Role of microRNA dysregulation in childhood acute leukemias:Diagnostics, monitoring and therapeutics: A comprehensive review
Joanna Szczepanek
Joanna Szczepanek, Centre for Modern Interdisciplinary Technologies, Nicolaus Copernicus University, Toruń 87100, Poland
Abstract MicroRNAs (miRNAs) are short noncoding RNAs that regulate the expression of genes by sequence-specific binding to mRNA to either promote or block its translation; they can also act as tumor suppressors (e.g., let-7b, miR-29a, miR-99,mir-100, miR-155, and miR-181) and/or oncogenes (e.g., miR-29a, miR-125b, miR-143-p3, mir-155, miR-181, miR-183, miR-196b, and miR-223) in childhood acute leukemia (AL). Differentially expressed miRNAs are important factors associated with the initiation and progression of AL. As shown in many studies, they can be used as noninvasive diagnostic and prognostic biomarkers, which are useful in monitoring early stages of AL development or during therapy (e.g., miR-125b,miR-146b, miR-181c, and miR-4786), accurate classification of different cellular or molecular AL subgroups (e.g., let-7b, miR-98, miR-100, miR-128b, and miR-223),and identification and development of new therapeutic agents (e.g., mir-10, miR-125b, miR-203, miR-210, miR-335). Specific miRNA patterns have also been described for commonly used AL therapy drugs (e.g., miR-125b and miR-223 for doxorubicin, miR-335 and miR-1208 for prednisolone, and miR-203 for imatinib),uncovering miRNAs that are associated with treatment response. In the current review, the role of miRNAs in the development, progression, and therapy monitoring of pediatric ALs will be presented and discussed.
Key words: MiRNome, MicroRNA; Acute leukemia; Acute myeloid leukemia; Acute lymphoblastic leukemia; Biomarker; Classification; Prognosis; Drug resistance
INTRODUCTION
Acute leukemias
Childhood acute leukemias (ALs) are a group of diseases with varied immunophenotypes and specific genetic abnormalities[1]. Acute lymphoblastic leukemia (ALL), the most common type of childhood leukemia, develops from early forms of B- or T-cells at different stages of maturity[2]. Most of the remaining cases account for acute myeloid leukemia (AML), which develops from myeloid cells that form white blood cells[3](other than lymphocytes), red blood cells, or platelets[1]. The 5-year survival rate for children with AL has greatly increased over time and is now more than 90% overall for ALL and in the range of 60% to 70% for AML[4,5]. Survival rates vary depending on the subtype of AL and other prognostic factors[1]. Relapse risk can be predicted by clinical and pharmacogenetic features, early treatment response to tailor chemotherapy intensity, and genetic characteristics of leukemic cells[6-8].
MicroRNA
MicroRNAs (miRNAs, miRs) are a family of small (about 22-nucleotide), endogenous,noncoding RNAs that negatively regulate gene expression in a sequence-specific manner[9,10]. It has been predicted that miRNAs, which account for at least 1% of human protein-coding genes, can control more than a third of the protein-coding genes[11-13]. At the posttranscriptional level, miRNAs exhibit temporally and spatially regulated expression patterns[9,14]. Discovering miRNA molecules, identifying their targets, and predicting their functional and regulatory mechanisms are critical for understanding biological processes (e.g., diverse development, cell growth control proliferation, differentiation, and apoptosis) and their roles in the etiopathology of disease[15,16].
Great interest has emerged in modulating miRNA expression for therapeutic purposes. Many scientific studies have shown that these molecules play an important role as regulators of genes in many organisms, and miRNAs have already been implicated in a growing number of human diseases, including cancers[12,16,17]. miRNAs can act as either oncogenes and/or tumor suppressors, contributing to malignant transformation in solid and hematological tumors[18,19]. miRNAs that typically target a tumor suppressor are classified as oncomiRs and are generally upregulated in different types of cancer. MiRNAs, which downregulate oncogenes, are defined as tumor suppressor miRs[19]. Promoter methylation, mutation, or deletion or defective miRNA processing often leads to loss of suppressor miRs in cancer[20]. Some miRNAs may exert contrasting oncogenic/tumor-suppressive effects on cancer-modifying extrinsic factors and the leukemic cells themselves (Figure 1).
Deregulation of miRNA expression patterns is a hallmark of hematological malignancies and can be useful for the classification of AL genetic subtypes, which are important for differential diagnosis, prognosis, and treatment monitoring. These molecules control the levels of potentially large numbers of proteins, many of which might be important drug targets, and are useful in the development of new therapeutic regimens[21]. Cancer-specific miRNA signatures correlated with diagnosis,progression, prognosis, and response to treatment were determined for many cancers,including childhood leukemia. Although the importance of miRNA molecules in the genetic basis of leukemia is documented in an increasing number of publications, the role of miRNAs in pediatric AL still needs to be established. Profiling the expression levels of miRNA molecules at several levels with unprecedented resolution, depth,and speed is possible thanks to the development of high-throughput technologies,such as next-generation sequencing, microarrays, mass spectrometry, and new bioinformatics tools[15,22].
In the current review, the role of miRNAs in the development, progression, and therapy monitoring of pediatric ALs will be presented and discussed.
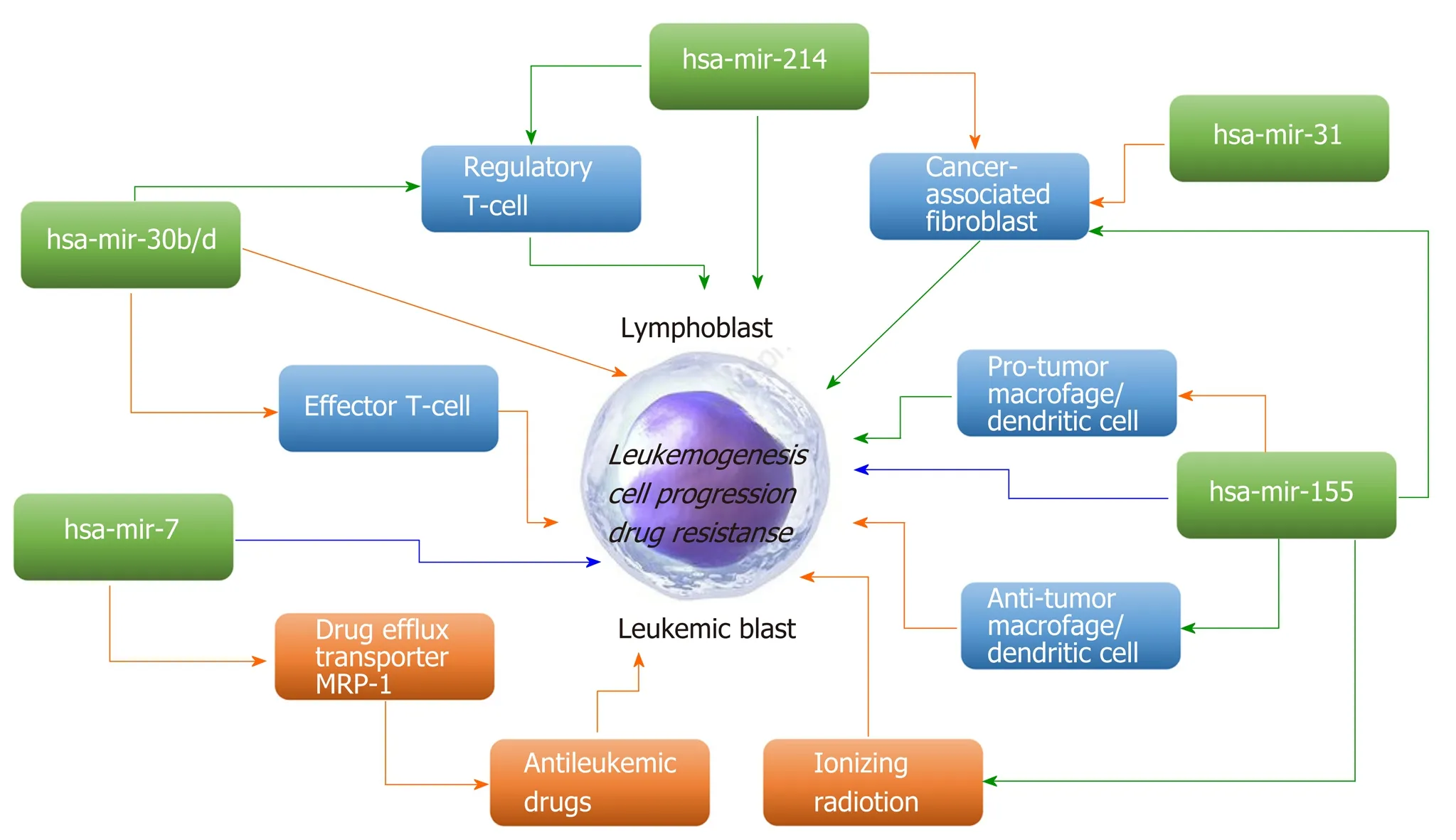
Figure 1 Examples of microRNAs that may exert contrasting oncogenic/tumor-suppressive effects on tumor-modifying extrinsic factors and the leukemic cells themselves (modified from Svoronos et al[19], with the permission of the author). Green arrows, positive regulation; orange arrows, negative regulation;blue arrows, either positive or negative regulation. A narrow extension directly from a microRNA to the central leukemic cell refers to promotion/inhibition of leukemic cell progression/survival through the microRNA’s direct regulation of leukemic cell-endogenous mRNAs.
ROLE OF MICRORNA IN DIAGNOSIS AND CLASSIFICATION
It is very important in the treatment of AL to treat patients in the right risk groups,which allows both maximizing the effectiveness of therapy and minimizing its toxicity. At present, the exact stratification of patients into relevant genetic subclasses,and subsequent risk groups, is still a laborious and expensive diagnostic process,including comprehensive morphology, immunophenotype, cytogenetics, response to induction therapy, and genetic testing[23-26].
Currently, several types of classification can be specified in AL. The main ones are:(A) French-American-British (FAB) morphological classification, distinguishing three ALL subtypes (L1-L3) and eight AML subtypes (M0-M7); (B) Cytochemical classification; (C) Immunological classification; (D) Cytogenetic classification; and (E)Molecular classification[27-29]. The World Health Organization system divides ALL into several groups, the most common of which are B-cell ALL (B-ALL) and T-cell ALL (TALL). B-ALL can be further subclassified according to distinct patterns of genomic alterations and gene expression signatures (most frequent abnormalities:ETV6-RUNX1 fusion, TFPT-PBX1 fusion, BCR-ABL1 fusion; and MLL fusions)[30,31]. In T-ALL, several subgroups have been recognized,e.g., immature/LYL1, TAL1, HOX11,HOX11L2, and HOX. The World Health Organization classification includes several AML subtypes with recurrent genetic abnormalities [e.g., AML with t(8;21)(q22;q22)RUNX1-RUNX1T1, AML with inv(16)(p13.1;q22) or t(16;16)(p13.1;q22)CBFB-MYH11,APL with t(15;17)(q22;q12)PML-RARA, and AML with t(9;11)(p22;q23)MLLT3-MLL][32,33]. In addition, molecular changes cannot be detected by classical karyotyping,such asWT1,NPM1,CEBPA, andDNMT3Amutations. Based on genetic classifications for subgroups of AML, it is supposed that two of the major subgroups of AML patients are most important: Those with disruptions of theCBFcomplex and those with disruptions of theFLT3gene[34,35]. Classification systems are still unsatisfactory because most of the recurrences occur in the intermediate and standard risk groups[26].
It is supposed that the cancer-specific miRNA patterns rather than mRNA signatures are a more accurate method of classifying cancer subtypes[36,37]. Molecular classification based on expression alterations (genes or miRNA) allows the determination of the genetic profile of myeloid and lymphoid lineage, distinguishing B-linear and T-linear ALL tumors, as well as listing subclasses within leukemia subtypes. Based on miRNA analyses, a correlation between immunophenotype and chromosome disorders and various miRNA expression patterns in all subtypes of AL was demonstrated. A top-ten discriminative miRNA set was proposed for AL types and their main subgroup, as shown in Figure 2 and 3.

Figure 2 A top ten discriminative microRNA set for the main acute lymphoblastic leukemia subgroups. The subtypes display a unique discriminating microRNA (except where overlap is shown) that distinguishes each subgroup from each other (modified from Grobbelaar et al[3], with the permission of the author). ALL: Acute lymphoblastic leukemia.
The type and subtype of childhood leukemia plays a major role in both treatment options and patient prognosis. Determining the type (ALL or AML) and subtype of the leukemia is performed by testing samples of the blood, bone marrow, and sometimes lymph nodes or cerebrospinal fluid[23]. In recent years, there has been significant progress in the use of comprehensive transcriptomic and genomic methods(expression, single nucleotide polymorphism, and comparative genomic hybridization arrays; whole exome, whole transcriptome, and whole genome sequencing) to identify leukemia subtypes and inherited and somatic genomic changes, but nevertheless, much work is needed to define the intrinsic and extrinsic determinants of leukemia progression, prognosis, and drug response[8,38,39]. A summary of miRNA marker functions, based on the example of pediatric ALL, is presented in Table 1.
Certain miRNA profiles have been identified to associate specifically with AL types. Zhanget al[40]showed a general expression pattern and assigned 21 upregulated and 11 downregulated miRNAs for primary ALL and 17 upregulated and 18 downregulated miRNAs for primary AML, but only 17 miRNAs showed convergent expression between two types of pediatric AL. By defining sets of characteristic genes for each leukemia subtype, including miR-34a, miR-128a, miR-128b, and miR-146a in ALL and miR-100, miR-125b, miR-335, miR-146a, and miR-99a in AML, they emphasized that these miRNA sets are significantly different from those selected for adult leukemias. This observation is consistent with reports of another group of researchers. Zhanget al[40]selected lineage-specific markers for the most common cytogenetic and FAB classification subtypes of AML. The overexpression of several miRs was significantly related to the degree of cell maturity and differentiation. High expression levels of miR-335 in M1, miR-126 in M2 (andAML1-ETO+), and miR-125b in M3 (andPML/RARAstatus) were observed[40]. In that study, the authors indicated that miR-125b and miR-126 may serve as favorable prognosticators for M3 and M2 patients, respectively. Miet al[41]demonstrated, based on a genome-wide miRNA expression analysis on AL samples, that the expression signatures of only two miRNAs could accurately discriminate ALL from AML (accuracy rate > 95%). They proposed the possibility of using such lineage discriminatory miRNAs to develop a rapid and accurate diagnostic test of ALLvsAML in the future; however, the possibility of a more accurate diagnosis of childhood AL based on the miRNA signature remains to be realized. Miet al[41]used a large-scale genome-wide miRNA expression assay to identify 27 differentially expressed miRNAs as markers for diagnosis and treatment. Of these, miR-128a and miR-128b were significantly overexpressed in ALL compared to AML, whereas let-7b and miR-223 were significantly downregulated[41].
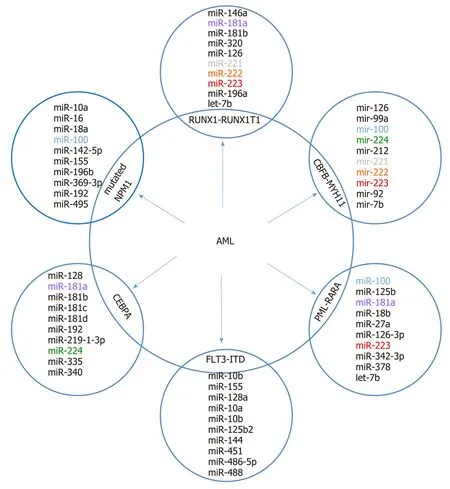
Figure 3 A top ten discriminative microRNA set for the main acute myeloid leukemia subgroups. The subtypes display a unique discriminating microRNA (except where overlap is shown) that distinguishes each subgroup from each other (based on data from Trino et al[69]). AML: Acute myeloid leukemia.
Schotteet al[42]revealed distinct miRNA expression profiles for seven different subtypes of pediatric ALL. They showed that the precursor B-ALL has a specific expression pattern characterized by low expression levels of miR-127 and miR-143.This feature allowed distinguishing this cell line from control and CD34+ cells. In turn, T-ALL cells showed differential expression of 28 miRNAs. High expression of various miRNAs (e.g., miR-223, miR-222/222*, miR-98, and miR-511) was observed in ALL with hyperdiploidy. Interestingly, the miRNA signature ofTEL-AML1-positive and hyper diploid cases partly overlapped, which may suggest a common underlying biology. Interestingly, specific classification profiles have been described for each of the pediatric AL subtypes, except forBCR-ABL1-positive and "B-other" ALL.
Almeidaet al[21]used massive parallel sequencing to describe novel sets of 16 miRNAs correlated with childhood ALL subtypes. Among the subtype discriminators, ten (miR-708-5p, miR-497-5p, miR-151a-5p, miR-151b, miR-371b-5p, miR-455-5p, miR-195-5p, miR-1266-5p, miR-574-5p, and miR-425-5p) were downregulated and six (miR-450b-5p, miR-450a-5p, miR-542-5p, miR-424-5p, miR-629-5p, and miR-29c-5p) were upregulated in pediatric T-ALL. Researchers also assigned individual molecules to six functional categories, of which three were associated with inducedmiRNAs and three with repressed miRNAs in T-ALL (encompassed 45 genes that were shared by induced and repressed miRNAs; 210 genes were targeted by overexpressed miRNAs, and 143 genes were targeted by downregulated miRNAs).Differentially expressed genes and their targets represented relevant biological pathways, including viral carcinogenesis, cell cycle, and B-cell receptor signaling pathways for induced miRNAs andTGF-βsignaling, apoptosis,NF-kappa Bsignaling,and cell differentiation and hematopoiesis processes[21]. Almeidaet al[21]also identified miR-29c-5p as the best discriminator of the pediatric leukemia cell line.

Table 1 Targets of microRNA associated with acute leukemia (based on data in Human microRNA Disease Database http://www.cuilab.cn/hmdd)
MiRNA: MicroRNA; ALL: Acute lymphoblastic leukemia; AML: Acute myeloid leukemia; Hsp90: Heat shock protein 90; TGF-β1: Transforming growth factor beta 1; UTR, Untranslated region; POLD1: Polymerase delta 1; MCM2: Minichromosome maintenance complex component 2; PLK-4: Polo-like kinase 4.
Liet al[43]showed that the downregulation of miR-100 and miR-99a expression levels was a significant feature of leukemic blasts and was strongly correlated with the patient's 5-year survival. For these two marker molecules, differences in expression patterns between ALL and AML were noted, and white blood cell (WBC)count, ALL type (T-cell or B-cell), theMLL-rearranged gene, and theBCR-ABLfusion gene were correlated with changes in miR-100 and miR-99a levels. In addition, it has been shown inex vivoexperiments that upregulation of the expression of these molecules inhibited the expression ofIGF1RandmTORand their downstream oncogeneMCL1[43]. de Oliveiraet al[44]focused on assessing expression selected based on previous studies of miRNA markers[45], including miR-92a, miR-100, miR-125a-5p,miR-128a, miR-181b, miR-196b, and let-7e. As noted in leukemic blasts, miR-100, miR-196b, and let-7e showed lower expression levels, and miR-128 and miR-181 showed higher expression levels than normal bone marrow cells. Overexpression of miR-196 was observed for T-ALL. A high expression level was characteristic for patients who presented a WBC count < 50000/mm3at diagnosis (P= 0.01) and was also associated with the presence of t(12;21) and the absence of a hyperdiploid karyotype. This suggests the possibility of a t(12;21)-specific regulation of miR-100. Differentiated expression of miR-181b and miR-128a was associated with the presence of t(4;11)[44].
Swellamet al[46]investigated the expression signature of miRNA-125b-1 and miRNA-203 among childhood ALL, and they proposed these miRNAs as useful molecular markers for the diagnosis of childhood ALL. They noticed that the expression level of miRNA-125b-1 was significantly higher in peripheral blood (PB)isolated from 43 newly diagnosed children with ALL, while the miRNA-203 level was significantly lower in childhood ALL compared to control samples. Moreover,miRNA-125-1 was increased in T-ALL compared to other ALL phenotypes, and the miRNA-203 expression level was high in T-ALL followed by pre-B-ALL[46].
Nabhanet al[47]focused on analyzing the involvement of just one marker molecule in the development of leukemia,i.e. miR-181a can both function as a tumor suppressor or an oncogene. The function of miR-181a is associated with cell metabolism and expression levels of target mRNAs. A decrease in the expression level of miR-181a was observed in the serum of children diagnosed with ALL.Investigators, as the first team, linked the miR-181a expression signature with Smad7 and TGF-β1 protein levels in the serum of childhood ALL. They found that miR-181a expression achieved a highly significant positive and a significant negative correlation with TGF-β1 and Smad7, respectively[47]. Furthermore, miR-181a was identified by Yanget al[48]as the most differentially downregulated miRNA (among the 17 identified miRNAs) in the PB of childhood ALL patients with the t(12;21)translocation. Nabhanet al[47]suggested that the diagnostic accuracy of pediatric ALL can be improved by using a small set of miRNA markers. Based on the data, they calculated that the combined use of miR-181a and Smad7 increased the sensitivity of diagnosis to 90%, whereas the combined use of miR-181a and TGF-β1 increased sensitivity to 100%.
It is supposed that differentiated miRNA expression profiles contribute to AML heterogeneity and have diagnostic and clinical significance. In numerous studies,comparisons of miRNA profiles between AML blasts and normal cells and among AML with recurrent genetic abnormalities were made[49-56]. In AML, miRNA signatures can also distinguish between cytogenetic subtypes[51-53,57]or molecular subtypes (e.g., NPM1, CEBPA, or FLT3 mutations)[52]. Most of the reported miRNAs in pediatric AML are presented in Figure 3. The prognostic significance of miRNA expression patterns in AML was confirmed[58-66].
Zhuet al[67]indicated that the 3-miRNA signature contributed to pediatric cytogenetically normal AML and can be a reliable prognostic biomarker. They found that high expression of miR-146b predicts poor prognosis and that miR-181c and miR-4786 are significantly related to favorable prognosis factors. The results indicate that the 3-miRNA-based signature is a reliable prognostic biomarker for pediatric AML.The upregulation of miR-10a, miR-10b, and miR-196b[52,68]and the downregulation of miR-192[51,68]can be biomarkers forNPM1mutations. Patients with t(8;21) had overexpression of miR-126[53,56]and miR-146a[49,57]. High expression level of miR-155 was strongly associated withFLT3-ITDalteration[50-52,69].
ROLE OF MICRORNA IN DIAGNOSIS AND CLASSIFICATION
The results of scientific research have led to the selection of biomarkers for diagnostic classification and prognostic evaluation, and have thus brought closer personalized medicine. Nevertheless, there is a constant need to search for sensitive and specific determinants for childhood AL progression[3,70].
One of the most commonly identified biomarkers associated with the development of AL is miR-125b, which is an important molecular regulator in normal cell homeostasis, cell metastasis, and disease pathogenesis and progression[71,72]. Playing either an oncogenic or a tumor-suppressive role through numerous target genes is crucial in abnormal proliferation, metastasis, and invasion of cells in hematological malignancies[71]. Soet al[73]reported that miR-125b overexpression (through repressingIRF4) promotes myeloid and B-cell leukemia by inducing tumorigenesis, immortality,and self-renewal of progenitor cells. By targetingARID3ain B-ALL cases with the t(11;14)(q24;q32) translocation, upregulation of miR-125b blocked differentiation and helped avoid apoptosis by blockade of caspase activation by a mechanism independent of p53 andBAK1. Moreover, high levels of miR-125b were linked to an increase in the expression of pluripotency-associated factors (OCT4,SOX2,KLF4, andNANOG)[74]. By suppressingTNFAIP3, theNF-κB-mediated increase in B-cell proliferation and dysregulation of glucose metabolism result in a reduction in apoptosis in T-ALL[75]. MiR-125b also blocks differentiation in myeloid progenitor cells with the t(2;11)(p21;q23) translocation[76]. In AML, the induction of leukemogenesis can be mediated by: (1) Pathways includingCDX2, miR-125b, andCBFβ(high levels ofCDX2activate miR-125b transcription, which in turn inhibitsCBFβtranslation)[77];(2) TargetingSTAT3transcription factors (alsoJUNDandBAK1)[78]; and (3)SuppressingABTBantiproliferative factors and deregulating genes involved in the p53 pathway, includingBAK1andTP53INP1[79].
One of the potential miRs related to important regulators in myeloid development is miR-223, a known regulator of myelopoiesis. Experimental works have shown that its expression increases with the degree of cell differentiation[80]. The high relative expression of miR-223 in AML1 samples was found by Ramsinghet al[81], but overexpression of miR-223 was not a common feature of leukemic cells. Danen-van Oorschotet al[82]observed levels of miR-29a, miR-155, miR-196a, and miR-196b in clinically relevant cytogenetic and molecular subgroups of 82 pediatric AML samples and observed higher expression of miR-196a/b in leukemic cells withMLLgene rearrangements,NPM1mutations, andFLT3-ITDin cytogenetically normal AML.Downregulation of miR-196a/b expression was observed inCEBPAmutated cases.Differentiated expression of these miRs was linked toHOXAandHOXBcluster genes involved in myeloid transformation. InFLT3-ITDandNPM1-mutated cases, miR-155 was overexpressed, and lower expression of miR-29a inMLL-rearranged pediatric AML was found[82]. A broad miRNA profiling experiment of cytogenetically distinct childhood AML cases was conducted by Daschkeyet al[49], where they selected miR-126, miR-146a, miR-181a/b, miR-100, and miR-125b as markers of theMLLrearranged AML subtype. Emmrichet al[83]pointed out that miR-9 is an important regulator of t(8;21)-mediated leukemogenesis. Low levels of miR-9 were observed in AML patients with t(8;21), but high expression levels were noted in cases withMLLrearrangements. As a potential mechanism of regulation of leukemic cell proliferation and differentiation, repression of the oncogenicLIN28B/HMGA2axis (including target genesCDH1,NFKB1,BACE1, andRES) was identified.
ROLE OF MICRORNA IN OUTCOME PREDICTION
Children with AL are often put into risk groups (low, intermediate, or high risk), with more intensive treatment given to higher risk patients. Generally, children at low risk have a better outcome than those at very high risk. However, it is important to know that even children in higher risk groups can often still be cured. AL is a heterogeneous disease; hence, different treatment results are observed in patients with similar histopathological diagnoses, stages of development, and similar treatment protocols.The correct stratification of patients into high-risk groups with recurrence (more intensive treatment) and low-risk patients (avoidance of therapy toxicity) is of great importance[84,85]. The previous stratification does not meet the abovementioned expectations, which is why high hopes have been placed on the possibilities resulting from research using genomics and transcriptomics methods[84].
More and more often in pediatric AL, the expression signature of the set of miRNA molecules has been correlated with well-known prognostic factors, which include:WBC count, age at diagnosis, cytogenetic and molecular genetic analyses of blast cells,prednisolone response on day 8, and immunophenotype. The possible model of diagnostic and prognostic management in pediatric ALL based on the miRNA signature is presented in Figure 4.
In recent years, several studies have been conducted to identify miRNAs as predictors of the risk of leukemia recurrence. One of the first verified biomarkers was miR-16, which is implicated in apoptosis induction by targetingBCL-2and cell cycle arrest[86-88]. Kaddaret al[89]obtained statistically significant relationships between low miR-16 levels and low WBC counts and good molecular markers. They found that, in the entire ALL group, miR-16 was significantly downregulated in the group of patients with leukocytes below 50 G/L and with hyperploidy or t(12;21). In T-ALL,overexpression of miR-16 expression was also related to corticosteroid resistance.After all, they could not assign a specific miR-16 expression profile to B-cell and TALL subgroups[89]. In the next study, Organista-Navaet al[90]conducted a multivariate analysis (including age at diagnosis, gender, and WBC of miR-24 expression, a wellknown promotor of the survival of hematopoietic cells). Targets of miR-24 are proapoptotic (FAF-1, caspase 9,Bim, andApaf-1) and cell cycle progression (enhancedMYC,E2F2,CCNB1, andCDC2or inhibitedp27Kip1andVH) proteins[91-93]. In a previous study, it was reported that miR-214 expression is associated with cytogenetic and molecular subtypes of adult AML (e.g., AMLs with t(8;21), t(15;17), inv(16),NPM1, andCEBPAmutations)[52]. Organista-Navaet al[90]indicate miR-214 as an independent marker for predicting the clinical outcome in both AML and ALL patients. Upregulation of miR-24 was significantly associated with poor prognosis,shorter overall survival (OS), and a high risk of leukemia relapse.
One of the most interesting research results in this field were presented by the team of Nemeset al[94]. Researchers collected PB and bone marrow samples from 24 ALL patients from all phases of treatment [collected at diagnosis, at conventional response checkpoints (on days 15, 33), and before beginning protocol M]. Differentiated expression of set miRNAs (miRNA-16, miRNA-21, miRNA-24, miRNA-29b, miRNA-128b, miRNA-142-3p, miRNA-155, and miRNA-223) with potential roles in hematologic malignancies was analyzed. Based on the results obtained, it was concluded that miR-223 (involved in the regulation of the cell cycle or different signaling mechanisms, such asE2F1, CEBPα, andE2A) and miR-128b (play roles in the regulation of thePI3K-AKT-mTORsignaling pathway through downregulation ofPTEN) expression signatures could be possible predictors of ALL relapse. Nemeset al[94]determined in general that high levels of miR-128b and low levels of miR-223 show a significant correlation with good prednisolone response and better prognosis in childhood ALL. In particular, they found an extreme high expression level of miR-128b at diagnosis, which significantly decreased as patients entered remission where normal levels of miR-128b expression for mononuclear cells were detected.Conversely, miR-223 expression can be undetectable at diagnosis, but during treatment and in remission, the level of miRNA is standard and then decreases again at relapse[94].
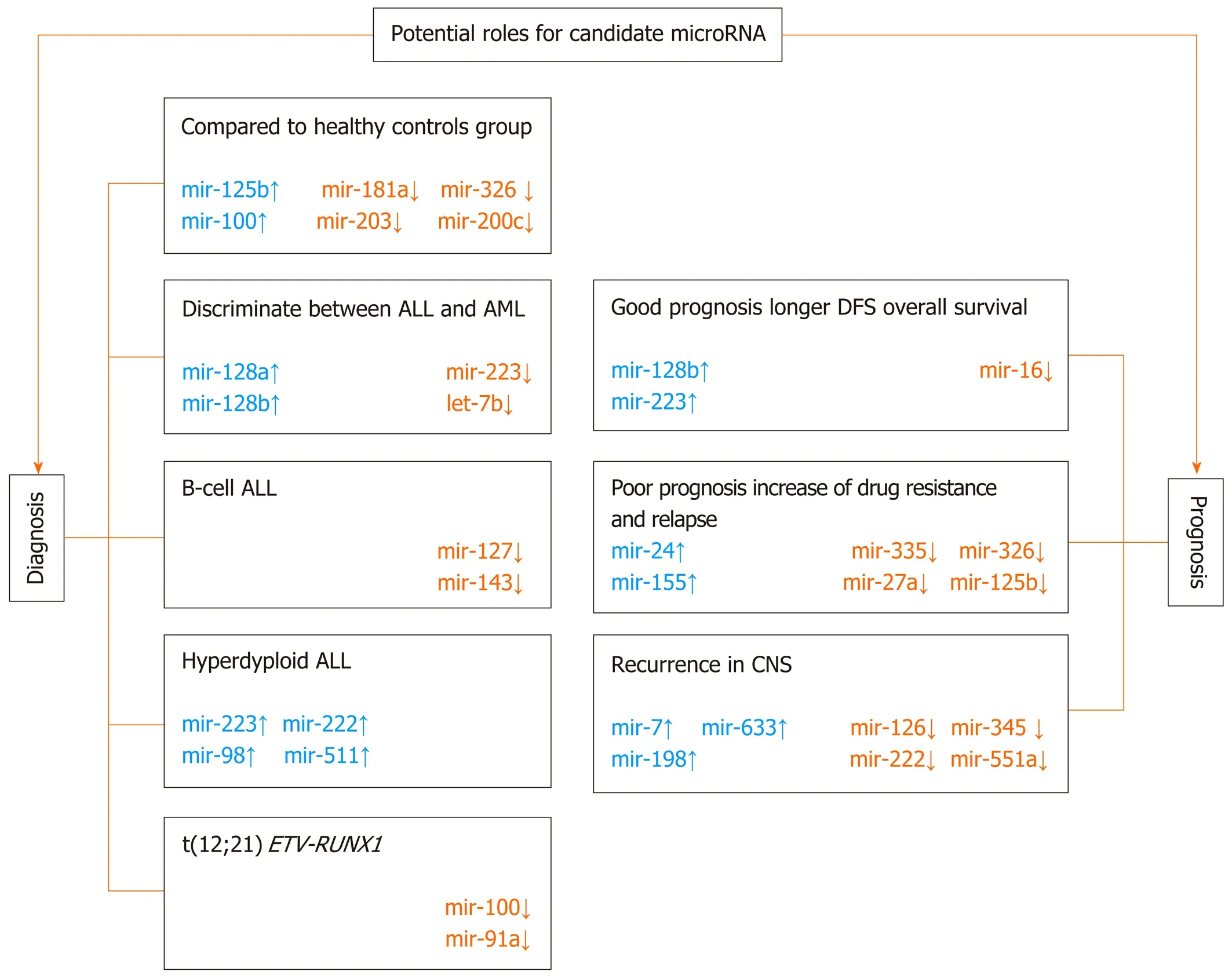
Figure 4 Possible roles of microRNAs in the diagnosis and prognosis of childhood acute lymphoblastic leukemia (modified from Grobbelaar et al[3], with the permission of the author). ALL: Acute lymphoblastic leukemia; AML: Acute myeloid leukemia.
Among the newly typed prognostic and therapeutic markers for pediatric AL is miR-335, which acts as a tumor suppressor and targets genes that participate in most of the important biological processes associated with human cancer and is involved in pathways such as p53, mitogen activated protein kinase, TGF-β, Wnt, epidermal growth factor, mammalian target of rapamycin, Toll-like receptor, and focal adhesion[95]. Using genome-wide miRNA microarray analysis, Yanet al[96]found that low expression level of miR-335 was associated with unfavorable prognosis, poorer 5-year event-free survival, and glucocorticoid resistance in ALL. Moreover, the study highlighted the potential mechanism of silencing the expression of miR-335.According to previous observations of recurrent leukemia[97-99], epigenetic silencing through DNA methylation was suggested. Yanet al[96]used the results ofin vitroexperiments to show that exogenous expression of miR-335 in leukemic cells increases sensitization to prednisolone-mediated apoptosis, and they concluded that reintroducing miR-335 expression or overridingMAPK1activity could become a promising therapeutic target for ALL treatment. The role of miR-335 in AML was analyzed by Zhouet al[100]. They found that: (1) Overexpression of miR-335(independent of its methylation) was negatively correlated with decreasedID4expression; (2) Aberrant miR-335 andID4(direct target) expression independently affected chemotherapy response and leukemia-free/OS in patients with AML; and (3)miR-335/ID4dysregulation facilitated leukemogenesis through the activation of thePI3K/Aktsignaling pathway. These results are similar to the Yanet al[96]study, which usedex vivoexperiments to show that it was possible to reduce pro-proliferative and antiapoptotic effects and restore the physiological role of mir-335 (through restoration ofID4expression)[100].
MiR-155 is another candidate predictive and prognostic marker of AL outcome.MiR-155 is evolutionarily conserved, and as an inhibitor of lineage differentiation, it is one of the most critical regulators of posttranscriptional gene expression in B cells[101,102]. It has also been confirmed to be associated with pathogenesis,aggressiveness and progression in CLL[103,104]; poor survival in adult and pediatric AML[105]; and poor clinical prognosis in Hodgkin's lymphomas[106]and B-cell-type Diffuse large B-cell lymphoma[107,108]. El-Khazragyet al[109]tried to connect the upregulation of miRNA-155a and miRNA-181a expression to ALL outcome. They found a significant correlation of high levels of minimal residual disease (MRD) and poor prognosis, but only overexpression of miRNA-155a was significantly related to high blast numbers (> 25%), unfavorable cytogenetic abnormality, total WBC, higher relapse rate, a higher MRD after 15 d, and poor prognosis[109]. Moreover, expression of both miR-155a and miR-181a was downregulated after chemotherapy, suggesting their potential use as biomarkers of therapeutic response in pediatric ALL.
A consistent and strong association of miR-155 overexpression with poor prognosis in pediatric AML was also confirmed[110-112]. An interesting relationship between the level of expression of miR-155, its biological function and prognosis in AML was noted by Narayanet al[105,111]. Researchers conducted experiments on cells obtained from patients diagnosed with AML and on a murine model, where they found that between 10- and 50-fold overexpression of miR-155 (plays the role of oncogene) is associated with poorer OS and increased tumor burden. However, when the miR-155 expression level is lower than 10-fold, outcomes may be favorable. When the level is higher than 50-fold (observed only in the murine model), miR-155 shows suppressor activity. Overexpression of miR-155 is associated with activation of B cells with an inflammatory stimulus of lipopolysaccharide andIL-4. The consequence of differentiated miR-155 expression is downregulation of the expression of target genes regulated by intermediate (CEBPB,SPI1, andTLE2) or high (MYBandKIT) miR-155 expression levels. Narayanet al[105,111]first described a novel dose-dependent function of miR-155 in the progression of AML and pointed out that it can have important therapeutic implications.
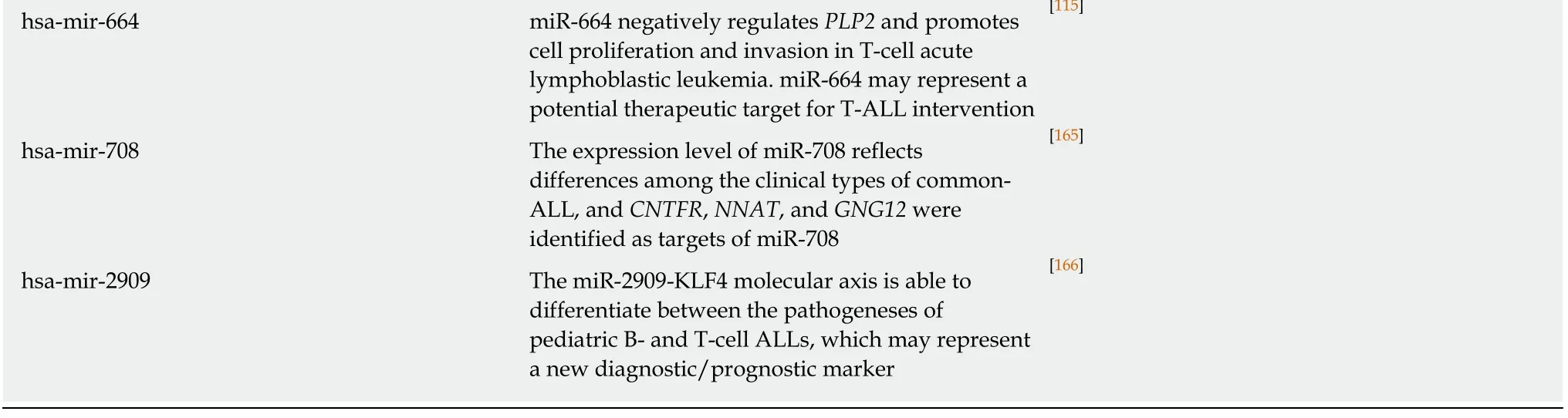
miRNAs can also act as independent prognostic factors to predict clinical outcomes for T-ALL patients. Miaoet al[113]identified highly expressed miR-590 as a candidate oncogenic marker for both adult and pediatric T-ALL. Through regulation of theRB1gene, miR-590 increased the proliferation and invasion of T-cells[113]. A common oncogenic marker in adult and pediatric T-ALL is miR-149. Overexpression of miR-149 inhibits apoptosis and enhances proliferationviathe target geneJUNB[114]. Another example is miR-664, whose overexpression inhibits the PLP2 gene in pediatric T-ALL.Deregulation ofPLP2results in changes in adhesion and migration of leukemic cells[115].
MiR-181 is often mentioned among AML prognostic markers. In adults, its relationship with prognosis in AML is quite well described[60,116]; however, only one published paper describes the correlation between miR-181 expression level and prognosis in the pediatric population. Liuet al[117]analyzed the physiological function and mechanism of miR-181 and found that these molecules are responsible for the G1/S transition and cell proliferationviathe tumor suppressorATM.
Zhanget al[65]found that high expression of oncogenic miR-99 is significantly related to the promotion of proliferation and apoptosis inhibition. Overexpression of miR-99a was observed in pediatric AML (FAB subtypes M1-M5). Interestingly, during complete remission, silencing expression of miR-99 was noted. Regulation of cell growth and differentiation was associated with modulation of the expression of such miR target tumor suppressor genes asCTDSPLandTRIB2.
Evolutionarily conserved miR-125b, as an important regulator of hematopoietic stem/progenitor cell apoptosis, confers a proliferative advantage to leukemic cells in both ALL and AML[118]. In AML, the level of miR-125b could be up to 90-fold higher in comparison to normal cells[76]. In patients with B-ALL carrying the t(11;14)(q24;q32)translocation, expression of miR-125b is also 30- to 600-fold higher in comparison to cases without the translocation[72,119]. In AML, as shown by Ufkinet al[120], miR-125a expression was downregulated in favorable and intermediate prognoses and associated with decreased survival. In addition,in vitroexperiments have identified a potential mechanism for regulating miR-125a expression, which is excessive methylation. They undertook effective attempts at global demethylation using decitabine, which resulted in an increase in miR-125a levels as well as, in consequence, inhibition of cell cycle proliferation and progression with increased apoptosis. In their study, they revealed that theErbBpathway is directly regulated by miR-125a. The authors suggested that further research intoErbBinhibitors and miR-125a molecules may contribute to the development of targeted AML therapy[120].
Linet al[61]indicated miR-370 as a potential noninvasive diagnostic and prognostic miRNA marker for childhood AML cases because its expression in bone marrow and serum samples at diagnosis was significantly decreased. The level of miR-370 was correlated with the FAB classification subtype M7 (P= 0.02) and unfavorable karyotype and with poor prognosis, unfavorable relapse-free survival (RFS), and shorter OS. Linet al[61]also noticed that serum miR-370 levels were more obvious in the subgroup of patients with intermediate-risk cytogenetics. Second, Linet al[62]described miR-335 as an independent prognostic marker of RFS and OS in pediatric AML. His high expression was related to the M7 subtype, unfavorable karyotype presence and shorter RFS and OS. The function of the biomarker unfavorable prognosis was confirmed for oncogene miR-183 by Wanget al[63]. High expression of miR-183 (especially in M7 AML) was associated with promotion of cell proliferation and G1/S transition and inhibition of apoptosis. Short RFS and OS were linked markedly with silencing ofPDCD6expression, a direct and functional target of miR-183. Wanget al[64]also proposed miR-375 as a prognostic factor for unfavorable cytogenetic risks in M7 ALL. Zhuet al[66]analyzed the clinical significance of miR-29a expression, a well-known gene that can act as either an oncogene or tumor suppressor. Low expression of this gene was characteristic of the M7 subtype of childhood AML and had shorter RFS and OS, while high expression was associated with good prognosis.
In summary, the most frequently identified prognostic miRNA markers of pediatric AL include the following: (1) miR-7, miR-16, miR-33, miR-100, miR-130b, miR-181,miR-215, miR-216, miR-369-5p, miR-496, miR-518d, miR-599, and miR-708 are unfavorable factors[40,42,89,121,122]; and (2) miR-10a, miR-23a, miR-27a, miR-128b, miR-134,miR-150, miR-191, miR-214, miR-223, miR-342, miR-484, miR-486, miR-487, miR-572 miR-580, miR-627, miR-624, let-7g, and let-7i are favorable factors[40,42,121,122].
Table 1 summarizes miRNAs involved in ALL prognosis, their impact in predicting relapse or progression, and their relationship with OS and treatment outcome.
ROLE OF MICRORNA IN THERAPY OUTCOME AND DRUG RESISTANCE
The correct assignment of patients to risk groups presents another challenge, which therapy is the most appropriate to use. Anticipating the response to the administered drug is a challenge in current oncology[123-126]. AL is curable in children in 60%-90% of cases[127,128]. Several research groups have attempted to search for sets of miRNAs involved in therapy failure. Assuming that the phenomenon of drug resistance is a primary feature of leukemia blasts, it is possible to predict the ineffectiveness of treatment[128]. MiRNA profiling is increasingly used in research on multifactorial phenomena, such as drug resistance, due to its ability to simultaneously analyze the most important genes for this process[129]. The mechanism of drug resistance consists of many complex processes that overlap one another, including: (1) A change in the function of receptors; (2) Heterogeneity of cancer cells; (3) Tissue microenvironment;(4) Intercellular interactions; (5) Disorders in transport and signal transduction; and(6) A change in the expression of key genes[130]. However, the genetic basis of chemotherapy resistance is still not well understood. Based on the genetic profile, it is possible to predict the response to the treatment used by establishing a correlation between the expression of specific sets of miRNAs and the sensitivity/resistance to a given anticancer agent or the effectiveness of the treatment (Table 2)[25,84].
Multidrug resistance followed by relapse is regarded as one of the most important clinical problems for effective treatment in patients diagnosed with AL and is still the main cause of cancer death in children[131]. MiRNAs are considered a relevant regulator response to drug administration. Ghodousiet al[132]conducted an analysis of the expression ofABCA2andABCA3transporters and their potential regulators, miR-326 and miR-200c. First, they confirmed that the expression levels of both miR-326 and miR-200c were significantly lower in patients with ALL diagnosis. They showed a significant downregulation of miR-326 in MRD+ and relapsed patients compared to the MRD- group, supporting the notion that decreased expression of miR-326 has an adverse impact on treatment response. A significant decrease in miR-200c expression level was observed between the MRD- patients and relapsed ALL patients. Ghodousiet al[132]pointed out that only the miR-326 level can be a negative prognostic biomarker that may discriminate between MRD+ and MRD- patients. The presented results(higher miR-128b expression correlated with good prednisolone response and better prognosis) suggest a correlation between miR-128b expression changes and steroid sensitivity of leukemic cells[94].
In several subsequent studies, miR-125b was selected as a potential biomarker for leukemia recurrence and was correlated with a high risk of therapy failure and poor survival prognosis[72,133-135]. Piatopoulouet al[135]evaluated the clinical significance of miR-125b for ALL prognosis and prediction of patients’ response to the Berlin-Frankfurt-Muenster (BFM) chemotherapy protocol. In their study, a low level of miR-125b was related to unfavorable prognosis, but after treatment with the BFM protocol,overexpression of this miR was detected. Higher expression on day 33/diagnosis was related to a higher risk for disease short-term relapse and worse survival, strongly suggesting that miR-125b could also be used as a clinically useful predictor of resistance to BFM chemotherapy[135]. Gefenet al[134]established upregulation of the miR-125b-2 cluster (miR-125b, miR-99a, and let-7c) inETV6/RUNX1-positive childhood ALL. Overexpression of the cluster was an independent leukemia event.Increased miR-125b-2 levels protected cells from death. The antiapoptotic activity can be associated with a marked inhibition of caspase 3 activation and the cleavage of its substratePARP. Differentiated expression of miR-125b-2 was also related to resistance to antileukemic agents staurosporine and doxorubicin.
In a study by Schotteet al[42], changes in the expression of miR-454 were only related to L-asparaginase resistance, but the strong overexpression of three miRs (miR-125b,miR-99a, and miR-100) was correlated with vincristine (VCR) and daunorubicin resistance. The strong association of miR-125b upregulation was especially evident inETV6-RUNX1+ patients, who were VCR-resistant[42]. The overexpression of miR-125b was linked to inhibition of VCR-induced apoptosis and induction of the proliferation of CD34+ cells. Schotteet al[42]noticed that the interference of miR-125b function mightprovide a way to sensitize patients to VCR. Moreover, researchers identified 14 miR signatures (independent of ALL subtype) useful for prognosis and prediction in ALL.Among others, miR-10a, miR-134, and miR-214 were correlated with a favorable outcome, which confirmed earlier reports linking these miRs with caspase-dependent proapoptotic activity of miR-10a[136], inhibition ofUSF2-mediated cell proliferation by miR-10a and miR-214[137,138], and oncogeneSOX2downregulation by miR-134[139].Differentiated expression of six miRNAs (miR-33, miR-215, miR-369-5p, miR-496,miR-518d, and miR-599) was related to an unfavorable long-term clinical outcome in ALL. Schotteet al[42]colleagues selected two miRs as potential targets for targeted therapy. They noted that it may be of clinical interest. The use of a demethylating agent increased the level of miR-10a in MLL-rearranged patients[42,99], and the application of antagomirs decreased the level of overexpressed miRNA in poor prognosis samples (e.g., miR-33)[42].

Table 2 MicroRNAs associated with resistance to commonly used drugs in childhood acute leukemia treatment
Akbari Moqadamet al[140]analyzed miR-125b, miR-99a, and miR-100 expression levels in correlation with VCR resistance inETV6-RUNX1+ Reh cells. Only the combination set of miRs influenced cell sensitivity to drug administration. MiRNA overexpression resulted in lower expression of their directly regulated target genes(DNTT,NUCKS1,MALAT1,SNRPE,PNO1,SET,KIF5B,PRPS2,RPS11,RPL38, andRPL23A) in VCR-resistant ALL cells[140].In vitro, the restoration of miR-100 and miR-99a in ALL cells suppressed cell proliferation and increased dexamethasone-induced cell apoptosis[43]. It has also been shown that the sensitivity ofBCR-ABL1(Ph+) cells to treatment with tyrosine-kinase inhibitors (such as imatinib) can be increasedin vitroby restoring miR-203[141]. Increasing the miR-203 expression level negatively regulates the expression level of target oncogenes (ABL1andBCR-ABL1) and thus inhibits cell proliferation[142].
One of the major problems in childhood AL is the risk of relapse, and the molecular mechanism is still poorly understood. Intensive scientific work is being conducted on cognition factors related to therapy response and the biology of recurrence. Hanet al[121]carried out a genome-wide miRNA array analysis to identify the miRNA expression patterns correlated with relapse or complete remission in childhood ALL.They identified a set of 70 differentially expressed miRNAs in samples at relapse or complete remission (CR) compared with the initial diagnosis of the same patients. The expression levels of miR-223, miR-23a, let-7g, miR-181, miR-708, and miR-130b were compared in samples at relapsevsdiagnosis and miR-27a, miR-223, miR-23a, miR-181,and miR-128b levels were compared in CR samples and diagnostic samples. In the relapse samples, strong downregulation of miR-223, miR-23a, and let-7g and upregulation of the miR-181 family, miR-708, and miR-130b were confirmed. Hanet al[121]found that miR-223 and miR-27a were overexpressed in patients during CR.Moreover, low levels of both miR-223 and miR-27a at the time of diagnosis were confirmed in patients who subsequently relapsed. Furthermore, Hanet al[121]identified a high expression level of miR-708 in both standard risk and middle risk ALL and a low expression level in high-risk patients. The lowest level of miR-708 expression at initial diagnosis was confirmed among the four immunophenotypes, pro-B-ALL, pre-B-ALL, common ALL, and T-ALL[121]. These results suggest that miR-708, miR-27a,and miR-223 expression levels at initial diagnosis could be independent and reliable prediction factors of the OS rate in childhood ALL and could also be used to predict the risk of relapse before patients undergo therapy. These miRNAs and their targets(e.g., IKZF1,IL-15, andCASP8AP2) might be helpful in the optimization of therapeutic protocols and novel targets for the development of new antileukemic agents[121].
A multifaceted study of miRNome was conducted by Zhanget al[40], who described specific expression patterns for both pediatric ALL and AML, selected prognostic markers, and determined the relationship of miRs with the risk of recurrence in the central nervous system and a lack of sensitivity to prednisone. Authors developed the signature of the overexpressed miR-7, miR-198, and miR-633 and downregulated miR-126, miR-345, miR-222, and miR-551a, in which changes in expression level were related to central nervous system relapse in ALL. Zhanget al[40]identified a set of eight differentiated genes, including miR-18a, miR-532, miR-218, miR-625, miR-193a, miR-638, miR-550, and miR-633, that differentiated patients according to a good or poor prednisone response. Interestingly, researchers were unable to confirm the association with ALs for mir-15 and mir-16, frequently mentioned regulators of apoptosis in hematopoietic cells and prednisolone resistance modulators[86,87,89,110,112]. Xuet al[143]performed a study to determine an early marker of relapse in pediatric ALL and found miR-7, miR-216, and let-7i (high expression) and miR-486, miR-191, miR-150,miR-487, and miR-342 (low expression).
ROLE OF MICRORNA IN NEW THERAPY DEVELOPMENT
Pharmacogenomics of tumors (a field dealing with the assessment of innate genetic determinants of different drug effects) enables the discovery of new anticancer drugs and their genetic targets on the basis of a well-defined mechanism of oncogenesis[126].The search for candidate molecules among miRNAs can also be used in the typing of a molecular target, drug design, determination of thein vitroandin vivoeffects of the drug on the global level of gene expression, identification of mechanisms of action,detection of potential toxicity, and determination of the pharmacodynamic effect[144-146].
miRNAs are one of the most promising molecules in the development of new antileukemic agents and therapeutic protocols. miRNAs, as differentially expressed molecules, can be regulated by two anti-ancient mechanisms. The first involves inhibition of miRNA activity by: (1) The use of miRNA inhibitors and oligomers,including RNA, DNA, and DNA analogues (miRNA antisense therapy), small molecule inhibitors, and miRNA sponges; or (2) miRNA masking. The second mechanism includes enhancement of miRNA function (miRNA replacement therapy)by: (1) The use of modified miRNA mimetics, such as plasmids; or (2) Lentiviral vectors carrying miRNA sequences[147](Figure 5).
Currently available therapeutic strategies based on the level of miRNA and its function in a cancer cell include transfection of mimic miRNAs or miRNA inhibitorsin vitro, which have the effect of increasing or decreasing the expression level of the candidate miRNA, respectively. Therapy based on modulation of miRNA levels is at a promising earlyex vivostage. An effective method of delivering inhibitors and activators of miRNA expression to leukemia cells has not yet been developed. The safety and efficacy of such a therapy have not been studied, and it is difficult to predict the long-term effects of such a treatment.
CONCLUSION
Research using miRNA profiling in disease is currently in an intensive phase, and this technique shows important and promising directions for future research. As our knowledge of the mechanisms and course of childhood AL increases along with advances in genetics and molecular biology and the development of sensitive analytical methods, the accurate assessment of their real relationship with long oncological results in the near future may be possible.
Research on miRNA patterns at individual stages of leukemogenesis can lead to a better understanding of the disease process itself, as well as to the development of modern classification and more effective therapy. As shown, miRNAs are differentially expressed in distinct stages of lymphopoiesis and myelopoiesis and provide a new look at the molecular pathways leading to AL development and the molecular pathogenesis of pediatric ALs. The aberrant miRNA signatures observed in AL can be used to define biomarkers for diagnosis, classification, prognosis, and therapy monitoring of this disease. MiRNA expression level monitoring carried out in the last two decades has shown that hematological cancers can be precisely classified based on a genetic signature that is associated with morphological,immunophenotypic, cytogenetic, molecular, and other cellular traits. Circulating miRNAs can be detected with the use of noninvasive and easily applicable methods with high accuracy and sensitivity.

Figure 5 Schematic strategy for the development of new therapeutic protocols to modulate the biological activity of microRNAs. This approach involves several mechanisms of the downregulation of oncomiRNAs or the upregulation/mimicking of oncosuppressor microRNAs (modified from Gokani et al[153], with the permission of the author). miRNAs: microRNAs.
MiRNAs are becoming increasingly popular among scientists and oncohematologists due to their important role in the etiology of AL. However, the prospect of their routine use in diagnostics requires further research to identify a small group of sensitive and specific biomarkers of diagnostic and prognostic significance. Undoubtedly, the results of numerous studies indicate a few promising candidates, but nevertheless, there is still no one single miRNA or small set that has an accuracy close to 100% to diagnose AL or differentiate its subtypes, regardless of other diagnostic factors. A much more promising direction for further research on miRNAs is to utilize their potential for effective and personalized medicine. In the case of miRNA, the therapeutic challenge is also to improve targeting (one miRNA usually regulates the expression of many genes) as well as to increase the circulation time of the molecule. The therapeutic use of miRNA therefore requires a combination with a suitable nanoparticle that directs miR as well as protects against inactivation and degradation[148].
One of the trends in modern diagnostics and antileukemia therapy is the exact use of nanotechnology, which primarily offers the opportunity to improve sensitivity,selectivity, and bioavailability and thus effectiveness[149]. The advantage of nanoparticles is their size, which allows them to cross biological barriers more effectively. Anticancer drug components include new classes of therapeutic agents such as small interfering RNA, miRs, and single strand DNA[148]. An additional improvement is the possibility of functionalizing the surface of the nanoconstruct with specific ligands[150]. Nevertheless, nanomedicine is the domain of solid tumors[149,151]. In the case of leukemia, however, nanoconstructions are used as noninvasive methods of diagnosis and treatment. An example would be the construction of a modern nanoparticle containing antagomiR-126 [nanoconjugate:LNP@antagomiR126@Anti-CD45.2 (lipopolyplex NPs)] for adult AML therapy[152].
Nevertheless, the introduction of miRNA assays into diagnostics and therapy is a challenge and requires further refinement, above all, of analysis standards. In this field, convergent results of analyses of various research teams obtained in independent groups of patients with AL diagnosis are a great achievement. This is possible despite conducting the experiment in different conditions and with different parameters in independent diagnostic laboratories. Therefore, the possibility of using miRNA in the classification and assessment of risk groups in the case of childhood AL does exist. Significant technical progress does not go hand in hand with clinical trials.There is a long way to go in understanding miRNA regulatory mechanisms in childhood AL. We still need to acquire and integrate data in the field of miRNAmRNA-protein interaction, phenotypic observation, posttranscriptional regulatory interactions, and functional analysis[15]. Much effort should also be made to standardize and validate laboratory procedures for determining miRNA levels.
Finally, although the survival rates for pediatric ALs has improved, there is still a need for identifying novel reliable, sensitive, and specific molecular markers, such as miRNAs, that can be used in a personalized approach to early diagnosis (perhaps even prevention), risk group stratification, and prediction of treatment response.
 World Journal of Clinical Oncology2020年6期
World Journal of Clinical Oncology2020年6期
- World Journal of Clinical Oncology的其它文章
- Angioimmunoblastic T-cell lymphoma accompanied by pure red cell aplasia: A case report
- Immune response activation following hyperthermic intraperitoneal chemotherapy for peritoneal metastases: A pilot study
- National Comprehensive Cancer Network guidelines compliance of a sarcoma service: A retrospective review
- Preoperative markers for the prediction of high-risk features in endometrial cancer
- Lingual lymph nodes: Anatomy, clinical considerations, and oncological significance
- Active surveillance in low risk papillary thyroid carcinoma
