菠萝磷转运蛋白1家族基因鉴定及特征分析
韩利红 刘潮 赵明玉 胡丽娟 胡玉霜





摘 要: 磷轉运蛋白1 (phosphate transporter protein 1, PHT1)家族在植物对磷的吸收及再利用过程中发挥重要作用。该研究对菠萝PHT1基因(AcoPHT1)进行全基因组鉴定,并对基因结构、编码蛋白保守功能域和基因表达进行了分析。结果表明:(1)共鉴定到9个AcoPHT1基因,位于基因组7个连锁群上,所有基因均含有1~3个内含子,内含子相位类型多样。(2)除AcoPHT1.8外,AcoPHT1蛋白均为碱性蛋白,所有蛋白属于亲水性蛋白,且含有10~13个跨膜功能域,均具有保守的PHT1蛋白标签序列GGDYPLSATIxSE,主要定位于叶绿体和细胞质中。(3)AcoPHT1蛋白聚类在单子叶植物组和单双子叶植物混合组中,相对于拟南芥,水稻PHT1与菠萝PHT1相似度更高。(4)AcoPHT1基因启动子区含有P1BS、W-box等与磷吸收和响应胁迫有关的多个顺式作用元件。(5)靶基因预测分析显示,基因AcoPHT1.2、AcoPHT1.8和AcoPHT1.9受多个miRNA调控。(6)AcoPHT1基因表达存在组织特异性和功能冗余性,不同PHT1基因可能在菠萝不同组织或发育阶段发挥作用。该研究结果为菠萝PHT1家族基因的功能鉴定和育种应用奠定理论基础。
关键词: 菠萝, 菠萝磷转运蛋白1(PHT1), 磷吸收, 启动子, miRNA, 组织表达
中图分类号: Q943
文献标识码: A
文章编号: 1000-3142(2021)12-1955-09
收稿日期: 2020-11-03
基金项目: 国家自然科学基金 (32060710,31860005);云南省地方本科高校基础研究联合专项(2017FH001-034) [Supported by the National Natural Science Foundation of China (32060710, 31860005); Yunnan Local Colleges Applied Basic Research Projects (2017FH001-034)]。
作者简介: 韩利红(1981-),博士,副教授,主要从事植物系统发育与进化研究,(E-mail)hanlihong9527@126.com。
通信作者: 刘潮,博士,副教授,主要从事植物逆境生物学研究,(E-mail)liuchao_80@163.com。
Genomic identification and characterization analysis of the
phosphate transporter protein 1 family gene in pineapple
HAN Lihong, LIU Chao*, ZHAO Mingyu, HU Lijuan, HU Yushuang
( Qujing Normal University, College of Biological Resource and Food Engineering, Key Laboratory of Yunnan Province Universities of the
Diversity and Ecological Adaptive Evolution for Animals and Plants on Yungui Plateau, Qujing 655011, Yunnan, China )
Abstract: Phosphate transporter protein 1 (PHT1) family plays pivotal roles in the uptake and re-mobilization of phosphate of plants. In this study, the analysis of whole genome-wide sequence of PHT1 genes in pineapple (Ananas comosus) was conducted, and the gene structure, encoding protein conserved domain and gene expression were further investigated. The results were as follows: (1) Nine PHT1 (AcoPHT1) genes of pineapple, which were located in seven linkage groups and contained one to three introns with various intron phase types, were identified. (2) Except for AcoPHT1.8, AcoPHT1 proteins were all basic proteins, and all the AcoPHT1 proteins were hydrophilic and contained 10 to 13 transmembrane domains, which conserved PHT1 protein tag sequence GGDYPLSATIxSE, and was mainly located in chloroplasts and cytoplasm. (3) AcoPHT1 protein clusters were found in monocotyledons group and monocotyledons and dicotyledons mixed group, and compared to Arabidopsis thaliana, AcoPHT1s proteins had higher similarity with rice PHT1 proteins. (4) The promoter region of AcoPHT1 gene contained a large number of P1BS, W-box and other cis-acting elements, which were related to phosphorus absorption and response to stress. (5) Prediction analysis showed that three genes, AcoPHT1.2, AcoPHT1.8 and AcoPHT1.9 were regulated by multiple miRNAs. (6) The expression of AcoPHT1 gene had tissue-specific and functional redundancy, and different PHT1 genes might play roles in different tissues or development stages. The results provide a theoretical reference for functional identification and breeding application of PHT1 family genes in pineapple.
Key words: pineapple, phosphate transporter protein 1 (PHT1), phosphorus absorption, promoter, miRNA, tissue expression
磷(phosphorus,P)是生命体重要组成元素之一,在植物生命代谢活动中发挥着必不可少的作用。植物通过根系从土壤中吸收磷酸盐形式的磷元素,土壤缺磷严重影响植物的生长代谢,是限制作物高产的重要因素(Abel et al., 2002)。为提高对磷的吸收能力,植物采取一系列策略进行应对,其中磷转运蛋白1 (phosphate transporter,PHT1)为高亲和力的磷转运因子,在低磷条件下对土壤磷的吸收起关键作用(Gu et al., 2016)。自拟南芥(Arabidopsis thaliana)PHT1基因首次被克隆鉴定(Muchhal et al., 1996)以来,目前拟南芥(Mudge et al., 2002)、水稻(Oryza sativa)(Liu et al., 2011)、大豆(Glycine max)(Fan et al., 2013)、番茄(Solanum lycopersicum)(Chen et al., 2014)、马铃薯(Solanum tuberosum)(Liu et al., 2017)、杨树(Populus trichocarpa)(Zhang et al., 2016)、苹果(Malus domestica)(Sun et al., 2017)等多个物种的PHT1家族基因已被鉴定。源自梨状孢子虫(Piriformospora indica)的高亲和力磷酸转运蛋白是首个被解析的PHT1晶体结构(Pedersen et al., 2013)。
系统发育研究表明,植物PHT1为主要促进子超家族(major facilitator superfamily,MFS)典型的phosphate: H+同向转运子(Liu et al., 2011),与酵母PHO84磷转运蛋白同源,存在糖转运功能域(PF00083),具有12个保守的跨膜功能域(transmembrane domains,TM)、亲水性N端和C端以及TM6和TM7之间的亲水环结构(Muchhal et al., 1996)。过表达拟南芥PHT1.1增加了植物对磷的吸收(Wang et al., 2014)。PHT1基因的上调表达增加了根际磷的吸收和转运(Raghothama, 1999)。低磷胁迫条件下,AtPHT1.1和AtPHT1.4在植物根与土壤接触面上的表皮、根毛细胞和根冠细胞中高度表达(Mudge et al., 2002),AtPHT1.8和AtPHT1.9在磷素由根到茎的转运过程中起作用(Lapis-Gaza et al., 2014)。OsPHT1.6编码高亲和磷轉运蛋白,在初生根、侧根的表皮和皮层细胞中表达,在水稻的磷吸收和转运中起作用,而OsPHT1.2编码低亲和力磷转运蛋白,定位于初生根和侧根的中柱中,负责磷的转运(Ai et al., 2009)。丛枝菌根真菌(arbuscular mycorrhizal fungi,AMF)通过诱导真菌PHT1和植物PHT1表达提高植物磷吸收(Javot et al., 2007;Walder et al., 2015)。
菠萝(Ananas comosus)属于凤梨科凤梨属多年生单子叶植物,是产量仅次于香蕉和芒果的第三大热带水果作物。目前,多个物种PHT1家族基因已被研究,而关于菠萝PHT1家族基因的研究鲜见报道。菠萝基因组数据的公布(Ming et al., 2015)为PHT1家族的全基因组鉴定提供了可能。本研究利用生物信息学方法对菠萝PHT1家族基因进行鉴定,并对PHT1家族成员基因结构、氨基酸保守基序、聚类和基因组织特异性表达进行分析,研究结果可为菠萝中磷利用机制的阐明和新品种选育提供参考。
1 材料与方法
1.1 菠萝PHT1家族成员的鉴定
以拟南芥和水稻PHT1家族成员蛋白序列作为查询序列,搜索菠萝基因组蛋白数据库,使用NCBI在线工具CDD和SMART软件进行蛋白功能域分析,剔除不含糖转运功能域(PF00083)的序列。
1.2 菠萝PHT1家族成员序列分析
使用ProtParam工具计算蛋白序列理化特征,使用TMpred程序预测蛋白跨膜区域;使用WoLF PSORT工具预测蛋白亚细胞定位;使用GSDS软件绘制基因外显子和内含子结构示意图;使用MEME在线程序预测氨基酸保守基序,搜索基序数目为10,其他参数默认。
1.3 miRNA靶标PHT1基因的预测
根据miRNA与其靶基因的互补性可预测miRNA的靶基因,使用在线软件psRNATarget对文献中菠萝miRNA(Zheng et al., 2016)的靶标PHT1基因进行预测。
1.4 PHT1家族序列比对及进化分析
使用MEGA 7.0选择最大似然法和邻接法构建系统聚类树,主要参数步长为1 000,替代模型为泊松模型,空位缺失数据的处理为部分删除;使用EvolView软件绘制聚类树;使用序列相似性分析工具Circoletto对拟南芥、水稻和菠萝PHT1蛋白序列相似性进行分析。
1.5 基因启动子区特征分析
从菠萝基因组数据库下载AcoPHT1基因转录起始位点上游1.5 kb序列,使用PLACE软件对基因启动子区顺式作用元件进行分析。
1.6 基因的组织表达分析
菠萝PHT1基因转录组数据于菠萝基因组数据库下载,基因组织特异性表达取样部位分别为根、叶的6个片段,花和果实的5个成熟阶段。使用HemI 1.0软件,根据log2(FPKM+1)值的转换数据绘制基因表达热图。
2 结果与分析
2.1 菠萝PHT1家族基因的鉴定
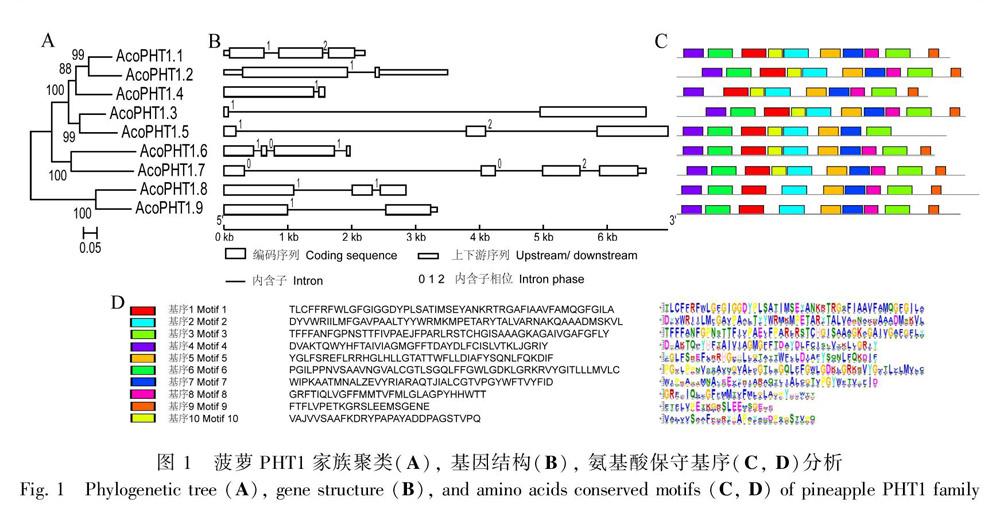
以拟南芥和水稻PHT1蛋白作为查询序列搜索菠萝蛋白数据库,共获得9个菠萝PHT1蛋白,均含有保守的糖转运功能域Sugar_tr (PF00083)。根据搜索比对得分,依次将基因命名为AcoPHT1.1-AcoPHT1.9。这9个基因分别位于连锁群(linkage groups,LG)1、3、6、8、14、20和22上。所有成员氨基酸数为499~602,包含10~13个TMD。除AcoPHT1.8为酸性蛋白外,其他蛋白均为碱性蛋白,等电点为7.99~9.03。所有蛋白平均疏水指数为0.113~0.403,均为正值,属亲水性蛋白。蛋白定位预测分析显示,AcoPHT1.3、AcoPHT1.6、AcoPHT1.7和AcoPHT1.9主要定位于叶绿体中,AcoPHT1.4、AcoPHT1.5和AcoPHT1.8主要定位于细胞质中,AcoPHT1.1和AcoPHT1.2主要定位于细胞膜上,部分蛋白可能定位于多个细胞器中(表1)。
2.2 miRNA对菠萝PHT1家族基因的调控分析
miRNAs在转录水平和转录后水平上调节基因表达,在植物生长、发育、成熟、生物和非生物胁迫反应等方面发挥重要作用(Chen, 2009)。本研究对miRNA及其PHT1家族靶基因的互作及调控关系进行预测。AcoPHT1.8可受到miR2275f的裂解抑制调控,受到miR2673h、miR2673i、miR2673k的转录抑制调控,AcoPHT1.2可受到miR2673h、miR2673i、miR2673k和miR399d的转录抑制调控,AcoPHT1.9可受到miR2673a、miR528a、miR528b的裂解抑制调控(表2)。
2.3 基因结构和氨基酸基序分析
利用MEGA 7.0 构建聚类树,发现菠萝PHT1主要分为三个聚类组,其中AcoPHT1.8和AcoPHT1.9聚在一组,AcoPHT1.6和AcoPHT1.7聚在一组, 其他5个菠萝PHT蛋白聚在一组 (图1:A)。基因结构分析显示,AcoPHT1基因含有1~3个内含子,其中含有1、2、3个内含子的基因数目分别为4、3和2,内含子相位类型(intron phase)多样(图1:B)。使用MEME软件分析发现菠萝PHT1蛋白含有10个保守的氨基酸基序,基序1含有保守的PHT1蛋白标签GGDYPLSATIxSE,存在于9个菠萝PHT1蛋白中。基序1~7编码跨膜功能域。AcoPHT1.4缺少基序6,其跨膜功能域相对较少,AcoPHT1.5缺少基序8和基序9,AcoPHT1.8和AcoPHT1.9缺少基序10,基序8~10功能未知(图1:C, D)。
2.4 聚类分析
为了解植物PHT1家族的聚类及进化,使用拟南芥、水稻、玉米、大豆、杨树和菠萝的PHT1全长蛋
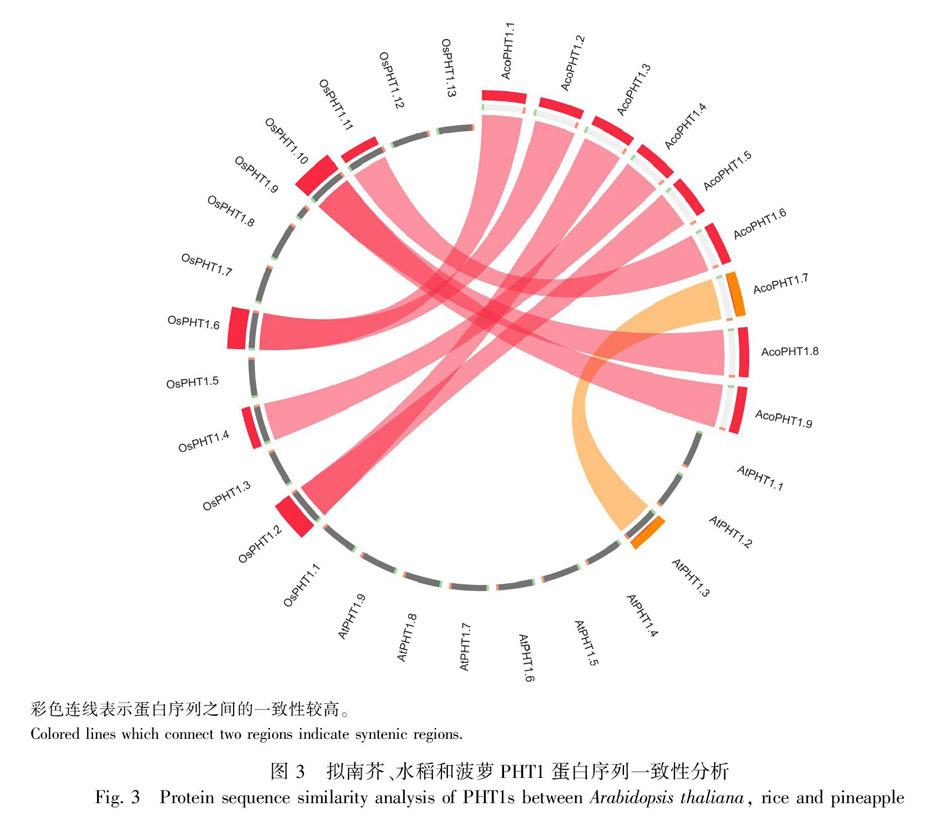
白序列,利用MEGA 7.0软件应用最大似然法和鄰接法构建系统发育树,两种进化树结果相似,文中选择最大似然法的结果进行展示(图2)。结果显示,植物PHT1蛋白共归为3组,分别为双子叶植物组、单子叶植物组和单双子叶植物混合组。菠萝PHT1主要分布在单子叶植物组中的亚组b、c和e以及单双子叶混合组的亚组b和c中。为了解菠萝PHT1蛋白的起源和进化,使用Circoletto软件对拟南芥、水稻和菠萝PHT1蛋白序列相似性进行分析,图3结果显示,绝大多数菠萝PHT1蛋白与水稻PHT1蛋白相似度较高,这些蛋白可能具有共同的起源,如AcoPHT1.1/AcoPHT1.2与OsPHT1.6,AcoPHT1.3/AcoPHT1.5与OsPHT1.2,AcoPHT1.8/AcoPHT1.9与OsPHT1.10,AcoPHT1.4与OsPHT1.4,AcoPHT1.6与OsPHT1.11具有较高的相似性,而AcoPHT1.7与AtPHT1.3具有较高的相似性。
2.5 菠萝PHT1家族基因启动子顺式作用元件分析
使用PLACE软件对AcoPHT1基因上游1.5 kb序列顺式作用元件进行分析(图4),结果发现180个顺式作用元件,这些元件参与植物生长发育、响应激素和胁迫信号等生物过程。AcoPHT1.1、AcoPHT1.5、AcoPHT1.6和AcoPHT1.7的启动子中存在2~4个P1BS元件(PHR1-binding site,GNATATNC)。OSE1ROOTNODULE(organ-specific elements of root nodules,AAAGAT)存在于AcoPHT1.1、AcoPHT1.3、AcoPHT1.4、AcoPHT1.6、AcoPHT1.7和AcoPHT1.8的启动子中,OSE2ROOTNODULE(CTCTT)存在于9个AcoPHT1基因启动子中。除AcoPHT1.5外,AcoPHT1基因均含有至少1个W-box。此外,菠萝PHT1家族基因启动子中还存在ABRELATERD1 (ACGTG)、CACGTGMOTIF (CACGTG)、LTRE1HVBLT49 (CCGAAA)、ELRECOREPCRP1(W-box,TTGACC)等胁迫响应和激素响应相关元件。
2.6 菠萝PHT1家族基因的组织表达分析
为了解菠萝PHT1家族基因在植物不同组织器官中的表达,从菠萝基因组数据库中下载并分析PHT1家族基因在根、叶、花和果实等器官的表达(图5)。发现除AcoPHT1.6外,其余8个AcoPHT1基因在不同组织中均有表达。其中,AcoPHT1.4和AcoPHT1.9在4类组织中均有表达,AcoPHT1.1在除部分叶片节段外的组织中有表达,AcoPHT1.7和AcoPHT1.8主要在叶组织中有表达,AcoPHT1.7在花组织中有表达,AcoPHT1.2仅在花组织中有较高表达,AcoPHT1.3和AcoPHT1.5主要在花、叶部分节段以及根和果实成熟不同阶段的组织中有表达。这些特征表明,菠萝PHT1家族基因具有组织表达特异性,且存在功能冗余,不同的PHT1基因可能在不同的组织或发育阶段发挥作用。
3 讨论与结论
植物根系通过PHT蛋白调控土壤磷素吸收,并转运到其他部分。本研究从菠萝基因组中鉴定到9个PHT1家族基因。菠萝中PHT1家族基因数目与拟南芥(9个)(Mudge et al., 2002)、番茄(8个)(Chen et al., 2014)相当,略少于水稻(13个)(Liu et al., 2011)、大豆(14个)(Fan et al., 2013)、苹果(14 个)(Sun et al., 2017)、杨树(14个)(Zhang et al., 2016)、陆地棉(Gossypium hirsutum)(17个)(晁毛妮等,2017),明显少于小麦(Triticum aestivum)(36个)(Teng et al., 2017)。一些物种中大量PHT1基因的存在,反映了在進化过程中发生的广泛的基因重复和多样化,是基因组不断扩展和重新排列的结果。系统发育可以作为推断物种间蛋白结构和功能变化的依据。本研究中,植物PHT1主要分为单子叶植物组、双子叶植物组和单双子叶混合组三类,植物PHT1家族进化分组与物种有明显的相关性。大多数菠萝PHT1蛋白与水稻PHT1蛋白相似度较高,亲缘关系较近。单双子叶植物混合组中的PHT1成员可能属于更古老的基因,在单双子叶植物进化分开后,通过基因复制进一步扩展出双子叶植物组或单子叶植物组新成员。
顺式作用元件通过与反式调节因子的相互作用调节基因的表达,在基因功能发挥中起决定作用。P1BS为植物基因上游的PHR1识别序列,控制磷胁迫响应基因在根中的表达和对低磷胁迫的响应(Rubio et al., 2001)。OSE1ROOTNODULE和OSE2ROOTNODULE为根瘤感染细胞中基因启动的两个器官特异性元件(Vieweg et al., 2004),这两个基序是根瘤感染细胞启动子活性所必需的(Stougaard et al., 1990)。WBOXNTERF3 (W-box,TGACY)在植物磷吸收和响应胁迫过程中起作用(Nishiuchi et al., 2004;Devaiah et al., 2007)。ABRELATERD1、CACGTGMOTIF、LTRE1HVBLT49、ELRECOREPCRP1属于胁迫响应和激素响应相关元件(Dunn et al., 1998;Eulgem et al., 1999;Chakravarthy et al., 2003)。本研究分析发现,一些AcoPHT1基因启动子区含有磷胁迫相关元件P1BS,这些基因受PHR1转录因子调控,可能在磷的吸收和利用过程中起作用(Rubio et al., 2001)。一些基因启动子区含有元件OSEROOTNODULE,这些元件存在于根瘤活化的基因启动子中,可能在菌根真菌与植物共生过程中促进植物营养元素吸收有关。大部分菠萝PHT1基因启动子区含有多个W-box,WRKY转录因子能通过结合W-box启动基因的表达,在植物的磷饥饿响应和应对多种胁迫过程中起作用(Devaiah et al., 2007;Xie et al., 2018)。菠萝PHT1家族基因启动子区含有大量与植物生长发育、响应激素和胁迫信号相关的顺式作用元件,说明该家族基因受多种信号调控,在植物生长发育、应对环境胁迫过程中发挥作用。
基因组织特异性表达与基因的功能密切相关。菠萝PHT1家族蛋白均含有多个跨膜结构域,可定位于多种细胞器膜上,其在特定组织中的表达,表明其功能的特异性与多样性。植物PHT1基因表达受环境磷素含量和水分条件影响(Mudge et al., 2002;Sun et al., 2017)。研究表明,PHT1家族基因受低磷诱导表达,在植物营养元素吸收过程中起作用。OsPHT1.3受OsPHR2直接调控,低磷胁迫下,其在幼叶和茎基部的表达高于根和老叶,OsPHT1.3介导了水稻对磷的吸收、转运和再利用,过表达OsPHT1.3增强了植物根和茎对磷的吸收(Chang et al., 2019)。OsPHT1.2和OsPHT1.3在植物体内存在物理互作关系(Chang et al., 2019)。AcoPHT1.1、AcoPHT1.3、AcoPHT1.5和AcoPHT1.9在菠萝根组织中检测到表达,这些基因可能在根的生长或某些功能的发挥中起作用,如磷的吸收。AcoPHT1.1、AcoPHT1.4和AcoPHT1.9在菠萝多个组织器官中均检测到表达,这些基因可能参与了植物多种生命过程。
参考文献:
ABEL S, TICCONI CA, DELATORRE CA,2002. Phosphate sensing in higher plants [J]. Physiol Plant, 115(1): 1-8.
AI PH, SUN SB, ZHAO JN, et al., 2009. Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation [J]. Plant J, 57(5): 798-809.
CHAKRAVARTHY S, TUORI RP, D’ASCENZO MD, et al., 2003. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements [J]. Plant Cell, 15(12): 3033-3050.
CHANG MX, GU M, XIA YW,et al., 2019. OsPHT1; 3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes [J]. Plant Physiol, 179(2): 656-670.
CHAO MN, ZHANG ZY, SONG HN, et al., 2017. Genome-wide identification and expression analysis of Pht1 family genes in cotton (Gossypium hirsutum L.) [J]. Cotton Sci, 29(1): 59-69. [晁毛妮, 張志勇, 宋海娜, 等, 2017. 陆地棉Pht1家族成员的全基因组鉴定及表达分析 [J]. 棉花学报, 29(1): 59-69.]
CHEN AQ, CHEN X, WANG HM, et al., 2014. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato [J]. Plant Biol, 14(1): 61.
CHEN XM, 2009. Small RNAs and their roles in plant development [J]. Ann Rev Cell Dev Biol, 25: 21-44.
DEVAIAH BN, KARTHIKEYAN AS, RAGHOTHAMA KG, 2007. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis [J]. Plant Physiol, 143(4): 1789-1801.
DUNN MA, WHITE AJ, VURAL S, et al., 1998. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) [J]. Plant Mol Biol, 38(4): 551-564.
EULGEM T, RUSHTON PJ, SCHMELZER E, et al., 1999. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors [J]. EMBO J, 18(17): 4689-4699.
FAN CM, WANG X, HU RB, et al., 2013. The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. [J]. Plant Biol, 13(1): 48.
GU M, CHEN AQ, SUN SB, et al., 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? [J]. Mol Plant, 9(3): 396-416.
JAVOT H, PUMPLIN N, HARRISON MJ, 2007. Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles [J]. Plant Cell Environ, 30(3): 310-322.
LAPIS-GAZA HR, JOST R, FINNEGAN PM, 2014. Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1; 8 and PHT1; 9 are involved in root-to-shoot translocation of orthophosphate [J]. Plant Biol, 14(1): 334.
LIU BL, ZHAO S, WU XF, et al., 2017. Identification and characterization of phosphate transporter genes in potato [J]. J Biotechnol, 264: 17-28.
LIU F, CHANG XJ, YE Y, et al., 2011. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice [J]. Mol Plant, 4(6): 1105-1122.
MING R, VANBUREN R, WAI CM, et al., 2015. The pineapple genome and the evolution of CAM photosynthesis [J]. Nat Genet, 47(12): 1435-1442.
MUCHHAL US, PARDO JM, RAGHOTHAMA KG, 1996. Phosphate transporters from the higher plant Arabidopsis thaliana [J]. Proc Natl Acad Sci USA, 93(19): 10519-10523.
MUDGE SR, RAE AL, DIATLOFF E, et al., 2002. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis [J]. Plant J, 31(3): 341-353.
NISHIUCHI T, SHINSHI H, SUZUKI K, 2004. Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves possible involvement of NtWRKYs and autorepression [J]. J Biol Chem, 279(53): 55355-55361.
PEDERSEN BP, KUMAR H, WAIGHT AB, et al., 2013. Crystal structure of a eukaryotic phosphate transporter [J]. Nature, 496(7446): 533-536.
RAGHOTHAMA KG,1999. Phosphate acquisition [J]. Ann Rev Plant Biol, 50(1): 665-693.
RUBIO V, LINHARES F, SOLANO R, et al., 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae [J]. Gene Dev, 15(16): 2122-2133.
STOUGAARD J, JRGENSEN JE, CHRISTENSEN T, et al., 1990. Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghemoglobin lbc 3 and N23 gene promoters [J]. Mol Gen Genet, 220(3): 353-360.
SUN TT, LI MJ, SHAO Y, et al., 2017. Comprehensive genomic identification and expression analysis of the phosphate transporter (PHT) gene family in apple [J]. Front Plant Sci, 8: 426.
TENG W, ZHAO YY, ZHAO XQ, et al., 2017. Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat [J]. Front Plant Sci, 8: 543.
VIEWEG MF, FRHLING M, QUANDT HJ, et al., 2004. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants [J]. Mol Plant Microb Interact, 17(1): 62-69.
WALDER F, BRUL D, KOEGEL S, et al., 2015. Plant phosphorus acquisition in a common mycorrhizal network: regulation of phosphate transporter genes of the Pht1 family in sorghum and flax [J]. New Phytol, 205(4): 1632-1645.
WANG H, XU Q, KONG YH, et al., 2014. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation [J]. Plant Physiol, 164(4): 2020-2029.
XIE T, CHEN CJ, LI CH, et al., 2018. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress [J]. BMC genomics, 19(1): 490.
ZHANG CX, MENG S, LI MJ, et al., 2016. Genomic identification and expression analysis of the phosphate transporter gene family in poplar [J]. Front Plant Sci, 7: 1398.
ZHENG Y, LI T, XU ZN, et al., 2016. Identification of microRNAs, phasiRNAs and their targets in pineapple [J]. Trop Plant Biol, 9(3): 176-186.
(責任编辑 周翠鸣)

