seed-setting rate and defect floral organs)for grain yield were concurrent in some mutants of panicle-related genes.For example,in the mutants ofOsMADS34/PAP2,although the number of SBs increases sharply,the development of spikelet is affected and the seed-setting rate decreases(Gao et al,2010;Kobayashi et al,2010).Therefore,while the excellent natural variations in the genes such asGn1a,TAW1,DEP1andIPA1had been used to largely benefit grain yield in rice,rebuilding and utilization of the genes pros and cons coexisting likeSB1andOsMADS34/PAP2by the molecular design might be a future direction with quite development potentiality for the panicle architecture and grain yield in rice breeding.
Furthermore,the phenotypes ofsb1-1/2/3/4/5showed difference to some degree,such as the numbers of branches,spikelets and grains per panicle.Among the five mutants,sb1-1andsb1-3generated more branches and spikelets on panicle but less grains.Premature translation termination insb1-1caused the missing of the FRCVR domain and IGGH domain of SB1,and the frameshift mutation insb1-3caused severe destruction of the SB1 structure,which may lead to the loss of SB1 protein function.By contrast,sb1-4andsb1-5only had single amino acid substitution and 17-amino acid deletion in the IGGH domain,respectively,and the phenotype was significantly weaker thansb1-1/3.Interestingly,the deletion of 29-bp on the first exon insb1-2led to the frameshift mutation,but the new protein was highly similar to SB1,and its phenotype was similar tosb1-4/5.That is,the integrity of SB1 protein is necessary for the normal development of rice panicle,and the severity of domain deletion or destruction is positively correlated with the mutant phenotype.
SB1 might accelerate transition from BM to SM as a broad-spectrum repressor
In thesb1panicles,as more SBs and TBs were produced,the expression of some genes related to initiate and maintain BM fate was up-regulated,such asRCN1,MOC1,MOC3,TAW1,IPA1andDEP1.It was reported that enhancing the function of most of these genes can promote branch development in rice(Nakagawa et al,2002;Li et al,2003;Huang et al,2009;Jiao et al,2010;Miura et al,2010;Yoshida et al,2013).These results together suggested thatSB1might be responsible for accelerating the transition from BM to SM in panicles,and the mutations ofSB1prolonged the BM fate and then resulted in more secondary and tertiary branches instead of producing spikelets.
SB1gene encodes a SHI/STY/SRS family protein and is allelic toOsSHI1.OsSHI1/SB1gene regulates branch development in rice by acting as a repressive partner of IPA1 protein,a transcription activator ofDEP1,to down-regulate the expression ofDEP1(Duan et al,2019).Inshi1,the mutant of completedeletion ofOsSHI1gene,the numbers of secondary branches and spikelets increase sharply,and theDEP1gene is also up-regulated significantly,similar with thesb1mutants.In the present study,we showed a more precise mechanism ofOsSHI1/SB1regulating branch development.Firstly,the data of ChIP-seq and ChIP-qPCR showedSB1can directly bind to the promoters of multiple panicle-related genes includingTAW1,MOC1andIPA1,besidesDEP1.Secondly,we further revealed that SB1 can interact with two proteins of the SEU-LUG co-repressing complex,OsSEU1 and OsLUG1.SEU-LUG is known as a plant Gro/Tup1 co-repressor complex and responsible for transcriptional repressing in multiple development events.InArabidopsis,the LUG-SEU complex with HDA19 can repress theAGMOUS(AG)expression in outer two whorls of floral organs via removing the acetylation mark from theAGlocus(Conner and Liu,2000;Liu and Karmarkar,2008;Guo et al,2015).Next,we also found the H3K9Ac level was increased in multiple sites of the ChIP-target genes.Therefore,our results together supported that SB1 can interact with the co-repressing complex OsSEU1-OsLUG1 and then directly repressed the expression ofIPA1,DEP1,TAW1andMOC1through affecting the histone H3 acetylation level on target regions.In particular,the result supported thatSB1acted as a broadspectrum transcriptional repressor on multiple targets in regulating panicle architecture in rice.Then,we speculated,it was the releasing of multiple branch development genes includingIPA1,DEP1,TAW1andMOC1that eventually resulted in extremely prolonging BM fate and then dramatically increasing the numbers of spikelets in thesb1andshi1mutants(Fig.7).
METHODS
Rice materials
Thesb1-1/2/3/4/5mutants were derived from an ethyl methane sulfonate(EMS)mutagenesis of anindicarice cultivar XIDA1B.Thesb1mutant was crossed with a sterile line 56S(indica)to yield the F1population and subsequently generate the F2mapping population.All rice materials were grown in experimental fields at the Rice Research Institute of Southwest University,China under natural conditions.
Phenotype characterization of sb1 mutant
During the maturity period,the complete panicles of thesb1-1/2/3/4/5and wild type plants were photographed using a single lens reflex camera(Canon,Tokyo,Japan).Ten main panicles of thesb1-1/2/3/4/5and wild type plants were then selected respectively to investigate phenotypical data,including the numbers of primary/secondary/tertiary branches,the number of spikelets/grains per panicle and the seed-setting rate.The development progress of young panicles(sb1and wild type plants)was examined using a SU3500 scanning electron microscope(Hitachi,Tokyo,Japan)at -20 °C under a lowvacuum environment.
Map-based cloning of SB1
The 693 F2plants that exhibited a mutant phenotype were selected as a mapping population.Locus polymorphic SSR markers that developed by our laboratory were employed for gene mapping ofSB1.Candidate genes were predicted using the Rice Genome Brower website(http://rice.plantbiology.msu.edu)and cDNAs of the genes in the fine-mapped region were amplified from both the wild type and thesb1mutant,and the PCR products were confirmed by sequencing in the TSINGKE biological technology company(Chongqing,China).The primers used in the mapping and candidate gene analysis were listed in Table S2.

Fig.7.Model of SB1 gene regulating rice panicle architecture.
Vector construction and transformation
To construct a complementary plasmid,a 4091-bp genomic fragment that contained the 3070-bp upstream sequence and 1021-bpSB1coding sequence coupled withGFPreporter gene coding sequence was amplified using the primers SB1-comF and SB1-comR(Table S2).The fragment was inserted into a binary vector pCAMBIA1300 using the pEASY®-Uni Seamless Cloning and Assembly Kit(TRANSGENE,Beijing,China)to construct the pCAMBIA1300-SB1-GFP vector.The recombinant plasmids were transformed into thesb1mutant using theAgrobacterium tumefaciens-mediated transformation method as described previously(Xiao et al,2009).
RNA isolation and qPCR analysis
Total RNAs from the root,shoot,leaf,sheath,tiller bud and panicles of the wild type plant were isolated using the RNAprep Pure Plant Kit(Tiangen,Beijing,China).The first-strand complementary cDNA was synthesized from 2 μg total RNA based on the PrimeScript®Reagent Kit with gDNA Eraser Kit(Takara,Dalian,China).qPCR was performed using the SYBR®Premix Ex Taq™ II Kit(Takara,Dalian,China)in an ABI 7500 Real-time PCR System(ABI,Carlsbad,USA).Relative gene expression was quantitated based on three biological replicates andOsActin(OsRac1,LOC_Os01g12900)was used as an endogenous control.The sequences of primers used in qPCR were listed in Table S2.
Subcellular localization
We confirmed the subcellular localization of SB1 according to the SB1-GFP fluorescence in the positive complementary transgenic plants.At the booting stage,single branch or spikelet of young panicles were transferred to a glass slide,then examined the fluorescence signal under an Olympus FluoView 1000-Confocal laser scanning microscope(ZEISS,Jena,Germany).
Protein sequence and phylogenetic analysis
The SB1-related protein sequences were downloaded from GenBank(http://www.ncbi.nlm.nih.gov/genbank/)using the full-length amino acid sequence of SB1 as a query.A phylogenetic tree was constructed using the maximum likelihood method based on the Jones-Taylor-Thornton matrix-based model with the lowest Bayesian information criterion scores by MEGA 5.0(Tamura et al,2011).Statistical support for the tree topology was assessed by means of a bootstrap analysis with 500 replicates.Protein motifs were predicted using the motif-based sequence analysis tool MEME(http://alternate.meme-suite.org/).
Yeast two-hybrid(Y2H)assay
The full-lengthSB1coding region was cloned into a pGBKT7 vector at theEcoRI andBamHI restriction sites.Then the pGBKT7-SB1 construct was transformed into the yeast strain Y2HGold.Transformants were selected on SD/-Trp and SD/-Ade/-His/-Trp media to test the auto-activation of SB1.FZP protein having strong auto-activation was used as a positive control and the empty pGBKT7 vector as a negative control.A young inflorescences cDNA library constructed by Shanghai OE Biotechnology Co.,Ltd in China was used to perform the Y2H screening and positive clones were identified by sequencing.The full-lengths ofOsSEU1andOsLUG1coding regions were cloned into the pGADT7 vector at theEcoRI andBamHI restriction sites,respectively.Then the pGADT7-OsSEU1 and pGADT7-OsLUG1 were used as preys and separately transformed into the yeast strain Y2HGold together with the bait construct pGBKT7-SB1.The selective medium SD-Leu/-Trp/-His/-Ade was used to test the interaction between bait(SB1)and preys(OsSEU1 and OsLUG1).The sequences of primers used in Y2H assay were listed in Table S2.
Bimolecular fluorescent complimentary(BiFC)
To conduct BiFC assay,the coding sequences of SB1,OsLUG1 and OsSEU1 were amplified by gene-specific primers(Table S2)and then ligated into BiFC vectors,including pSCYNE(SCN,modified)and pSCYCE(SCC,modified).The recombinant vector pairs were co-transformed intoN.benthamianaleaves.The fluorescence signals were captured under a confocal laser scanning microscope(LSM 800,Zeiss,Germany).
ChIP-Seq and ChIP-qPCR
The young panicles(<3 cm)from the SB1P:SB1:GFP complementary transgenic plants were collected to do the ChIP-seq using a GFP-antibody.The sequencing and data analysis were carried out by the Wuhan Tovin Biotechnology Co.,Ltd in China.ChIP-qPCR was used to confirm the direct binding ofSB1with its target genes and detect the acetylation level insb1.The young panicles(<3 cm)of SB1-GFP transgenic plants,sb1mutant and wild type plants were collected for isolation of chromatin extracts.ChIP assays were performed using the EpiQuik™ Plant ChIP Kit(P-2014-48,Epigentek,USA),anti-GFP antibody(ab290,Abcam,USA)and anti-H3K9Ac antibody(ab10812,Abcam,USA).All qPCR experiments were conducted with the progress:40 cycles of 95 °C for 5 s,60 °C for 30 s and 72 °C for 30 s in a reaction mixture containing 10 pmol of forward primer,10 pmol of reverse primer and 1 μL of DNA from ChIP/control or 1 μL of input DNA diluted 20-fold(per biological replicate)as a template.More than three biological repeats with three technical repeats each were used for statistical analysis.The recovered DNA was used as the template for ChIP-qPCR with the method described previously(Zhuang et al,2020).The primer sequences were listed in Table S2.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China(Grant No.31971919),the National Key Program for Research and Development of China(Grant No.2017YFD0100202),the Project Sponsored by Natural Science Foundation of Chongqing,China(Grant No.cstc2020jcyjjqX0020)and Chongqing Graduate Research and Innovation Project funding in China(Grant No.CYS20123).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science;
http://www.ricescience.org.
Fig.S1.Scanning electron microscope analysis of young panicles ofsb1-1and wild type.
Fig.S2.Sequence of motifs predicted by MEME.
Fig.S3.Protein sequence analysis of SHI/LRP/SRS protein family in rice,maize andArabidopsis thaliana.
Fig.S4.Subcellular location of SB1 by observing GFP signal in SB1P:SB1:GFP transgenic plants.
Table S1.Interacting proteins of SB1 identified by yeast twohybrid screening.
Table S2.Primers used in this study.
- Rice Science的其它文章
- Editing of Rice Endosperm Plastidial Phosphorylase Gene OsPho1 Advances Its Function in Starch Synthesis
- Valuation of Rice Postharvest Losses in Sub-Saharan Africa and Its Mitigation Strategies
- Rice Bran Oil:Emerging Trends in Extraction,Health Benefit,and Its Industrial Application
- Combined Drought and Heat Stress in Rice:Responses,Phenotyping and Strategies to Improve Tolerance
- OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1
- Genomic Predictio of Arsenic Tolerance and Grain Yield in Rice:Contribution of Trait-Specific Markers and Multi-Environment Models