Identification of microRNAs involved in crosstalk between nitrogen,phosphorus and potassiumunder multiple nutrient deficiency in sorghum
Zhenxing Zhu,Dan Li, Ling Cong, Xiaochun Lu
Crops Molecular Improving Lab,Liaoning Academy of Agricultural Sciences,Shenyang 110161,Liaoning,China
Keywords:
ABSTRACT Nitrogen (N), phosphorus (P), and potassium (K) are important for plant growth and development.MicroRNAs (miRNAs) play important roles in regulating plant response to nutrient (N, P, and K) deficiencies.Several miRNAs have been identified under nutrient deficiency conditions in many plant species.However, the manner in which miRNAs regulate the interaction between NPK signaling pathways under multiple nutrient deficiency remains largely unknown.We systematically compared and identified microRNAs involved in both single and triple NPK nutrient deficiency responses.We identified 32 shoot and 17 root miRNAs differentially expressed under potassium deficiency.Several NP starvation-associated miRNAs including miR169s and miR399s,were also regulated by K deficiency.Several identified miRNAs including miR5565c,miR5564, and miR1432 have not previously been associated with respectively N, P, and K deficiency (−N, −P, and −K).Expression correlation analysis between miRNAs and their predicted targets showed that miR169, miR172, and miR160 displayed expression trends exactly opposite to those of their corresponding predicted targets.Of 550 predicted novel miRNAs, novel_mir_42 was upregulated in shoots under −K but was downregulated under−N and −P.The effects of combined NPK starvation were not a simple addition of the individual stresses on sorghum seedlings.The identified common and specific differentially expressed miRNAs were observed under single and combined NPK deficiencies.These findings will help to further elucidate the functions of microRNAs and their interactions under multiple nutrient deficiency.
1.Introduction
MicroRNAs (miRNAs) are small noncoding RNAs with mature lengths of 20-24 nucleotides.miRNAs control gene expression by degrading target mRNAs.miRNA biogenesis involves several processes.To generate mature miRNAs,primary miRNA is cut by a DICER-LIKE 1(DCL1)complex and other processes,after which the mature miRNAs are transported to the cytoplasm from the nucleus.miRNAs are incorporated into an ARGONAUTE (AGO)complex to degrade target mRNAs [1].miRNAs are involved in most biological processes in plants, from germination to fruit development, and in responses to many abiotic and biotic stresses[2-6].NPK are the three most important macronutrients required for plant growth and crop yield.Many miRNAs involved in N,P and K starvation responses have been characterized[7-9].
miRNAs such as miRNA169, miRNA156, miRNA393, and miRNA172 function in plant responses to N stress.miRNA169 is downregulated under nitrogen- and phosphate-deficient conditions.miRNA169 reduces the size of flowers and delays flowering by repressing NFYA expression.miRNA169 also has a role in increased nitrate uptake in plants [10].miRNA156 is induced strongly under N starvation and targets the Squamosa Promoter Binding protein (SBP) or SBP-like (SPL)gene family,whose members are involved in plant flowering,flower architecture, and crown root development in rice[11-13].miRNA393 is a nitrate-responsive miRNA whose targets are bHLH transcription factors and auxin receptor AFBs, which are involved in controlling root system architecture, especially the primary root and lateral root growth response to nitrate [14].miRNA167 interacts with ARF8 to regulate root adaptive response in a nitrate-dependent manner [15].miRNA171, which is promoted by N starvation,targets SCL6-II, SCL6-III, and SCL6-IV and is linked to primary root elongation [16,17].miRNA172 is induced in maize leaves following low-nitrate treatment[18].miRNA172 is involved in the regulation of the transition from vegetative growth to flowering.miRNA172 targets the floral homeotic gene APETALA2 through translational inhibition[19].
miRNA399 and miRNA827 are two well-known inorganic phosphate (Pi) starvation-responsive miRNAs in plants[20,21].miRNA399 targets PHO2, an E2 conjugating enzyme,which is a key regulator of Pi signal-induced downstream gene expression in plants [22].miRNA827 is strongly induced under Pi starvation conditions;its target gene in Arabidopsis is AtNLA, a RING-type ubiquitin E3 ligase, which is involved in nitrate-dependent phosphate homeostasis [23].Other miRNAs are also involved in regulating phosphate starvation response.Expression levels of the wheat miRNA taemir408 are increased under Pi starvation and salt stress in wheat,demonstrating its important role in plant resistance to Pi starvation and salt stress [24].Wheat TamiRNA1139 is also responsive to Pi-deficient conditions and is involved in plant tolerance to Pi starvation [25].miRNA395 regulates several genes of the sulfur assimilation pathway and is induced by low-S treatment [26].miRNA395 is also induced strongly in shoots of Pi-deficient white lupin plants [27].However,whether miRNA395 is involved in the regulation of Pi starvation signaling is still unknown.
How miRNAs are directly affected by the uptake and physiological processes of potassium in plants is far from clear.There have been few reports of K starvation-responsive miRNAs in plants.The monocot-specific miRNA444, which functions in nitrate and phosphate uptake, targets the MADS23 gene, which is significantly induced under K starvation conditions,suggesting its function in the K pathway[28].
Research on the roles of miRNA in mineral nutrition has been focused mainly on plant responses to individual mineral deficiencies.Further complexity arises from the fact that in the soil,several nutrients can be deficient at the same time and in different combinations.However, miRNA regulation following combined deficiencies of N, P and K has not been reported.Sorghum is usually planted with little fertilizer and few other inputs by smallholder farmers in many countries.To date,miRNA regulation under multiple nutrient deficiencies in sorghum has not been studied systematically, limiting our understanding of sorghum tolerance to infertile soil.We compared miRNAs under single N, P, and K (−N, −P, and −K,respectively) and triple NPK (−NPK) deficiencies in sorghum,with the objective of identifying miRNAs with nutrient-specific and common responses to −N,−P,−K,and −NPK deficiencies.
2.Materials and methods
2.1.Plant materials and treatments
Seeds of the sorghum cultivar BTX623 were incubated overnight at 30 °C on wet filter paper, and germinated seeds were sown on a mesh tray floating in modified Hoagland nutrition solution.The hydroponic culture solution was adjusted to pH 5.8 with 1 mol L−1NaOH.Plants were grown in a greenhouse with an approximately 14 h light(30°C)/10 h dark (22 °C) photoperiod.Plants were grown in nutritionsufficient solution (0.5 L plant−1, 20 plants/treatment) for 7 days and then transferred to control (nutrient-sufficient conditions, CK), −N, −P, −K, and −NPK hydroponic culture for another 7 days.The composition of the culture solution was as follows: macronutrients (mmol L−1): KNO3(2), Ca(NO3)2·4H2O (2.5), MgSO4·7H2O (2), NH4NO3(0.75), and KH2PO4(0.5); micronutrients (μmol L−1): H3BO3(46), MnCl2·4H2O (9),ZnSO4·7H2O (0.8), Na2MoO4·2H2O (0.5), CuSO4·5H2O (0.2), and Fe-EDTA (100).KCl and CaCl2were substituted for KNO3and Ca(NO3)2·4H2O, respectively, and NH4NO3was removed (−N);KCl was used to substitute KH2PO4(−P); NaNO3and NaH2PO4were used to substitute KNO3and KH2PO4, respectively (−K);CaCl2was used to substitute Ca(NO3)2·4H2O, and KNO3,NH4NO3and KH2PO4were removed(−NPK).
2.2.RNA preparation and small RNA high-throughput sequencing
Shoots and roots of plants were harvested separately at the indicated times.Six sorghum plants were pooled as one sample for total RNA extraction.TRIzol reagent (Invitrogen,Carlsbad, CA, USA) and DNase I (Tiangen Biotech, Beijing,China) were used for total RNA extraction and genomic DNA elimination, respectively.The yield and quality of total RNA were measured by 1.2% agarose electrophoresis, a NanoDrop(Thermo Fisher Scientific,Waltham,MA,USA)and an Agilent 2100 Bioanalyzer(Agilent Technologies,Santa Clara,CA,USA).Quality-filtered RNAs were sent to the Beijing Genomics Institute(BGI,Shenzhen,China)for sequencing(one biological replicate).The sequencing process was as follows:small RNAs 18-30 nt in length were separated,and 5′and 3′adaptors were added.After the first strand of cDNA was synthesized and amplified, the PCR products were purified and subjected to high-throughput sequencing[29].All RNA-Seq data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA574791 (https://www.ncbi.nlm.nih.gov/sra/PRJNA574791).
2.3.Analysis of sequencing data
The 50-nt sequence from high-throughput sequencing was filtered to remove low-quality reads and contaminants.Clean reads were mapped to the sorghum genome with SOAP [30].Perfectly matching reads were used for further analysis.Small RNAs were classified into rRNA, scRNA, snoRNA, snRNA, and tRNA with annotations from GenBank and Rfam(10.1),and then unannotated reads were aligned to precursor and mature miRNA from miRbase (miRbase version 19, http://www.mirbase.org/).Transcript per million (TPM) values were used to normalize the expression of miRNAs in different libraries.Fold changes and Pvalues were calculated from the normalized expression (the differentially expressed miRNA threshold was a log2fold change≥1 and P-value <0.05).A Venn diagram was created with an online tool(http://bioinformatics.psb.ugent.be/webtools/Venn/).
2.4.Novel miRNA identification and target gene prediction
BGI developed prediction software, Mireap (http://sourceforge.net/projects/mireap/),to predict novel miRNAs from the secondary structure, and the minimum free energy of unannotated small RNA tags that were mapped to the genome.The candidate target genes of miRNAs were also predicted with Mireap.To study the function of the candidate target genes of miRNAs, Gene Ontology (GO) (http://www.geneontology.org/) and KEGG (http://www.genome.jp/kegg/)enrichment analyses were performed.
2.5.Real-time quantitative PCR validation of mature miRNA expression
qRT-PCR was performed using the stem-loop quantitative reverse transcription PCR method[31].First-strand cDNA was generated using the stem-loop RT primers listed in Table S1.qRT-PCR was performed with three technical and three biological replicates using a LightCycler480II (Roche, Switzerland) and SYBR Premix Ex Taq reagent (Takara Biomedical Technology(Beijing)Co.,Beijing,China).The primers used for real-time PCR are listed in Table S1.The reverse primer was universal and the forward primers were specific to the miRNA sequence.U6 snRNA was used as the reference gene for miRNA validation in sorghum.
2.6.Correlation analysis of the expression of genes and miRNAs
Correlation coefficients between the expressions of miRNAs and their target genes were calculated (target gene expression data are not shown).Only pairwise coefficients >0.9 were chosen for the next step of the analysis.Cytoscape software[32]was used to show the relationships between miRNAs and their targets.
3.Results
3.1.Overview of small RNAs responsive to NPK deficiencies
The 10 libraries produced 18,528,598-24,676,478 raw reads,with a mean of 21,376,010 per library(Table S2).The proportion of clean reads among total reads was approximately 99%.The unique sRNAs covered 58.4% to 87.6% of the genome (Table S3) and included miRNA, siRNA, piRNA, rRNA, tRNA, snRNA, snoRNA,repeat-associated sRNA, degraded tags of exons or introns, and unannotated sequences(Fig.S1).To investigate the relationship between the length of small RNAs and the proportion of total reads, unique reads with sizes ranging from 13 to 31 bp were chosen for analysis.The distribution of sRNA read lengths showed that 21 and 24 nt sRNAs were dominant,with the latter being the most abundant in the 10 libraries (Fig.1).The least abundant (14.6%) 21-nt sRNA was found in the −P-treated root library,which,in contrast,had the most abundant(62.3%)24-nt sRNAs among the 10 libraries.The most abundant 21-nt and 24-nt sRNAs were found in libraries from shoots of −N-treated plants and roots of −K-treated plants,respectively(Fig.1).Thus,different NPK supplies led to different patterns in the size distribution of sRNAs.
3.2.Identification of known miRNAs responsive to NPK single and triple deficiencies
From 124 (−NR) to 151 (−PR) unique mature miRNAs were found in the 10 libraries (Table S4).Of 63 miRNA families,miR168 was the most abundant, followed by the miR156,miR166, miR167, miR5564, and miR172 families (Table S5).Although some miRNAs were from the same family, there was a diversity of abundant miRNAs.Taking the miR156 family as an example, the counts of most members were nearly one million, but that of miR156d was only 10,000, and in roots this number was less than 10,000.Even within the same miRNA family, members such as miR169 and miR399 showed different responses to N, P, and K (Table S5).These results suggested that members of the same family might play diverse roles in the response to NPK starvation.
The regulatory pattern of differentially expressed miRNAs in the shoots of the −N-treated sample was different from those in the shoots of plants under −P, −K and −NPK conditions.There were more downregulated than upregulated miRNAs in shoots of−N-treated plants.The greatest number of differentially expressed miRNAs was in the shoots of −NPK-treated plants(up 43, down 26, 15 families) and roots of −P-treated plants (up 18,down 58, 19 families).Fewer differentially expressed miRNAs were identified in −K-treated plants in both shoots and roots(in shoots,20 upregulated,12 downregulated,16 families;in roots 6 upregulated,11 downregulated,12 families)(Fig.2A,B).Based on the known miRNA response to −N, −P, and −K (Fig.3), new differentially expressed miRNAs associated with the NPK starvation response were identified.They included miR6224b-3p,miR5565c, and miR5568c-5p under N deficiency and miR6219-5p, miR528, miR408, miR390, miR5564, and miR5565 under P deficiency.Under K deficiency there were 32 and 17 differentially expressed miRNAs in shoots and roots, respectively, including miR156s and miR160s, miR1432, miR164c, miR169i/o, and miR399i/d.qRT-PCR confirmed the expression patterns of some of these miRNAs(Fig.S2).
3.3.Specific and common differentially expressed miRNAs under NPK single and triple deficiencies
Fig.2C,D shows the overlap between differentially expressed genes under NPK single and triple deficiency conditions We identified 30 and 5 specific differentially expressed miRNAs in shoots and roots,respectively,under −NPK conditions(Fig.2C,D;Tables S6, S7).The 30 miRNAs differentially expressed only in shoots of −NPK-treated plants included sbi-miR5564b, sbimiR166a/b/c/d/h/i/j, sbi-miR437a/b/c/e/f/g/i/j/k/o/p/s/t, sbimiR396d/e, sbi-miR393a/b, and sbi-miR171a/b/d/i/k (Table S6).The five differentially expressed miRNAs found specifically in roots included sbi-miR396a/b,sbi-miR6223-3p,sbi-miR172b,and sbi-miR437x-3p (Table S7).NPK single deficiency-responsive miRNAs were also identified in shoots and roots.For example,six differentially expressed miRNAs (sbi-miR6223-3p, sbimiR169o, sbi-miR437r, sbi-miR5568d-5p, sbi-miR437l, and sbimiR156d)were specific to shoots and two(sbi-miR1432 and sbimiR319a) were specific to roots under K deficiency conditions only(Tables S6 and S7).
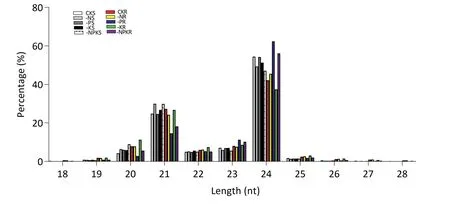
Fig.1– Size distribution of small RNAs.BTX623 plants were sampled under the following conditions: CK,control;−N,N deficiency;−P,P deficiency;−K, K deficiency;−NPK,combined NPK deficiency.S, shoot;R,root.
Five differentially expressed miRNAs, sbi-miR169p/q/r-3p,sbi-miR171c and sbi-miR528, were responsive to −N, −P, −K,and −NPK conditions in roots(Tables 1,S7),but with different expression patterns under single and triple NPK deficiencies.For example, miR169p/q/r-3p were downregulated significantly both in shoots and roots, especially in roots under −N,−P, and −NPK conditions.These three miR169 members were also downregulated in −K-treated roots but showed no differential expression in the shoots of −K-treated plants.Ten differentially expressed miRNAs, sbi-miR156a/b/c/f/g/h/i,sbi-miR528, sbi-miR408 and sbi-miR164c, were responsive to−N, −P, −K, and −NPK conditions in shoots (Tables 1, S6).All seven members of the miR156 group were expressed in distinct patterns in shoots under −N, −P, −K, and −NPK conditions and were downregulated only in roots under −P conditions.sbi-miR164c was upregulated by −N, −P, −K, and especially by −NPK conditions (Table 1), showing an additive effect of the individual stresses on the expression of this miRNA under combined NPK starvation(−NPK).
3.4.The effect of nutrient interactions on miRNA expression under multiple nutrient deficiency
To directly examine the interaction between N, P, and K deficiency responses,we compared the miRNA expression profile under single nutrient and triple nutrient deficiencies(Tables S8-S13).From the Venn diagram,it is clear that most N starvationresponsive miRNAs in shoots also occurred in −NPK-treated plants,and only four differentially expressed miRNAs were found in N-treated plant shoots (Fig.2C, D; Table S8).Thirty-two miRNAs were differentially expressed under both −N and −NPK conditions in shoots, accounting for 89% of the differentially expressed miRNAs in the shoots of −N-treated plants(Table S8).These results strongly suggest that most differentially expressed miRNAs in −N-treated plants were not affected in the shoots of−NPK-treated plants, indicating that −P and −K have little effect on miRNA expression profiles under −N.Similar behavior was also observed in −K-treated plants,especially in roots(Fig.3).The number of miRNAs commonly differentially expressed in the roots of −K and −NPK-treated plants accounted for 71% of the total differentially expressed miRNAs under −K (Table S13),suggesting that the expression of most −K responsive miRNAs was unaffected by −N and −P conditions.Commonly differentially expressed miRNAs between the roots of −P and −NPK-treated plants accounted for only 43%of the total differentially expressed miRNAs in −P-treated plants, suggesting that more than 50% of the differentially expressed miRNAs were affected by −N and −K.For example, Pi starvation induced miR399, and the expression levels of most miR399 members were lowered both in shoots and roots of −NPK-treated plants compared with plants under −P conditions (Fig.3), indicating that −N and −K conditions have a strong effect on the expression of miR399.Two miRNAs,miR156s and miR167s, were repressed during Pi starvation and this repressed expression was not observed under −NPK conditions(Fig.3-B).These results showed the effects of −N and −K on −P responsive miRNAs.It appeared that the miRNA expression profile in −N-treated plants was least affected under −NPK,followed by −K and −P conditions.
3.5.Combined correlation analysis of the expression of miRNAs and their targets
To investigate the function of miRNAs, targets of miRNAs were predicted(Table S14, Fig.S3).GO analysis of the targets showed enrichment of functional terms in all kinds of biological processes: “cellular response to stimulus”, “anion transport”,“regulation of developmental process”, and “reproductive process”, especially in transcription, RNA related metabolism, and macromolecule biosynthetic processes(Fig.S3).

Fig.2– Overview of changes in transcripts of miRNAs under N,P, K,and NPK deficiency conditions.(A)and(B)Numbers of differentially expressed miRNAs in shoots(A)and roots(B)under various NPK treatments.(C)and(D)Venn diagrams showing the overlap of the responsive miRNAs in shoots(C)and roots(D)of −N-,−P-,−K-,and −NPK-treated plants.BTX623 plants were sampled under the following conditions:CK,control;−N,N deficiency;−P,P deficiency;−K,K deficiency;−NPK,combined NPK deficiency.S,shoot;R, root.
The expression correlation between miRNAs and their predicted target genes was calculated by a combined analysis of the expression profiles of miRNAs and their targets.Thirtyone pairs of miRNAs and targets showed high correlation(with absolute values of correlation >0.9) in their expression(Fig.S4).Among them,three miRNAs,sbi-miR169,sbi-miR172,and miR160, displayed expression trends exactly opposite to those of their corresponding predicted targets (Fig.4, Table S15).For two predicted targets of sbi-miR169, Sb02g002110.1 and Sb01g011220.2, the predicted annotations were respectively protein kinase domain and CCAAT-binding transcription factor (CBF-B/NF-YA) subunit B.The expression of Sb01g011220.2 was specifically induced 2.59 and 2.99-fold in the shoots and roots,respectively,of −N-treated plants(Table S15).One predicted target of sbi-miR172 was Sb02g007000.1,which was annotated as a putative indeterminate spikelet 1,AP2 domain protein.This gene was significantly suppressed in roots under −N, −P, −K, and −NPK conditions (Table S15).One target of miR160, Sb06g022810.1, was predicted to be auxin-responsive factor 10 in sorghum.Sb06g022810.1 expression was strongly induced in shoots of −N-treated plants.The expression of the targets was positively correlated with that of several miRNAs.For example, the target of sbi-miR164c was Sb06g019010.1, which was induced under −N and −P conditions in shoots(Table S15).It is likely that the spatiotemporal expression of these miRNAs does not completely overlap with that of their targets.
3.6.Identification of novel miRNAs
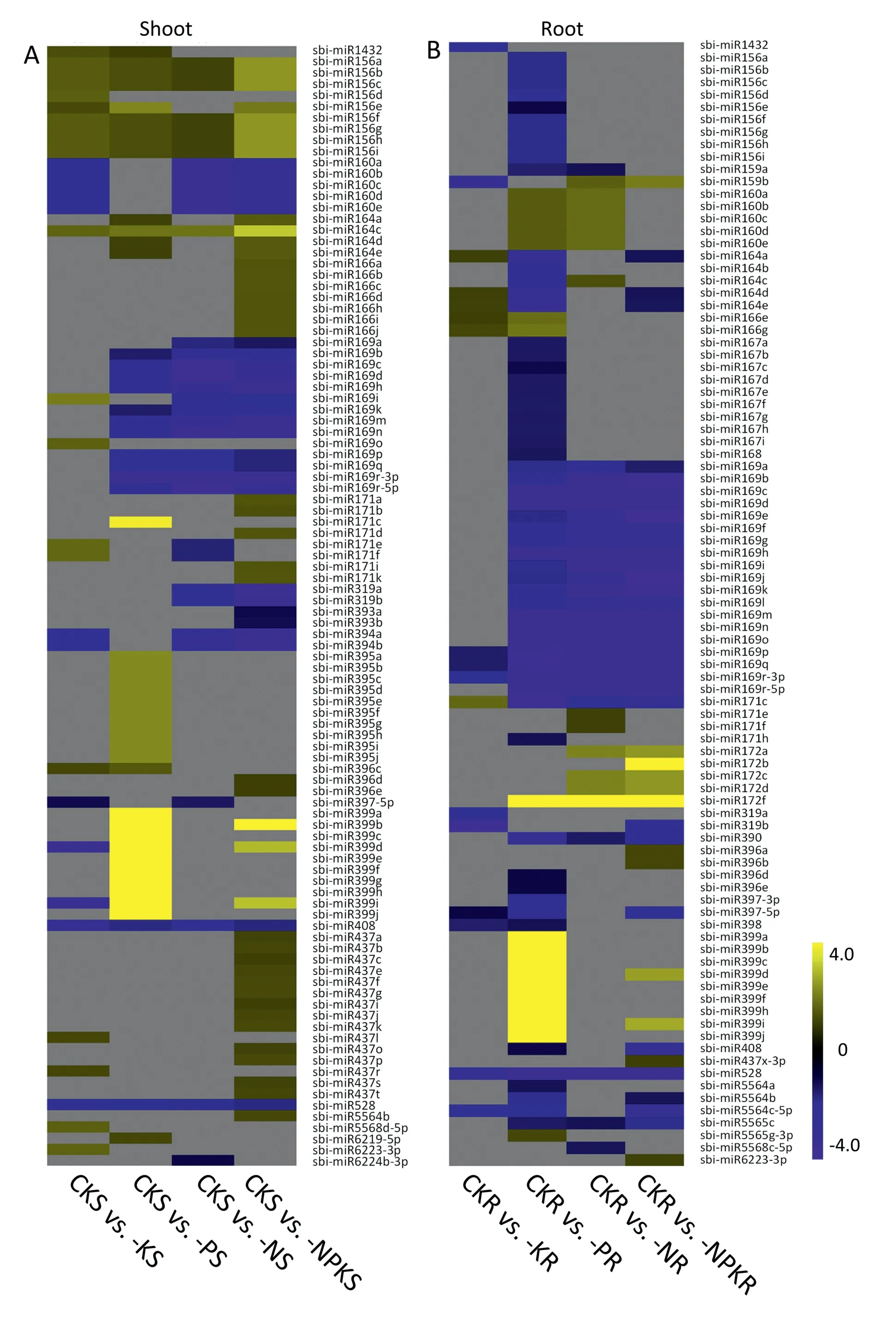
Fig.3–Identification of differentially expressed miRNAs in shoots(A)and roots(B) responsive to N,P,K, and NPK deficiency.BTX623 plants were sampled under the following conditions: CK,control;−N,N deficiency; −P, P deficiency;−K, K deficiency;−NPK,combined NPK deficiency.S,shoot;R, root.
Based on the characteristic hairpin structure of miRNA precursors, we identified 550 novel predicted miRNAs in the10 libraries.The expression of most of the novel predicted miRNAs was very low, and counts of only 48 novel miRNA reads were ≥100(Table S16).GO analysis of targets(Table S17;Fig.S5) showed that only a few functional terms were enriched: development-related terms, response to endogenous stimulus, transport, and chromosome organizationrelated terms(Fig.S5).
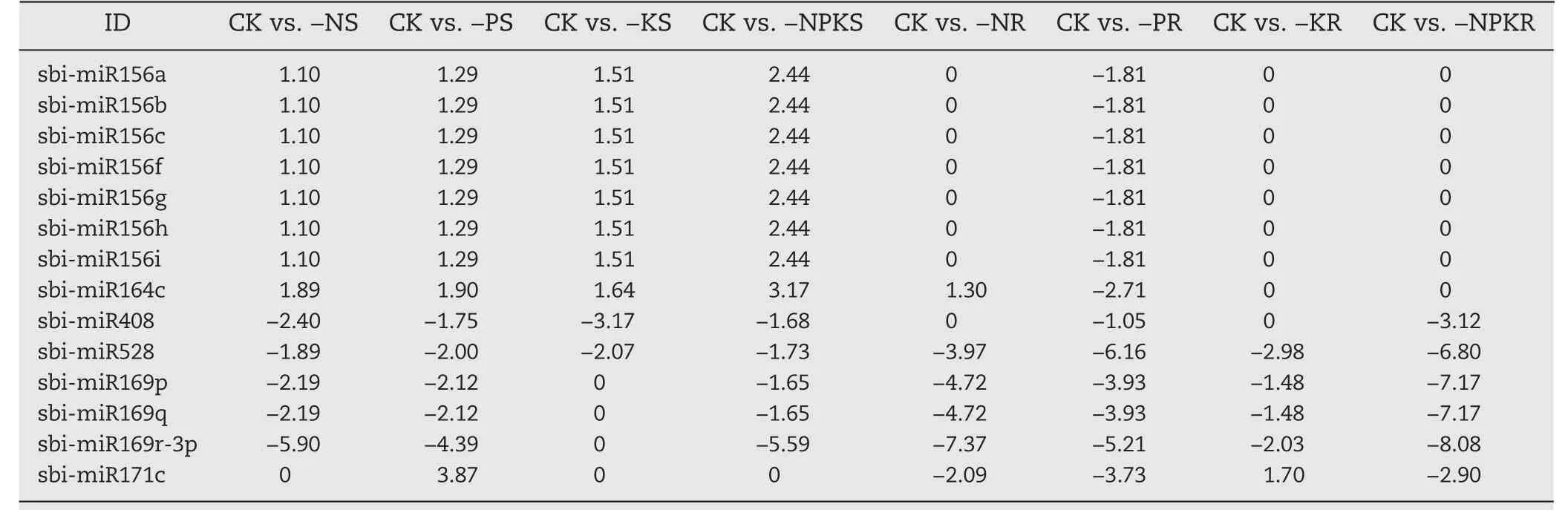
Table 1–Expression of common differentially expressed miRNAs under −N,−P, −K, and −NPK conditions.
Mature sequences of all the novel miRNAs were subjected to BLASTN search in miRbase [33].Small number of novel miRNAs had high similarity with miRNAs found in miRbase.A total of 14 novel miRNAs were found to have likely homologs in the other species (Table S18).The expression patterns of two novel miRNAs are shown in Table 2.For example,novel_mir_74 was the homolog of miR827, and its predicted mature sequence was the same as that of OsmiR827 from rice.Based on the sequence,novel_mir_74 was expressed predominantly in shoots,but the increase in expression was stronger in roots than in shoots under Pi starvation conditions.The predicted targets of novel_mir_74 were Sb04g031920 and Sb06g025950, which were predicted as SPX-MFS-domain proteins.The expression pattern of Sb06g025950 displayed a direct negative correlation with that of novel_mir_74 under P starvation conditions.In contrast, Sb04g031920 expression displayed a positive correlation with that of novel_mir_74(Table S15).It is likely that Sb06g025950 is the target of novel_mir_74 in sorghum.novel_mir_42 expression was increased in roots of plants under −K conditions, whereas it was significantly decreased both in shoots and roots under −N and −P conditions.qRT-PCR results validated the expression patterns of novel_mir_74 and novel_mir_42 (Fig.S2).novel_mir_42 is the homolog of miRNA169, differing from sorghum miRNA169 family members by mutations at the 14 and 15 nt positions (Fig.5).Three candidate targets of novel_mir_42, Sb01g045500, Sb01g032710, and Sb02g038960 were predicted as CCAAT-binding transcription factor (CBFB/NF-YA) subunit B.Expression of the three genes was increased under −N conditions in both shoots and roots(Fig.5).
4.Discussion
Previous studies have reported differential expression miRNAs under single N, P, and K deficiencies in many plant species [7-9]; however, miRNA expression profiles under combinations of NPK deficiencies were lacking.In the present study, we identified common and specific miRNAs under single N, P, and K and triple NPK deficiencies.These data provide important information about miRNAs involved in crosstalk during the N, P, and K deficiency responses in sorghum.
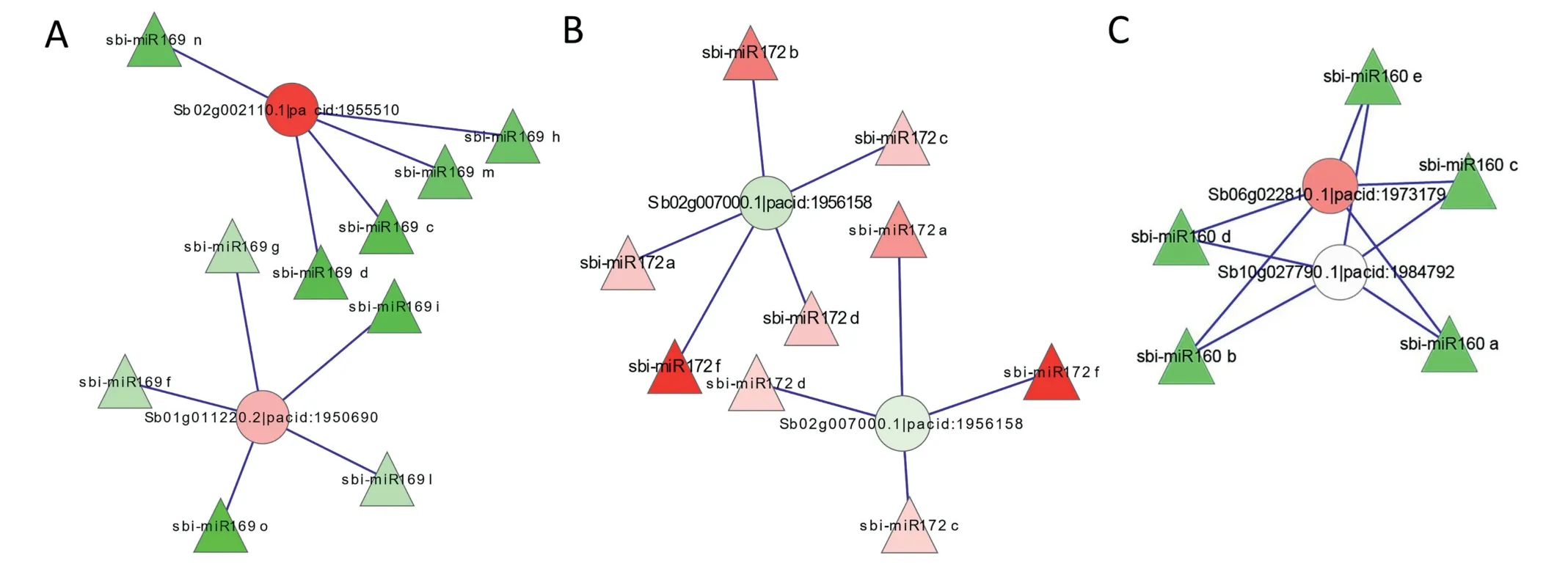
Fig.4–Selected results of expression correlation analysis between miRNAs and their targets.(A)Interaction of miR169 with its target;(B) Interaction of miR172 with its target;(C)Interaction of miR160 with its target.
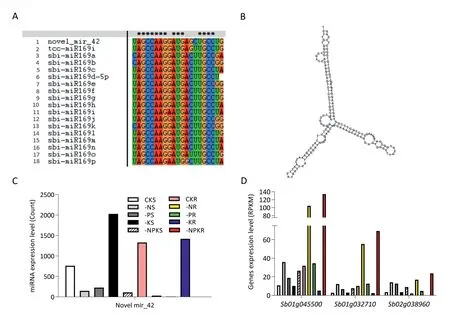
Fig.5–Overview of novel mir_42.(A)Sequence alignment of novel mir_42,the miR169 family from sorghum and tcc-miR169i from Theobroma cacao(the best hit in the miRbase).(B) Predicted secondary structure of novel mir_42.(C)Expression of novel mir_42 under N,P,K,and NPK starvation conditions.(D)Expression of three predicted targets of novel mir_42 under N,P,K and NPK starvation conditions.BTX623 plants were sampled under the following conditions: CK,control;−N,N deficiency;−P,P deficiency;−K,K deficiency;−NPK,combined NPK deficiency.S,shoot;R, root.
Under −NPK conditions,30 specific differentially expressed miRNAs, including miR166, sbi-miR171 and sbi-miR437, were found in plant shoots (Table S6).This differential expression indicated a direct interaction among N,P,and K at the miRNA level.miR166 and its target HD-ZIP transcription factors regulate shoot, flower, and vascular development [34].In barley, miR171 is involved in the transition from the vegetative to the reproductive phase by activating the miR156 pathway [35].These results indicate that regulation of shoot growth and development via miR166 and miR171 is a strategy helping plants cope with extreme nutrient supplies (−NPK).miR437 is a monocot-specific miRNA whose function is unknown.Two predicted targets of miR437 were Sb01g007855 and Sb03g003980 (Table S14).Differentially expressed sbi-miR171e/f were specifically increased in the roots of −N-treated plants(Fig.3,Table S7).Overexpression of miR171c and triple scl6 results in pleiotropic phenotypes,including primary root elongation [17].This result suggests that sbi-miR171e/f is involved in the regulation of root elongation under −N conditions.Interestingly, miR395 was found to be responsive to Pi starvation only in shoots.miR395 is involved in regulating sulfate assimilation and transport[36].miR395 was also upregulated in the leaves of lupin under phosphate starvation conditions but was suppressed by Pi starvation in Arabidopsis [27,37].Although the functions of miRNAs are conserved in plants, this finding suggests that miR395 has different functions in sorghum.Several members of miR156 were responsive to Pi starvation only in roots(Fig.3); in contrast, most miR156 members were upregulated in shoots during −N, −P, and −K conditions and especially under −NPK conditions, indicating the tissue-specific functional differences of miR156.miR156 was also found in roots of white lupin treated with −P [27].Although the targets of miR156 are SQUAMOSA PROMOTER BINDING PROTEIN-LIKE(SPL) transcription factors, these miRNAs function as regulators of shoot development, delaying the vegetative phase change [11].Thus, the function of miR156 in rootsawaits further study.Unlike the findings of responsive miRNAs linked to −N and −P conditions, specific miRNAs were identified sporadically under −K conditions, consistent with a few previous miRNA studies on K deficiency [38].The miRNA transcriptome under NPK triple-nutrient deficiency conditions was not solely explained by the independent additive effects of the single deficiencies.Thus, additional combinations of NPK deficiency experiments should be performed.Some miRNA members are responsive to different nutrient deficiencies, and these miRNAs might play important roles under different nutrient deficiencies.Despite the implication of miR169 involvement in the N starvation response [10], most members of the miR169 family were downregulated by −N and −P conditions,in agreement with a previous study [37].miR169 was also downregulated under−NPK conditions,but no additive effect on the expression of miR169 was observed under combined NPK starvation (Fig.3).−K-induced miR169o/i, especially miR169o was not responsive to −N and −P in shoots, whereas 3 miR169 members were suppressed in −K-treated roots (Fig.3-B),suggesting that these miR169 members play an important role in K starvation response.These observations also highlighted the diverse functions of the miRNA members.Most members of miR399 were specifically strongly upregulated in both shoots and roots under Pi starvation conditions,but this increased expression under −P was alleviated by−NPK (Fig.3).Nitrogen starvation could repress the Pi starvation response in plants [39], our results may demonstrate that N starvation represses Pi starvation responses at the miRNA level.miR399 regulates Pi signaling by degradation of PHO2,an E2 conjugating enzyme[22].miR399d/i were also downregulated in shoots by −K, showing their function in K deficiency response.

Table 2– Expression of two predicted novel miRNAs.
sbi-miR171c and sbi-miR528 were both responsive to −N,−P,−K,and −NPK in roots.sbi-miR171c was downregulated by−N, −P, and −NPK, whereas it was upregulated in roots by −K(Fig.3).This finding suggests that miR171c has a different function in −K than in −N and −P responses.miR528 was downregulated in both shoots and roots under −N,−P,−K,and−NPK (Fig.3).miR528 is a monocot-specific small RNA that is involved in regulating flowering, ROS accumulation and antiviral defense in rice, lodging resistance under high nitrogen conditions in maize,and resistance to salinity stress and nitrogen starvation in creeping bentgrass [40-42].sbimiR408,sbi-miR156,and sbi-miR164c were also responsive to−N, −P, −K, and −NPK in shoots.miR408 regulates many biological processes including biotic and abiotic stress responses, fertility, flowering time, and biomass production[43-46].Thus, miR528 and miR408 may represent a common pathway for plant responses to NPK starvation, such as ROS signaling [47].miR156 is a regulator of shoot and flower development [11].Although miR164 is a negative regulator of N remobilization during leaf senescence,it is also involved in lateral root initiation [48,49].In roots, five miR164 members were downregulated by −P, whereas one and three members of miR164 were upregulated by −N and −K,respectively(Fig.3),indicating their functional diversity in response to P,N,and K starvation in roots.
Although we predicted 550 novel miRNAs, most novel miRNAs showed low expression levels, in agreement with previous reports in grape and sorghum[50,51].A small proportion of these novel miRNAs are likely homologs of known miRNAs reported in miRbase(Table S18).It is possible that novel_mir_74 is miR827 in sorghum because of its expression pattern (Table 2).Possibly because of the lack of updated database results,miR827 was not detected in sorghum.Relatively low expression levels in−NPK-treated plants indicated that the expression of novel_mir_74 was also affected by −N and −K.miR827, a Pispecific starvation-responsive miRNA,is involved in maintaining nitrate-dependent phosphate homeostasis by targeting the NLA gene in Arabidopsis [21,23].Two SPX-MES domain protein genes were the predicted targets of miR827 in sorghum(Table S15).The SPX-MES domain protein genes are likely involved in the phosphate-starvation response [52].miR827 targets AtNLA,which is a RING-type ubiquitin ligase.Unlike what has been demonstrated in Arabidopsis,in rice,the predicted target genes of osa-miR827 were not homologs of AtNLA.Although OsNLA1, a homolog of AtNLA,is also involved in the regulation of phosphate signaling in rice,several pieces of evidence suggest that OsNLA is not the target of OsmiRNA827;the function of miRNA827 differs between Arabidopsis and rice[53].In addition,novel_mir_74 was predominantly expressed in shoots, in contrast to wits expression in Arabidopsis.Thus,the functions of miR827 in sorghum and Arabidopsis are likely different.It is possible that the function of miR827 is conserved between rice and sorghum.
Novel_mir_42 showed high similarity with miRNA169 but with two base pair mutations in the 14- and 15-nt positions compared with the sorghum miRNA169 family members(Fig.5).This predicted novel miRNA was downregulated under −N and −P conditions and was upregulated in roots of−K-treated plants only.miRNA169o/i were also induced in shoots under −K- conditions, although most members of miR169 were downregulated under −N and −P conditions(Fig.3).Three predicted targets of novel_mir_42 were CCAATbinding transcription factor(CBF-B/NF-YA)subunit B genes,in agreement with results from Arabidopsis [10].The expression of these three predicted targets also showed an exact inverse correlation with that of novel_mir_42, especially in shoots of−N− and −P-treated plants (Fig.5).Thus, novel_mir_42 is probably a new member of the miR169 family.miR169 may be involved in the NPK starvation response in plants.Further studies should be conducted to investigate miR169s involved in P and K starvation.Other predicted novel miRNAs should also be validated in future experiments.
CRediT authorship contribution statement
Zhenxing Zhu and Xiaochun Lu designed the experiment.Zhenxing Zhu, Dan Li, and Ling Cong performed the experiment.Zhenxing Zhu analyzed the data and wrote the manuscript.Zhenxing Zhu and Xiaochun Lu revised the article.All authors read and approved the final manuscript.
Declaration of competing interest
Authors declare that there are no conflicts of interest.
Acknowledgments
This research was supported by the National Key Research and Development Program of China (2018YFD1000701,2018YFD1000700), the China Agriculture Research System(CARS-06-13.5-A3), and the National Natural Science Foundation of China(31301393).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2020.07.005.
- The Crop Journal的其它文章
- Brief Guide for Authors
- Short Communication Seed-specific overexpression of cotton GhDGAT1 gene leads to increased oil accumulation in cottonseed
- Waterlogging stress in cotton:Damage,adaptability,alleviation strategies,and mechanisms
- Dominant early heading without yield drag in a sister-line BC breeding progeny DEH_229 is controlled by multiple genetic factors with maineffect loci
- Genome-wide alternative splicing variation and its potential contribution to maize immature-ear heterosis
- RNA interference targeting ω-secalin genesdifferentially affects the processing quality in a wheat T1BL·1RS translocation line

