光对园艺植物花青素生物合成的调控作用
王峰,王秀杰,赵胜男,闫家榕,卜鑫,张颖,刘玉凤,许涛,齐明芳,齐红岩,李天来
光对园艺植物花青素生物合成的调控作用
王峰,王秀杰,赵胜男,闫家榕,卜鑫,张颖,刘玉凤,许涛,齐明芳,齐红岩,李天来
(沈阳农业大学园艺学院/设施园艺省部共建教育部重点实验室/北方园艺设施设计与应用技术国家地方联合工程研究中心(辽宁),沈阳 110866)
花青素是植物中一类重要的类黄酮化合物,在植物花朵、果实等器官色泽形成和抗氧化过程中起着重要作用。植物组织中花青素的形成依赖于光信号,但是光信号对花青素生物合成的调控机制及信号网络很大程度上还不清晰。本文简述了花青素生物合成及运转过程的研究进展,简要归纳了MYB、bHLH、WDR三类主要因子对花青素合成的转录调控作用,重点阐释光信号(光强、光质、光照时长)对植物花青素合成的调控作用。研究表明,光环境(光强、光质、光照时长)主要通过不同的光受体(UVR8、CRYs、PHOTs、PHYs)影响光信号通路重要因子COP1的泛素化能力和HY5的稳定性,以及其他光信号转录因子如光敏色素互作因子PIFs的稳定性,进而调控花青素的生物合成过程。这些光信号因子一方面直接结合到调控花青素合成的MYB、bHLH、WDR三大类转录因子上,转录激活或抑制它们的表达进而调控花青素的合成;另一方面,这些光信号因子通过与MYB、bHLH、WDR三大类转录因子蛋白互作,影响它们形成的MBW复合体稳定性,进而调控花青素的合成。此外,这些光信号因子还可以通过不依赖于MBW复合体的通路调控花青素的合成,如HY5通过调控影响花青素的生物合成;另外,一些未知的光响应因子可能以不依赖MBW通路的方式直接或间接地调控花青素合成基因和液泡膜上的运转蛋白,改变液泡酸化,调节花青素的合成。同时,光信号会影响光合电子传递,光合电子传递链中的一些因子也会通过依赖和不依赖MBW的途径影响植物花青素的合成。这些途径如何协调以及哪些信号因子优先受光环境(光强、光质、光照时间)调控?本文为深入研究光信号对花青素生物合成的调控机理提供参考,以探索光调控花青素积累的有效途径及靶标分子,为利用基因工程、代谢工程和光环境调控手段改良园艺植物花青素积累提供理论基础。
光;花青素;转录因子;转录调控;园艺植物
花青素是自然界中广泛存在于植物中的水溶性天然色素,在植物的花、叶片和果实等器官中均有积累。花青素可以通过其艳丽的色彩帮助植物吸引昆虫和飞鸟等媒介,帮助植物传粉和散播种子。另外,花青素具有较强的抗氧化能力,可以减轻植物组织免受活性氧(ROS)等胁迫的伤害,同时,在抗衰老[1]及防治心血管疾病[2]等方面对人体也有保健功效。光环境作为影响植物花青素合成的重要环境因子,受到越来越多的关注,解析光对花青素生物合成的调控作用,对精准调控园艺植物不同器官花青素的含量、提高园艺产品营养成分及人体保健等方面具有重要的研究和应用价值。
1 花青素的生物合成代谢
花青素生物合成途径是植物类黄酮途径的一个分支途径,主要在内质网表面进行。一般来说,花青素由3个丙二酰辅酶A分子和一个4-香豆酰辅酶A分子在细胞质中结合,经由查尔酮合酶(CHS)作用产生查尔酮(图1)。查尔酮经查尔酮异构酶(CHI)催化形成黄烷酮柚皮素,黄烷酮-3-羟化酶(F3H)催化黄烷酮柚皮素形成二氢黄酮醇(DHK)。DHK一方面直接被二氢黄酮醇还原酶(DFR)催化形成无色天竺葵素;另一方面,DHK经由二氢黄酮醇-3′-羟化酶(F3′H)作用形成二氢槲皮素(DHQ),DHQ和DHK在二氢黄酮醇-3′, 5′-羟化酶(F3′5′H)作用下形成二氢杨梅素(DHM)。DHQ和DHM被DFR催化后分别形成无色矢车菊素和无色飞燕草素。3种无色花青素在花青素合成酶(ANS)催化下形成天竺葵素、矢车菊素和飞燕草素。不稳定的花青素通过类黄酮-3-O-葡萄糖基转移酶(UFGT)催化形成稳定的花青苷。矢车菊素苷经由甲基转移酶(OMT)催化形成芍药花苷,飞燕草素苷经由OMT催化形成牵牛花素苷和锦葵素苷(图1)。
花青素苷合成后需要在转运蛋白和转运囊泡的协助下有效地向液泡中转运并贮藏(图1),从而防止花青素苷自身被氧化变性及对细胞造成毒害。由于花青素苷最终所处的液泡环境会影响花青素苷的呈色,如酸性环境会使其颜色红移,而碱性环境会使其颜色蓝移[3],因此,明确花青素苷的液泡运转机制极其重要。研究发现,花青素苷液泡转运的主要方式为:在GST协助下被靶向定位到液泡附近,液泡膜上的MRP类转运蛋白识别后将其跨膜转运至液泡[4];由液泡膜上的MATE类转运蛋白将其跨膜转运到液泡中,这个过程需要如H+-ATPase和H+-pyrophosphatase(V-PPase)质子泵将质子运输到液泡中对液泡酸化[5];由囊泡包裹,细胞膜导出的囊泡以膜融合的方式直接将花青素运输到贮藏液泡中,这个过程不依赖GST和MRP3蛋白的协助[4]。另外,ABC运转蛋白也参与花青素苷的液泡吸收和外排过程[6]。
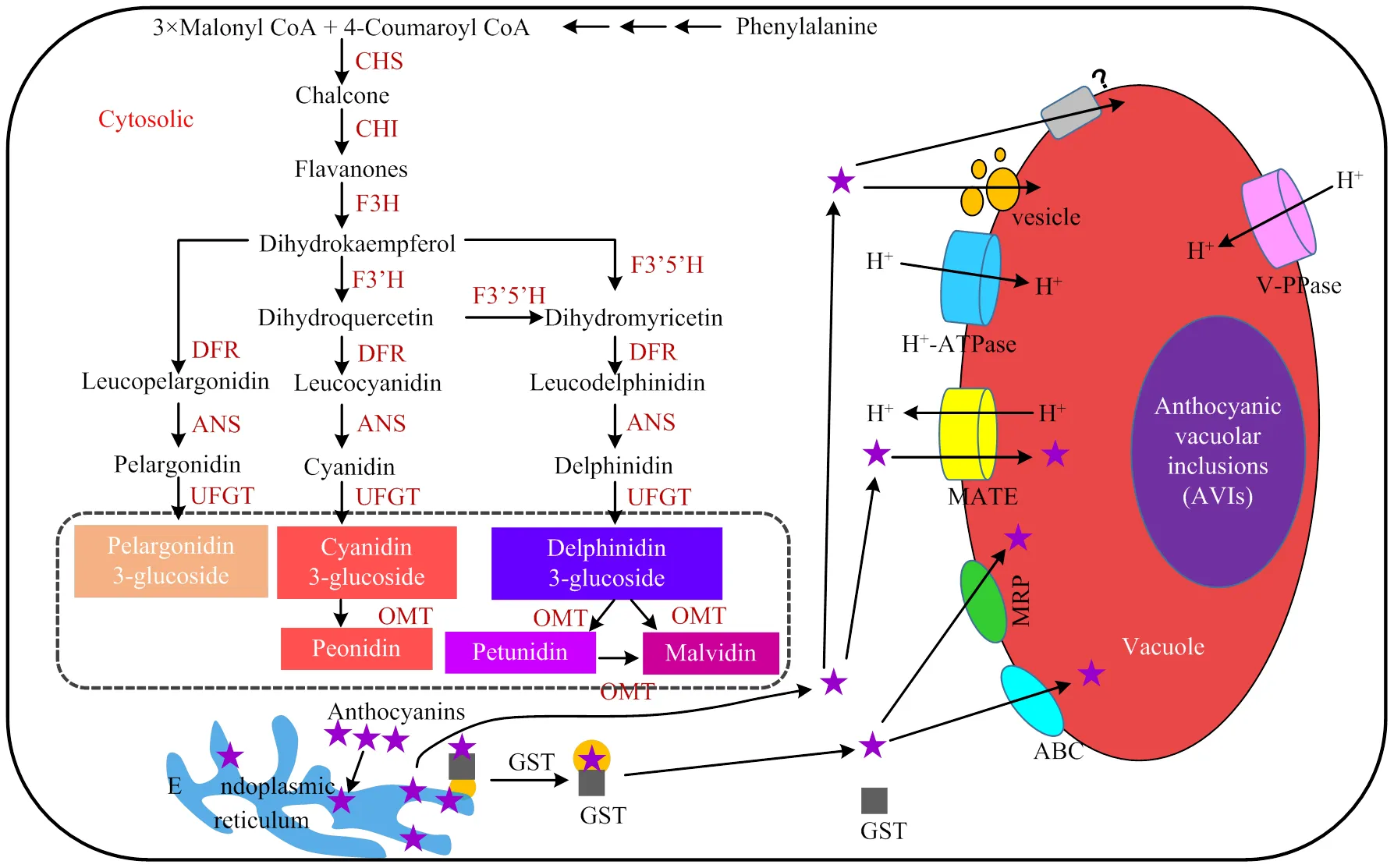
CHS:查尔酮合酶Chalcone synthase;CHI:查尔酮异构酶Chalcone isomerase;F3H:黄烷酮-3β-羟化酶Flavanone-3β-hydroxylase;F3′H:二氢黄酮醇-3′-羟化酶Dihydroflavonoid-3'-hydroxylase;F3′5′H:二氢黄酮醇-3′, 5′-羟化酶Dihydroflavonoid-3', 5'-hydroxylase;DFR:二氢黄酮醇还原酶Dihydroflavonol reductase;ANS:花青素合成酶Anthocyanin synthase;UFGT:尿苷二磷酸-葡萄糖-类黄酮-3-葡糖基转移酶UDP-glucose flavonoid 3-glucosyltransferase;OMT:甲基转移酶O-methyltransferases;GST:谷胱甘肽转移酶Glutathione S-transferase;MRP:多药耐药相关蛋白Multidrug resistance-associated protein;MATE:多药和有毒化合物排出家族蛋白Multidrug and toxic compound extrusion;ABC:C型的ATP结合蛋白C type of ATP-binding cassette;AVIs:花青素苷液泡内涵体Anthocyanic vacuolar inclusions;Phenylalanine:苯基丙氨酸;3×Malonyl CoA:3×丙二酰辅酶A;4-Coumaroyl CoA:4-香豆酰辅酶A;Chalcone:查尔酮;Flavanones:黄烷酮;Dihydrokaempferol(DHK):二氢黄烷酮;Dihydroquercetin(DHQ):二氢栎精;Dihydromyricetin(DHM):二氢杨梅酮;Leucopelargonidin:无色天竺葵素;Leucocyanidin:无色矢车菊素;Leucodelphinidin:无色飞燕草素;Pelargonidin:天竺葵素;Cyanidin:矢车菊素;Delphinidin:飞燕草素;Pelargonidin 3-glucoside:天竺葵素苷;Cyanidin 3-glucoside:矢车菊素苷;Delphinidin 3-glucoside:飞燕草素苷;Peonidin:芍药花青素苷;Petunidin:牵牛花素苷;Malvidin:锦葵素苷;Anthocyanins:花青素;Cytosolic:细胞质;Endoplasmic reticulum:内质网
2 花青素生物合成的转录调控
花青素生物合成途径的主要基因在转录水平上受多种转录因子的调控,主要包括MYB、bHLH和WDR转录因子,这3类转录因子可以形成MBW复合体[7]。拟南芥中花青素合成基因分为早期合成基因(early biosynthetic genes,EBGs)和后期合成基因(later biosynthetic genes,LBGs),EBGs包括、和′,LBGs包括和[8]。EBGs的表达不受MBW复合物调控,而LBGs的表达受MBW复合物调控[9]。
2.1 MYB转录因子
MYB蛋白家族N端含有可与DNA结合的保守MYB结构域,C端在不同物种间序列变异较大,但常有转录激活或抑制结构域的作用[10]。MYB结构域由1–4个R基序(R repeat)组成,根据含有R基序的数量分为1R-MYB、R2R3-MYB、3R-MYB和4R-MYB。其中有2个R基序的R2R3-MYB是最常见的MYB转录因子,也是参与调控类黄酮代谢和花青素生物合成的重要转录因子。MYB蛋白功能异常或靶基因启动子上MYB识别序列[MYB-recognizing elements,MRE;识别位点ANCNN(C/A)C]异常,都可能导致花青素的生物合成不正常[11]。
玉米中C1(Colorless-1)是植物中首个被发现的R2R3-MYB转录因子,它正调控玉米中和的表达,从而促进玉米花青素的合成[12]。矮牵牛中MYB型转录因子PhAN2(anthocyanin 2)的C端具有与玉米C1相似的结构域,与PhAN4均正调控的表达,从而促进矮牵牛花青素的合成[13]。与矮牵牛PhAN2同源的亚洲杂交百合的LhMYB6和LhMYB12同样可以促进白百合花青素的积累[14]。最近研究发现,野生番茄SlAN2-like转录因子可以激活的表达,诱导番茄果皮中花青素的积累[15],但SlAN2-like转录激活的会反馈抑制的表达[16];另外,SlAN2-like的选择性剪切对番茄花青素的积累也有影响[17]。研究发现,拟南芥MYB转录因子AtPAP1(production of anthocyanin pigment 1)不仅正调控和等基因的表达,而且促进糖基转移酶基因和的表达,诱导矢车菊素大量积累[18]。豆科植物紫花苜蓿、蒺藜苜蓿和白三叶中与AtPAP1同源的LAP1(legume anthocyanin production 1)同样能够有效地促进矢车菊素积累[19]。苹果MdMYB10与AtPAP1有较高同源性[20],MdMYB10结合到自身启动子上游23 bp的串联重复序列上,激活自身转录,进而促进的表达,使苹果果肉花青素积累,而白色果肉苹果中启动子上则缺少这段序列,导致的转录水平较低,破坏了花青素的积累[21]。研究发现,MYB10在果实花青素形成中是保守的,草莓[22]、甜樱桃[23]、油桃[24]和梨[25]等果实花青素形成均与MYB10相关。此外,植物中一些各异的MYB也能够影响花青素的积累。如拟南芥AtMYB113和AtMYB114[9],龙胆中的GtMYB3[26],菊花中的CmMYB6[27],葡萄中的VvMYBA、VvMYB5a和VvMYB5b[28],苹果的MdMYB9、MdMYB11和MdMYB110[29-30],以及番茄SlMYB75[31]。
为保证花青素在植物体内的代谢平衡,一些MYB在花青素生物合成中起着负调控因子的作用。如拟南芥AtMYB2负调控、和的表达[32];葡萄VvMYB4-like抑制和的表达[33];苹果MdMYB6过表达会抑制和等基因的表达[34];智利草莓FcMYB1会抑制的表达[35]。这些抑制型MYB一方面直接转录抑制花青素合成途径相关基因的表达;另一方面通过与调控花青素合成的正调控因子结合,抑制它们的表达或破坏MBW复合体的形成。如具有独特TLLLFR抑制结构域的AtMYBL2转录因子,通过与AtTT8结合进而抑制和的表达[36];AtSPL9(squamosa promoter binding protein-like 9)转录因子通过破坏MBW复合体的稳定性抑制的表达[37];葡萄中的VvMYBC2-L1和VvMYBC2-L3与VvAN1结合,破坏MBW复合体的稳定性,抑制花青素的积累[38]。研究发现,矮牵牛PhMYB27一方面通过与PhAN1互作,激活其C末端EAR基序抑制的表达[39];另一方面通过与bHLH型转录因子PhJAF13和PhAN11结合互作,破坏MBW复合体形成,使MBW复合物从激活状态变为抑制状态,进而抑制花青素合成[40]。AtCPC(CAPRICE)与PhMYB27不同,它是C末端不含抑制基序的R3-MYB,不能直接与MBW复合物结合,而是间接减少MBW复合物的活性,从而抑制花青素的生物合成[41]。
2.2 bHLH转录因子
bHLH(basic helix-loop-helix)转录因子具有与DNA结合的高度保守的bHLH结构域,这个结构域的N端由13—18个亲水氨基酸残基组成碱性区(basic),负责与DNA接触,C端为HLH域,主要参与同源或异源二聚体的形成,其中的环状区(Loop,L)连接着2个螺旋(helix,H)。bHLH转录因子决定了MBW复合物识别靶基因启动子上的转录结合位点及激活靶基因转录的特异性[42]。研究表明,花青素相关的bHLH对靶基因的有效识别序列(bHLH-recognizing elements,BRE)为CACN(A/C/T)(G/T)[11]。bHLH蛋白功能异常或靶基因启动子上BRE异常,都可能导致花青素合成异常。
最早发现的一批调控花青素合成的bHLH家族转录因子是玉米中的R1、B1、Lc(Leaf color)和Sn[43-44]。ZmLc能够激活和的表达,促进花青素的生物合成[13]。与玉米同源的拟南芥bHLH转录因子为AtTT8(transparent testa 8)、AtGL3(glabra 3)和AtEGL3(enhancer of glabra 3),它们通过调控的表达来促进花青素的生物合成[45]。蒺藜苜蓿MtTT8是与拟南芥AtTT8同源的bHLH型转录因子,它能与MtLAP1和MtWD40-1互作形成MBW复合体,促进花青素的合成[46]。研究发现,矮牵牛中PhJAF13和PhAN1是调控花青素生物合成的主要bHLH类调控因子,它们均能与PhAN2互作激活的表达,且PhAN1还可以直接激活的表达[42]。同样,烟草中的NtAN1a与NtAN2互作激活和的表达[47]。非洲菊bHLH调控因子GMYC1与MYB转录因子AN2和GMYB10互作,特异地在花冠和心皮部位促进的表达[48];紫色天葵的bHLH型基因和MYB转录因子GbMYB1共同表达能激活和启动子,诱导花青素合成[49]。另外,苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作,促进果实颜色变红[20];荔枝LcbHLH1 和LcbHLH3 转录因子与LcMYB1共同作用,促进花青素积累[50];龙胆GtbHLH1蛋白能与GtMYB3互作,促进龙胆花瓣花青素合成[26];亚洲百合LhbHLH2能与LhMYB6和LhMYB12相互作用,激活、、的表达促进花青素的合成[14]。以上结果表明,bHLH转录因子除了直接调控的表达外,还可以与MYB转录因子互作,调控花青素的合成。
2.3 WDR蛋白家族
WDR(WD40 repeat proteins,WDR)蛋白家族具有保守而特异的二肽重复基序,每个重复的WD基元大概有40—60个氨基酸残基组成,WDR在蛋白质互作时作为支架起固定作用。矮牵牛的PhAN11是第1个被发现调控花青素生物合成的WDR蛋白,它作用于PhAN2的上游,通过激活的表达调控花青素的合成[51]。拟南芥AtTTG1(transparent testa glabra 1)与矮牵牛PhAN11同源,它一方面与bHLH型转录因子AtGL3蛋白互作,直接促进及的表达;另一方面,AtTTG1与AtPAP1及AtGL3/AtEGL3互作,形成MYB/bHLH/TTG1转录复合体,进而调控花青素的生物合成[9]。玉米中约有20个基因影响其花青素的产生,其中ZmPAC1(pale aleurone color 1)是类似于矮牵牛PhAN11和拟南芥AtTTGl的WDR蛋白,的缺失导致花青素含量降低[52]。研究发现,杨梅MrWD40-1与MrMYB1和MrbHLH1相互作用,参与杨梅花青素的积累[53];苹果MdTTG1与bHLH转录因子和MYB 转录因子相互作用形成复合物,激活花青素合成基因和的转录[54];蓝莓WD40转录因子VcWDL2与VcMYBL1和VcbHLHL1互作,参与调控蓝莓果实花青素的合成[55];此外,辣椒果实中的沉默后,辣椒的花青素含量明显减少[56]。尽管WDR属于广谱性表达蛋白、组织特异性较弱,但其一般无冗余拷贝,所以它的突变会影响MBW复合体的形成或复合体的定位及信号传递,最终破坏了MBW复合体对众多靶基因的表达调控。
2.4 其他类型调控因子
除MYB、bHLH和WDR外,miRNA、JAZ蛋白、bZIP蛋白等也参与了植物花青素生物合成的调控。研究发现,miR828/TAS4-siR81(-)能与花青素正调控因子PAP1、PAP2、MYB113结合,抑制它们的表达,进而负调控拟南芥花青素的积累[57]。miR156靶向定位的SPL9转录因子可使MBW转录复合体变得不稳定,最终负调控拟南芥花青素的生物合成[37]。另外,番茄miR858可以抑制R2R3-MYB的表达,减少花青素的合成[58]。拟南芥茉莉酸信号通路中的AtJAZ蛋白不仅可以与bHLHs(AtGL3、AtEGL3、AtTT8)和R2R3-MYBs(AtPAP1、AtPAP2)蛋白结合,阻碍MBW复合体的形成,抑制花青素的生物合成,而且可以作为正调控因子参与JA诱导的花青素积累[59]。另外,bZIP型转录因子,如HY5不仅可以直接调控和等花青素结构基因的表达[60],而且可以结合到、、等转录因子启动子上影响它们的表达,同时还可以通过调控的表达,间接调控花青素相关基因的表达[61-63]。此外,HY5还能与PIF3(phytochrome interacting factor 3)等bHLH蛋白互作,共同调控花青素合成途径的下游靶基因,从而促进花青素的合成[64]。
3 光调控植物花青素的生物合成
光是影响植物花青素合成的最重要环境因子之一。在植物绿色组织或细胞中,光通过光受体及光合电子传递调节植物花青素的合成与积累,从而保护植物组织免受活性氧(ROS)等的胁迫及调控植物色泽的形成。因此,本文对光信号调控植物花青素合成及代谢进行总结(图2),具体内容如下。
3.1 光强调控植物花青素的生物合成
强光可以刺激许多植物花青素的形成与积累[65]。番茄()果实暴露在光下部分比遮阴部分的花青素含量高[66];非洲菊花序进行黑暗处理后,其花青素被抑制[67]。主要原因是光可以提高苯丙氨酸解氨酶(PAL)、查耳酮合酶(CHS)、二氢黄酮醇4-还原酶(DFR)、类黄酮葡萄糖苷转移酶(UFGT)等花青素合成途径中关键合成酶的活性,进而促进花青素的合成和积累。研究发现,强光可以促进花青素生物合成途径结构基因和调节基因的表达。如强光促进拟南芥花青素合成结构基因、及调节基因的表达,从而促进植物花青素的合成与积累[68]。强光促进矮牵牛和的表达,而弱光或黑暗致使矮牵牛和紫苏等植物的花青素结构基因表达量下调甚至不表达,使植株出现白花或浅色花[69-70]。草莓果实中和等结构基因及转录因子FaMYB10、FaMYB1的表达量会随着光照强度的降低而降低,弱光会使草莓红色减弱、花青素含量降低[71]。强光可以诱导番茄和辣椒等植物R2R3-MYB转录因子SlAN2和CaMYBa的表达,抑制含抑制基序的矮牵牛的表达,从而促进花青素的积累[39]。另外,强光可以促进番茄SlAN11、SlTT8和SlAN2蛋白结合,形成MBW复合体,进而促进番茄花青素结构基因的表达及花青素的积累,而SlMYBL2则反馈抑制花青素合成基因的表达[72]。强光可以增加苹果的表达,从而诱导其下游基因的表达,促进果皮中花青素的积累[73]。另外,强光可以通过蓝光受体CRY1(crytochrome 1)诱导的表达,进而促进植物花青素的积累[74]。此外,光合电子传递可能在植物花青素合成中也起着重要的作用[75],因此,光信号对植物花青素合成的调控是多通路协作的复杂过程。
3.2 光质调控植物花青素的生物合成
3.2.1 紫外光(UV)和蓝光 在植物花青素合成和积累的过程中,不同光质对植物花青素形成的调控作用不同。对于大多数植物,紫外线(UV)是花朵成色、花青素积累的重要因子[76]。UV按波长由短到长依次分为UV-C、UV-B和UV-A。研究发现,UV-C可以促进紫甘蓝花青素酰基转移酶基因以及R2R3-MYB家族转录因子MYB114和PAP1的表达,进而促进花青素合成[77]。另外,UV-C可以提高杨梅果实中的PAL、CHI、4-香豆酰辅酶A连接酶(4CL)和肉桂酸氢化酶(C4H)的活性,从而增加花青素和类黄酮物质的合成与积累[78]。研究发现,一定程度的UV-B可以通过加强蓝莓中CHI酶活性提高其花青素的含量[79]。UV-B在植物体内主要通过两种形式诱导花青素的积累。植物UVR8(UV resistance locus 8)感知UV-B辐射后,由二聚体形式转化为单体形式[80]。单体形式UVR8和COP1、SPA聚合产生UVR8-SPA- COP1复合物[81],并聚集在细胞核中。这种复合体一方面引起MYB、bHLH和WDR 3种转录因子响应,促进MBW复合体形成,直接或间接促进花青素合成途径中各基因的表达[82]。如UV-B可以诱导莴苣叶片中的表达[83],以及甘蓝和的表达[84],这可能与UVR8和MYB13的互作相关[85]。另一方面,UVR8-SPA-COP1复合物可稳定HY5蛋白[86](图2),HY5转录因子可以激活R2R3-MYB转录因子,促进植物花青素合成基因的表达,从而诱导花青素的积累[61]。如UV-B下,苹果中MdHY5与的启动子结合,激活其转录,进而促进苹果花青素的合成[64]。研究发现,温室中生长的茄子由于UV照射较少,果实着色不良,当对茄子进行UV-A补光后,茄子颜色加深[87]。研究报道,与白光相比,UV-A可以提高番茄幼苗和果实中花青素的积累[88],同时,UV-A可以诱导芜菁和的表达[89]。
植物通过隐花色素CRY感受UV-A和蓝光[90]。研究发现,拟南芥突变体中表达下调,花青素积累减弱[76]。蓝光下,CRY1和CRY2可以与COP1和WD40相互作用,但二者作用机制不同。CRY1与SPA1(suppressor of phyA1)互作,将SPA1与COP1隔离,从而阻碍COP1-SPA1蛋白复合体的形成[91];而CRY2-SPA1互作则增强了CRY2-COP1的互作,CRY1/2-SPA1相互作用均减弱了COP1的E3泛素连接酶活性,从而使COP1下游转录因子的蛋白处于稳定状态[92]。研究发现,CRY1和CRY2可以激活光信号转录因子HY5和COL5,进而促进花青素生物合成结构基因及的表达,促进花青素的积累[86]。研究发现,茄子SmCRYs在光下抑制SmCOP1的活性,促使SmHY5和SmMYB1结合到和的启动子上激活它们的表达,进而促进茄子花青素的积累;而黑暗下,SmCRYs不能抑制SmCOP1的活性,SmHY5和SmMYB1被SmCOP1靶定并通过26S泛素蛋白酶体途径降解,阻止了依赖SmMYB1激活的花青素合成途径[93]。最近研究发现,拟南芥转录共激活因子AtAN3(angustifolia 3)可以结合到的启动子上,负调控的表达;缺失后,植物的花青素含量明显降低[94](图2),但是否通过光受体影响植物花青素的合成尚不清楚。研究发现,增加蓝光的比例可以提高番茄幼苗的花青素含量[95]。蓝光促进杨梅果实MrMYB1及花青素合成相关结构基因和的表达,从而促进果实中花青素的合成[96]。樱桃在蓝光和白-蓝-绿光的照射下增加了PAL酶的活性,使果实中的花青素含量升高[97]。另外,蓝光可以通过PHOT和CRY受体增加草莓中花青素的积累[98-99]。以上结果表明,短波长的UV和蓝光可以通过UVR8和CRY,在转录和转录后方面影响MBW复合体或调控下游光信号转录因子的表达,进而影响植物花青素的生物合成,但其作用机制还待进一步深入研究。
3.2.2 红光和远红光 红光和远红光在植物花青素的生物合成中也起着重要作用。研究发现,红光诱导茄科蔬菜中的表达及覆盆子果实中黄酮类物质的合成,进而促进花青素的合成[100]。另外,高红光/远红光比例下生长的番茄,其花青素正常合成,但低红光/远红光照射下,番茄花青素的合成却受到严重抑制[101-102],这主要是高比例的红光可以促进DFR酶和花色素双氧酶(LDOX)活性,进而促进花青素的积累,而远红光增多(低比例的红光/远红光)抑制DFR酶和LDOX酶的催化作用[101]。光敏色素是感受红光和远红光的主要光受体。研究发现,红光受体phyB主要通过两种方式发挥作用(图2):红光使phyB以活性的Pfr形式从细胞质进入细胞核,一方面通过光敏色素-转录因子途径直接调控基因表达;另一方面通过光敏色素-COP1途径间接调控基因表达[103]。但是远红光信号调控花青素合成的机制尚不清楚,因为远红光受体光敏色素A(phyA)和红光受体phyB均能够抑制COP1的活性,而COP1负调控花青素的合成[104],但增加远红光的比例(低红光/远红光比例)却降低花青素的积累,这可能是由于远红光对phyB的钝化作用大于远红光对phyA的激活作用。
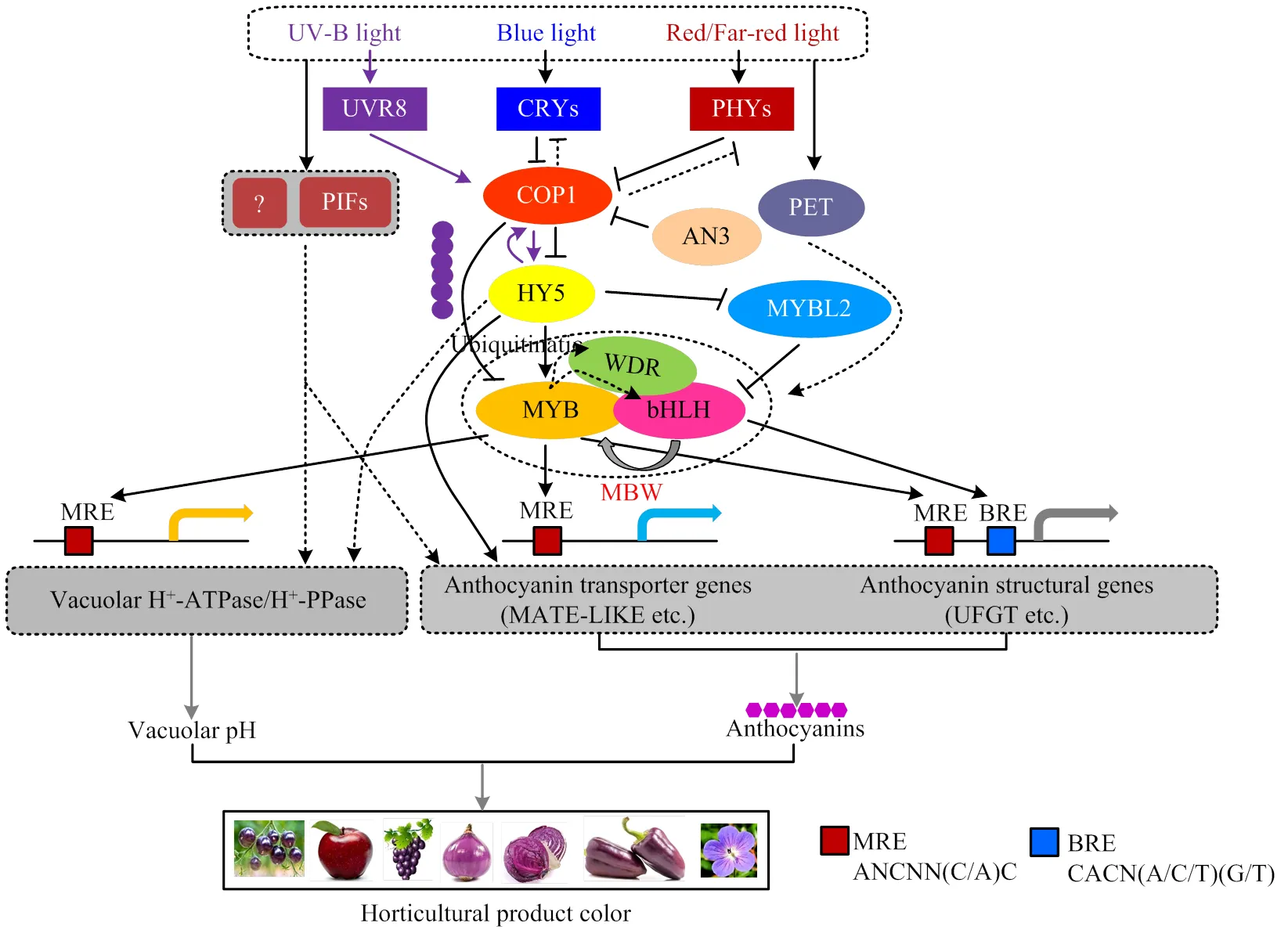
UVR8:UV-B光受体;CRYs:隐花色素;PHYs:光敏色素;MBW:MYB-bHLH-WDR构成的复合体;COP1:暗形态建成中的E3泛素连接酶蛋白;HY5:光信号转录因子;PIFs:光敏色素互作因子;AN3:花青素合成调控因子;PET:光合电子传递;MRE:MYB识别序列;BRE:bHLH识别序列UVR8: UV resistance locus 8; CRYs: crytochrome; PHYs: phytochromes; MBW: MYB-bHLH-WDR complex; COP1: constitutively photomorphogenic 1; HY5: elongated hypocotyl 5; PIFs: phytochrome interacting factors; AN3: ANGUSTIFOLIA3; PET: photosynthetic electron transport; MRE: MYB-recognizing elements; BRE: bHLH-recognizing elements
3.3 光周期(光照时长)对植物花青素生物合成的调控
光周期也会影响植物花青素的生物合成。研究表明,利用补光技术延长光照时间能显著提高植物花青素的积累[105]。光照可以提高烟草花青素生物合成基因的表达量,增加花青素的合成,而黑暗则作用相反[8]。对芜菁地下根进行光照处理时,表达量随着光照时间的延长逐渐增加[106]。另外,延长光照时间显著增强紫叶李叶片中PAL酶的活性,促进花青素的生物合成,使其叶色变红[107]。黑暗下植物花青素含量较低可能与COP1有关,因为突变体在黑暗中能产生花青素[104]。黑暗下,COP1在细胞核中大量积累,它可能通过E3泛素途径将其下游光信号及花青素正调控转录因子降解[92]。另外,黑暗中,SPA下调PAP1和PAP2转录水平[108]。因此,COP1-SPA复合体是光照或黑暗调控植物花青素生物合成的中心调节因子[104]。然而,并非光照时间越长,花青素积累越多。如亚洲杂交百合‘Vivaldi’在黑暗条件下生长时,花青素含量很低,光照后迅速增加,但光照时间过长后,花青素含量又逐渐降低[14]。因此,明确光照时长对植物花青素积累的影响,对合理利用LED光谱技术调控植物花青素的积累有重要意义。
光环境及光受体(UVR8、CRYs、PHOTs、PHYs)主要通过COP1、HY5、PIFs等光信号因子来调控植物花青素的合成,这些光信号因子一方面直接结合到调控花青素合成的MYB、bHLH、WDR三大类转录因子上(图2),转录激活或抑制它们的表达[61-63];另一方面,这些光信号因子通过与MYB、bHLH、WDR三大类转录因子蛋白互作,影响它们形成的MBW复合体的稳定性[93,104],进而调控花青素的合成。此外,这些光信号因子还可以通过不依赖MBW的通路调控花青素的合成[64-65],如HY5通过调控影响花青素的合成[58,60];另外,一些未知的光响应因子可能以不依赖MBW通路的方式直接或间接地调控花青素合成基因和液泡膜上的运转蛋白,改变液泡酸化,调节花青素的合成[5-6]。
4 光环境和生物工程改良园艺植物的花青素
4.1 通过调控光环境改良园艺植物花青素
花青素既影响植物的色泽,也作为抗氧化物质对人体健康有利,因此,提高园艺作物花青素含量成为改善果实营养的热点。光强、光质、光照时长等光环境,可以改变园艺植物花青素的含量。如增加光照可以促进苹果果皮积累花青素,使果皮变红[109]。番茄()果实暴露在光下部分比遮阴部分的花青素含量高[66]。强光诱导番茄和辣椒的和表达,促进番茄和辣椒花青素的积累[39]。当对茄子进行UV-A补光后,茄子颜色加深[87]。增加光环境中蓝光的比例可以促进番茄中花青素的含量[95]。研究表明,光环境的调控是改良园艺作物花青素的重要手段,因此,LED补光技术成为调控园艺作物色泽的关键技术。
4.2 通过调控关键基因进行生物工程改良
传统杂交和诱变为主的育种技术对园艺植物花青素的改良周期长。通过改变调控花青素关键基因的表达水平进而改变花青素积累,是花青素生物工程和代谢工程的一个重要策略。如金鱼草和转入番茄后,转基因番茄的果皮和果肉呈现深紫色,花青素含量远高于蓝莓[110],这样的番茄对提高人体抗氧化能力等有重要的功效。玫瑰、康乃馨和菊花中过量表达,花朵合成飞燕草素呈现蓝紫色,解决了传统育种技术培育不出富含飞燕草素蓝色花朵的难题[111-112]。将转入烟草中可产生1种具有药用价值的花青素Cyanidin 3-O-rutinoside,且这种成分占总花青素含量的98%[113]。这一研究使工业上提取大量具有药用功能的花青素成为可能,同时以烟草作为原料降低了生产成本,极大地促进了花青素代谢工程和生物工程的发展。
5 展望
近年来,人类比以往更加关注花青素在植物色泽形成、抗氧化、人体保健等方面的作用,而光作为花青素合成的重要因子也受到越来越多的关注。解析光对花青素生物合成的调控网络对精准调控园艺植物不同器官花青素的含量有重要的生物学意义。尽管人类利用模式植物(矮牵牛和拟南芥)对花青素的生物合成途径有了较深的理解,并且陆续克隆和鉴定了一些其他物种花青素合成和运转途径的基因,但关于光信号对植物花青素生物合成及运转调控机制的研究才刚刚起步,仍有许多问题值得进一步思考:(1)不同光受体与MBW复合体的互作调控关系尚不清晰。目前除了HY5对花青素积累的调控机制取得一定进展外,与MBW复合体互作的其他光信号因子及其功能的解析,对构建光信号调控花青素的信号网络意义重大;(2)尽管已经发现了许多调控花青素生物合成的正调控因子,但其负调控因子研究相对较少,尤其是光对这些负调控因子的作用机制严重匮乏;(3)甲基化、羟基化、糖基化及酰基化等催化作用对花青素的形成和运输有重要作用,但光信号是否影响这些修饰过程尚不清楚;(4)园艺植物果实中花青素的积累通常受光照、温度等环境因素共同作用的影响,探究光环境与其他环境因子在调控花青素中的互作机制,对合理利用设施环境调控手段,精准调控园艺植物花青素的形成,进而改良观赏植物花色、提高园艺产品的营养价值有重要的指导意义。
[1] WEI J Y, WU H J, ZHANG H Q, Li F, CHEN S R, HOU B H, SHI Y H, ZHAO L J, DUAN H J. Anthocyanins inhibit high glucose-induced renal tubular cell apoptosis caused by oxidative stress in db/db mice.,2018, 41(3): 1608-1618.
[2] ISAAK C K, PETKAU J C, BLEWETT H, KARMIN O, SIOW Y L. Lingonberry anthocyanins protect cardiac cells from oxidative-stress- induced apoptosis., 2017, 95(8): 904-910.
[3] YOSHIDA K, KONDO T, OKAZAKI Y, KATOU K. Cause of blue petal colour., 1995, 373: 291.
[4] POUSTKA F, IRANI N G, FELLER A, LU Y, POURCEL L, FRAME K, GROTEWOLD E. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route inand contributes to the formation of vacuolar inclusions., 2007, 145(4): 1323-1335.
[5] HU D G, SUN C H, MA Q J, YOU C X, CHENG L L, Hao Y J. MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples., 2016, 170(3): 1315-1330.
[6] SHITAN N, YAZAKI K. New insights into the transport mechanisms in plant vacuoles., 2013, 305: 383-433.
[7] RAMSAY N A, GLOVER B J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity., 2005, 10(2): 63-70.
[8] LIU Y, TIKUNOV Y, SCHOUTEN R E, MARCELIS L F M, VISSER R G F, Bovy A. Anthocyanin biosynthesis and degradation mechanisms invegetables: A Review., 2018, 6: 52.
[9] GONZALEZ A, ZHAO M, LEAVITT J M, LLOYD A M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex inseedlings., 2008, 53(5): 814-827.
[10] DUBOS C, STRACKE R, GROTEWOLD E, WEISSHAAR B, MARTIN C, LEPINIEC L. MYB transcription factors in, 2010, 15(10): 573-581.
[11] ZHU Z X, WANG H L, WANG Y T, GUAN S, WANG F, TANG J Y, ZHANG R J, XIE L L, LU Y Q. Characterization of theelements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation., 2015, 66(13): 3775-3789.
[12] PAZ-ARES J, GHOSAL D, WIENAND U, PETERSON P A, SAEDLER H. The regulatorylocus ofencodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators., 1987, 6: 3553-3558.
[13] QUATTROCCHIO F, WING J F, LEPPEN H, MOL J, KOES R E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes., 1993, 5: 1497-1512.
[14] YAMAGISHI M, SHIMMOYAMADA Y, NAKATSUKA T, MASUDA K. TwoGenes, homologs of, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic Hybrid Lily., 2010, 51(3): 463-474.
[15] SUN C L, DENG L, DU M M, ZHAO J H, CHEN Q, HUANG T T, JIANG H L, LI C B, LI C Y. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh., 2020, 13(1): 42-58.
[16] YAN S S, CHEN N, HUANG Z J, LI D J, ZHI J J, YU B W, LIU X X, CAO B H, QIU Z K.encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription ofto fine-tune anthocyanin content in tomato fruit., 2020, 225(5): 2048-2063.
[17] COLANERO S, TAGLIANI A, PERATA P, GONZALI S. Alternative splicing in thegene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits., 2020, 1(1): 100006.
[18] TOHGE T, NISHIYAMA Y, HIRAI M Y, YANO M, NAKAJIMA J, AWAZUHARA M, INOUE E, TAKAHASHI H, GOODENOWE D B, KITAYAMA M, NOJI M, YAMAZAKI M, SAITO K. Functional genomics by integrated analysis of metabolome and transcriptome ofplants over-expressing an MYB transcription factor., 2005, 42: 218-235.
[19] PEEL G J, PANG Y, MODOLO L V, DIXON R A. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in., 2009, 59(1): 136-149.
[20] ESPLEY R V, HELLENS R P, PUTTERILL J, STEVESON D E, KUTTY-AMMA S, ALLAN A C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10., 2007, 49(3): 414-427.
[21] ESPLEY R V, BRENDOLISE C, CHAGNé D, KUTTY-AMMA S, GREEN S, VOLZ R, PUTTERILL J, SCHOUTEN H J, GARDINER S E, HELLENS R P, ALLAN A C. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples., 2009, 21(1): 168-183.
[22] MEDINA-PUCHE L, CUMPLIDO-LASO G, AMIL-RUIZ F, HOFFMANN T, RING L, RODRíGUEZ-FRANCO A, CABALLERO J L, SCHWAB W, MUñOZ-BLANCO J, BLANCO-PORTALES R. MYB10plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening offruits., 2014, 65(2): 401-417.
[23] STARKEYIč P, PAUKšTYTė J, KAZANAVIčIūTė V, DENKOVSKIENė E, STANYS V, BENDOKAS V, ŠIKšNIANAS T, RAžANSKIENė A, RAžANSKAS R. Expression and anthocyanin biosynthesis-modulating potential of sweet cherry (L.) MYB10and bHLHgenes., 2015, 10(5): e0126991.
[24] RAVAGLIA D, ESPLEY R V, HENRY-KIRK R A, ANDREOTTI C, ZIOSI V, HELLENS R P, COSTA G, ALLAN A C. Transcriptional regulation of flavonoid biosynthesis in nectarine () by a set of R2R3 MYB transcription factors., 2013, 13: 68.
[25] WANG Z G, MENG D, WANG A D, LI T L, JIANG S L, CONG P H, LI T Z. The methylation of thepromoter is associated with green-skinned sport in Max Red Bartlett pear., 2013, 162(2): 885-896.
[26] NAKATSUKA T, HARUTA K S, PITAKSUTHEEPONG C, ABE Y, KAKIZAKI Y, YAMAMOTO K, SHIMADA N, YAMAMURA S, NISHIHARA M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers., 2008, 49(12): 1818-1829.
[27] LIU X F, XIANG L L, YIN X R, GRIERSON D, LI F, CHEN K S, YIN X R. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation inflowers., 2015a, 194: 278-285.
[28] COSTANTINI L, MALACARNE G, LORENZI S, TROGGIO M, MATTIVI F, MOSER C, GRANDO M S. New candidate genes for the fine regulation of the colour of grapes., 2015, 66(15): 4427-4440.
[29] CHAGNé D, WANG K L, ESPLEY R V, VOLZ R K, HOW N M, ROUSE S, BRENDOLISE C, CARLISE C M, KUMAR S, DE SILVA N, MICHELETTI D, MCGHIE T, CROWHURST R N, STOREY R D, VELASCO R, HELLENS R P, GARDINER S E, ALLAN A C. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes., 2013, 161: 225-239.
[30] AN X H, TIAN Y, CHEN K Q, LIU X J, LIIU D D, XIE X B, CHENG C G, CONG P H, HAO Y J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples., 2014, 56(4): 650-662.
[31] JIAN W, CAO H H, YUAN S, LIU Y D, LU J F, LU W, LI N, WANG J H, ZOU J, TANG N, XU C, CHENG Y L, GAO Y Q, XI W P, BOUZAYEN M, LI Z G. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits., 2019, 6: 22.
[32] DUBOS C, LE GOURRIEREC J, BAUDRY A, HUEP G, LANET E, DEBEAUJOI I, ROUTABOUL J M, ALBORESI A, WEISSHAAR B, LEPINIEC L. MYBL2 is a new regulator of flavonoid biosynthesis in., 2008, 55(6): 940-953.
[33] PéREZ-DíAZ J R, PéREZ-DíAZ J, MADRID-ESPINOZA J, GONZáLEZ-VILLANUEVA E, MORENO Y, RUIZ-LARA S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco., 2016, 90: 63-76.
[34] GAO J J, SHEN X F, ZHANG Z, PENG R H, XIONG A S, XU J, ZHU B, ZHENG J L, YAO Q H. The MYB transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic.,2011, 106(2): 235-242.
[35] SALVALLINI A, PIMENTEL P, MOYA-LEóN M A, HERRERA R. Increased accumulation of anthocyanins infruits by transient suppression ofgene., 2013, 90: 25-36.
[36] MATSUI K, UMEMURA Y, OHME-TAKAGI M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in., 2008, 55(6): 954-967.
[37] GOU J Y, FELIPPES F F, LIU C J, WEIGEL D, WANG J W. Negative regulation of anthocyanin biosynthesis inby a miR156- targeted SPL transcription factor., 2011, 23: 1512-1522.
[38] CAVALLINI E, MATUS J T, FINEZZO L, ZENONI S, LOYOLA R, GUZZO F, SCHLECHTER R, AGEORGES A, ARCE-JOHNSON P, TORNIELLI G B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine., 2015, 167(4): 1448-1470.
[39] ALBERT N W, LEWIS D H, ZHANG H, SCHWINN K E, JAMESON P E, DAVIES K M. Members of an R2R3-MYB transcription factor family inare developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning., 2011, 65(5): 771-784.
[40] ALBERT N W, DAVIES K M, LEWIS D H, ZHANG H B, MONTEFIORI M, BRENDOLISE C, BOASE M R, NGO H, JAMESON P E, SCHWINN K E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots., 2014, 26(3): 962-980.
[41] ZHU H F, FITZSIMMONS K, KHANDLWAL A, KrANZ R G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in., 2009, 2(4): 790-802.
[42] SPELT C, QUATTROCCHIO F, MOL J N, KOES R.ofencodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes., 2000, 12(9): 1619-1632.
[43] CHANDLER V L, RADICELLA J P, ROBBINS T P, CHEN J, TURKS D. Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation ofutilizinggenomic sequences., 1989, 1(12): 1175-1183.
[44] CONSONNI G, VIOTTI A, DELLAPORTA S L, TONELLI C. cDNA nucleotide sequence of, a regulatory gene in maize., 1992, 20(2): 373.
[45] BAUDRY A, HEIM M A, DUBREUCQ B, CABOCHE M, WEISSHAAR B, LEPINIEC L. TT2, TT8, and TTG1 synergistically specify the expression ofand proanthocyanidin biosynthesis in., 2004, 39(3): 366-380.
[46] LI P H, CHEN B B, ZHANG G Y, CHEN L X, DONG Q, WEN J Q, MYSORE K S, ZHAO J. Regulation of anthocyanin and proanthocyanidin biosynthesis bybHLH transcription factor MtTT8.,2016, 210(3): 905-921.
[47] BAI Y, PATTANAIK S, PATRA B, WERKMAN J R, XIE C H, YUAN L. Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b of tobacco have originated from two ancestors and are functionally active., 2011, 234(2): 363-375.
[48] ELOMMA P, UIMARI A, MEHTO M, ALBERT V A, LAITINEN R A, TEERI T H. Activation of anthocyanin biosynthesis in() suggests conserved protein-protein and protein- promoter interactions between the anciently diverged monocots and eudicots.,2003, 133(4): 1831-1842.
[49] SHIMIZU Y, MAEDA K, KATO M, SHIMOMURA K. Co-expression ofandinduces anthocyanin accumulation in roots of culturedDC. plantlet on methyl jasmonate treatment., 2011, 49(2): 159-167.
[50] LAI B, DU L N, LIU R, HU B, SU W B, QIN Y H, ZHAO J T, WANG H C, HU G B. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis inandduring anthocyanin accumulation., 2016, 7: 166.
[51] DE VETTEN N, QUATTROCCHIO F, MOL J, KOES R. Thelocus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants and animals., 1997, 11(11): 1422-1434.
[52] CAREY C C, STRAHLE J T, SELINGER D A, CHANDLER V L. Mutations in theregulatory gene of theanthocyanin pathway have distinct phenotypes relative to the functionally similargene in., 2004, 16(2): 450-464.
[53] LIU X F, YIN X R, ALLAN A C, LIN-WANG K, SHI Y N, HUANG Y J, FERGUSON I B, XU C J, CHEN K S. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry () during anthocyanin biosynthesis., 2013, 115(3): 285-298.
[54] AN X H, TIAN Y, CHEN K Q, WANG X F, HAO Y J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation., 2012, 169(7): 710-717.
[55] ZHAO M R, LI J, ZHU L, CHANG P, LI L L, ZANG L Y. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development., 2019, 10(7): 496.
[56] AGUILAR-BARRAGáN A, OCHOA-ALEJO N. Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins inpepper fruit., 2014, 58(3): 567-574.
[57] YANG F X, CAI J, YANG Y, LIU Z B. Overexpression of microRNA828 reduces anthocyanin accumulation in., 2013, 115(2): 159-167.
[58] JIA X Y, SHEN J J, LIU H, Li F, DING N, GAO C Y, PATTANAIK S, PATRA B, LI R Z, YUAN L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato., 2015, 242(1): 283-293.
[59] QI T c, SONG S S, REN Q C, WU D W, HUANG H , CHEN Y, FAN M, PENG W, REN C M, XIE D X. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in.2011, 23(5): 1795-1814.
[60] WANG Y L, WANG Y Q, SONG Z Q, ZHANG H Y. Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in., 2016, 9: 1395-1405.
[61] SHIN D H, CHOI M, KIM K, BANG G, CHO M, CHOI S B, CHOI G, PARK Y I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in., 2013, 587(10): 1543-1547.
[62] NGUYEN N H, JEONG C Y, KANG G H, YOO S D, HONG S W, LEE H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in., 2015, 84(6): 1192-1205.
[63] AN J P, QU F J, YAO J F, WANG X N, YOU C X, WANG X F, HAO Y J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple., 2017, 4: 17056.
[64] SHIN J, PARK E, CHOI G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in., 2007, 49(6): 981-994.
[65] MAIER A, HOECKER U. COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions., 2015, 10(1): e970440.
[66] MAZZUCATO A, WILLEMS D, BERNINI R, PICARELLA M E, SANTANGELO E, RUIU F, TILESI F,SORESSI G P. Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation., 2013, 72: 125-133.
[67] MENG X C, WANG X J. Regulation of flower development and anthocyanin accumulation in., 2004, 79: 131-137.
[68] COMINELLI E, GUSMAROLI G, ALLEGRA D, GALBIATIA M, WADEB H K, JENKINSB G I, TONELLIA C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in., 2008, 165(8): 886-894.
[69] QUATTROCCHIO F, VERWEIJ W, KROON A, SPELT C, MOL J, KOES R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway., 2006, 18(5): 1274-1291.
[70] ALBEA N W, LEWIS D H, ZHANG H B, IRVING L J, JAMESON P E, DAVIES K M. Light-induced vegetative anthocyanin pigmentation in., 2009, 60(7): 2191-2202.
[71] 邵婉璐, 李月灵, 高松, 李钧敏, 梁宗锁. 光照强度对成熟红颜草莓果实着色和花青素生物合成的影响及可能的分子机制. 植物研究, 2018, 38(5): 661-668.
SHAO W L, LI Y L, GAO S, LI J M, LIANG Z S. Effects of light intensity on the fruit coloration and anthocyanian biosynthesis inDuch.'Benihoppe' and the possible molecular mechanism., 2018, 38(5): 661-668. (in Chinese)
[72] ZHANG Y J, LI Y, LI W P, HU Z L, YU X H, TU Y, ZHANG M, HUANG J Y, CHEN G P. Metabolic and molecular analysis of nonuniform anthocyanin pigmentation in tomato fruit under high light., 2019, 6: 56.
[73] GU K D, WANG C K, HU D G, HAO Y J. How do anthocyanins paint our horticultural products?, 2019, 249: 257-262.
[74] KLEINE T, KINDGREN P, BENEDICT C, HENDRICKSON L, STRAND A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response ofto high irradiance., 2007, 144(3): 1391-1406.
[75] DAS P K, BANG G, CHOI S B, YOO S D, PARK Y I. Photosynthesis-dependent anthocyanin pigmentation in., 2011, 6(1): 23-25.
[76] 胡可, 韩科厅, 戴思兰. 环境因子调控植物花青素苷合成及呈色机理. 植物学报, 2010, 45(3): 307-318.
HU K, HAN K T, DAI S H. Regulation of plant anthocyanin synthesis and pigmentation by environmental factors., 2010,45(3): 307-318. (in Chinese)
[77] 袁利. UV-C处理对紫甘蓝花青素合成的影响以及花青素酰基转移酶的克隆[D]. 北京: 中国农业科学院, 2018.
YUAN L. Study on effect from UV-C treatment on anthocyanin biosynthesis and the cloning of anthocyanin acyltransferase of Purple cabbage [D]. Beijing: Chinese Academy of Agricultural Sciences, 2018. (in Chinese)
[78] 喻譞, 姜璐璐, 王焕宇, 金鹏, 郑永华. UV-C处理对杨梅采后品质及苯丙烷类代谢的影响. 食品科学, 2015, 36(12): 255-259.
YU X, JIANG L L, WANG H Y, JIN P, ZHENG Y H. Effects of UV-C treatment on quality and phenylpropanoid metabolism of postharvest Chinese bayberry fruit., 2015, 36(12): 255-259. (in Chinese)
[79] 杨乐, 杨俊枫, 侯智霞, 宫中志, 王冲, 史文君. UV-B对不同发育时期离体蓝莓主要果实品质及相关酶活性的影响. 西北植物学报, 2015, 35(12): 2477-2482.
YANG L, YANG J F, HOU Z X, GONG Z Z, WANG C, SHI W J. Effects of UV-B treatment on the major quality of Blueberry and related enzyme activities in different developmental stages., 2015,35(12): 2477-2482. (in Chinese)
[80] HUANG X, OUYANG X, YANG P, LAU O S, ChEN L, WEI N, DENG X W. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B., 2013, 110(41): 16669-16674.
[81] PASSERI V, KOES R, QUATTROCCHIO F M. New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles., 2016, 7: 153.
[82] LI Y Y, MAO K, ZHAO C, ZHAO X Y, ZHANG X L, SHU H R, HAO Y J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple., 2012, 160(2): 1011-1022.
[83] PARK J S, CHOUNG M G, KIM J B,HAHN B S,KIM J B, BAE S C,ROH K H,KIM Y H,CHEON C I,SUNG M K,CHO K J. Genes up-regulated during red coloration in UV-B irradiated lettuce leaves., 2007, 26(4): 507-516.
[84] 齐艳, 邢燕霞, 郑禾, 孙倩倩, 李殿波, 王晋芳, 石锦, 赵冰, 郭仰东. UV-A和UV-B提高甘蓝幼苗花青素含量以及调控基因表达分析. 中国农业大学学报, 2014, 19(2): 86-94.
QI Y, XING Y X, ZHENG H, SUN Q Q, LI D B, WANG J F, SHI J, ZHAO B, GUO Y D. UV-A and UV-B involved in induction and regulation of anthocyanin biosynthesis in cabbage., 2014, 19(2): 86-94. (in Chinese)
[85] QIAN C Z, CHEN Z R, LIU Q, MAO W W, CHEN Y L, TIAN W, LIU Y, HAN J P, OUYANG X H, HUANG X. Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses., 2020. doi: 10.1016/j.molp.2020.02.015.
[86] LIU C C, CHI C, JIN L J, ZHU J H, YU J Q, ZHOU Y H. The bZip transcription factor HY5 mediates CRY1a‐induced anthocyanin biosynthesis in tomato.,, 2018, 41: 1762-1775.
[87] MATSUMARU K, KAMIHAMA T, INADA K. Effect of covering materials with different transmission properties on anthocyanin content of eggplant pericarp., 1971, 9: 9-15.
[88] GUO J, WANG M H. Ultraviolet A-specific induction of anthocyanin biosynthesis andexpression in tomato (L.).. 2010, 62(1): 1-8.
[89] ZHOU B, LI Y H, XU Z R, YAN H F, HOMMA S, KAWABATA S. Ultraviolet A-specific induction of anthocyanin biosynthesis in the swollen hypocotyls of turnip ()., 2007, 58(7): 1771-1781.
[90] JIAO Y L, LAU O S, DENG X W. Light-regulated transcriptional networks in higher plants., 2007, 8(3): 217-230.
[91] LAU O S, DENG X W. The photomorphogenic repressors COP1 and DET1:20 years later., 2012, 17(10): 584-593.
[92] LIU B, ZUO Z C, LIU H T, LIU X M, LIN C T.cryptochrome 1 interacts with SPA1 to suppress COP1activity in response to blue light.,2011, 25(10): 1029-1034.
[93] JIANG Z H, CHEN C, WANG J, XIE W Y, WANG M, LI X, ZHANG X Y. Purple potato (L.) anthocyanins attenuate alcohol-induced hepatic injury by enhancing antioxidant defense., 2016, 70(1): 45-53.
[94] MENG L S. Transcription coactivatorANGUSTIFOLIA3 modulates anthocyanin accumulation and light-induced root elongation through transrepression of., 2015, 38: 838-851.
[95] HERNáNDEZ R, EGUCHI T, DEVECI M, KUBOTA C. Tomato seedling physiological responses under different percentages of blue and red photon flux ratios using LEDs and cool white fluorescent lamps., 2016, 213: 270-280.
[96] SHI L Y, CAO S F, CHEN W, YANG Z F. Blue light induced anthocyanin accumulation and expression of associated genes in Chinese bayberry fruit., 2014, 179: 98-102.
[97] KOKALJ D, ZLATIć E, CIGIć B, VIDRIH R. Postharvest light- emitting diode irradiation of sweet cherries (L.) promotes accumulation of anthocyanins., 2019, 148: 192-199.
[98] KADOMURA-ISHIKAWA Y, MIYAWAKI K, NOJI S, TAKAHASHI A. Phototropin 2 is involved in blue light-induced anthocyanin accumulation infruits., 2013, 126(6): 847-857.
[99] XU F, CAO S F, SHI L Y, CHEN W, SU X G, YANG Z F. Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit., 2014, 62(20): 4778-4783.
[100] KATZ A, WEISS D. Light regulation of anthocyanin accumulation and chalcone synthase gene expression inflowers., 1999, 47(4): 225-229.
[101] 陈静, 陈启林, 翁俊, 刘源, 程智慧, 徐春和.不同红光/远红光比例(R/FR)的光照影响番茄幼苗叶片中花青素合成的研究. 西北植物学报, 2004, 24(10): 1773-1778.
CHEN J, CHEN Q L, WENG J, LIU Y, CHENG Z H, XU C HEffect of illumination with different red/far-red ratios on anthocyanidin synthesis in tomato seedling leaves., 2004, 24(10): 1773-1778. (in Chinese)
[102] LIU Z J, ZHANG Y Q, WANG J F, LI P, ZHAO C Z, CHEN Y D, BI Y R. Phytochromeinteracting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light inseedlings., 2015, 238: 64-72.
[103] PFEIFFER A, NAGEL M K, POPP C,WüST F,BINDICS J,VICZIáN A,HILTBRUNNER A,NAGY F,KUNKEL T,SCHäFER E. Interaction with plant transcription factors can mediate nuclear import of phytochrome B., 2012, 109(15): 5892-5897.
[104] MCNELLIS T W, VON AMIM A G, ARAKI T, KOMEDA Y, MISER S, DENG X W. Genetic and molecular analysis of an allelic series ofmutants suggest functional roles for the multiple protein domains., 1994, 6(4): 487-500.
[105] 潘晓琴, 宋世威. 光环境影响植物花青素生物合成研究进展. 植物学研究, 2019, 8(2): 118-125.
PAN X Q, SONG S W. Research advance on the effects of light environment on anthocyanin biosynthesis in plants., 2019, 8(2): 118-125. (in Chinese)
[106] 闫海芳. 光环境影响花青素合成途径中相关基因表达的机制[D]. 哈尔滨: 东北林业大学, 2003.
YAN H F. Mechanism of light environment influencing the expression of correlated genes in biosynthesis pathway of anthocyanin [D]. Harbin: Northeast Forestry University, 2003. (in Chinese)
[107] 史宝胜. 紫叶李叶色生理变化及影响因素研究[D]. 哈尔滨: 东北林业大学, 2006.
SHI B S. Research on the physiological characters and the influence factors on leave color of cherry plum [D]. Harbin: Northeast Forestry University, 2006. (in Chinese)
[108] MAIER A, SCHRADER A, KOKKELINK L, FALKE C, WELTER B, INIESTO E, RUBIO V, UHRIG J F, H€ULSKAMP M, HOECKER U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in., 2013, 74(4): 638-651.
[109] TAKOS A M, JAFFE F W, JACOB S R, BOGS J, ROBINSON S P, WALKER A R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples., 2006, 142: 1216-1232.
[110] BUTELI E, TITTA L, GIORGIO M, MOCK H P, MATROS A, PETEREK S, SCHIJLEN EGWM, HALL R D, BOVY A G, LUO J, MARTIN C (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors., 2007, 26, 1301-1308.
[111] KATSUMOTO Y, FUKUCHI-MIZUTANI M, FUKUI Y, BRUGLIERA F, HOLTON T A, KARAN M, NAKAMURA N, YONEKURA- SAKAKIBARA K, TOGAMI J, PIGEAIRE A. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin., 2007, 48: 1589-1600.
[112] BRUGLIERA F, TAO G Q, TEMS U, KALC G, MOURADOVA E, PRICE K, STEVENSON K, NAKAMURA N, STACEY I, KATSUMOTO Y, TANAKA Y, MASON J G. Violet/blue chrysanthemums- metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors., 2013, 54: 1696-1710.
[113] HE X Z, LI Y, LAWSON D, XIE D Y. Metabolic engineering of anthocyanins in dark tobacco varieties., 2017, 159: 2-12.
Light Regulation of Anthocyanin Biosynthesis in Horticultural Crops
WANG Feng, WANG XiuJie, ZHAO ShengNan, YAN JiaRong, BU Xin, ZHANG Ying, LIU YuFeng, XU Tao, QI MingFang, QI HongYan, LI TianLai
(College of Horticulture, Shenyang Agricultural University/The State Education Ministry and Liaoning Provincial Key Laboratory of Protected Horticulture/National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang 110866)
Anthocyanins are among the most important flavonoid compounds in plants, which play significant roles in color formation of plant organ, such as flower and fruits, as well as antioxidant process. Light is one of the most important environmental factors affecting anthocyanin biosynthesis pathway, but it still remains unclear in the mechanism and signaling networks of light regulation of anthocyanin. This review briefly introduced the anthocyanin biosynthesis and transportation pathway, and summarized the molecular mechanism of anthocyanin transcriptional regulation by three kinds of transcription factors, including MYB, bHLH and WDR. In addition, it emphasized on the light signaling regulation of anthocyanin biosynthesis. The researches showed that the light environment (light intensity, light quality, and light duration) regulated the biosynthetic process of anthocyanin mainly through different light receptors (UVR8, CRYs, PHOTs, and PHYs), which affected the ubiquitination ability of COP1, the stability of HY5, and the stability of other light signal transcription factors, such as the phytochrome-interacting factors (PIFs). On the one hand, these light signal factors directly could bind to the promoters of,and, activate or inhibit these genes expression and then regulate the synthesis of anthocyanin. On the other hand, these light signal factors interacted with proteins of MYB, bHLH and WDR, affecting the stability of the MBW complex formed by them. In addition, these light signaling factors could also regulate anthocyanin synthesis through MBW independent pathways, such as HY5 also affect anthocyanin biosynthesis by regulating. In addition, some unknown light signaling factors might directly or indirectly regulate anthocyanin synthesis genes and interacting with some vacuolar membranes proteins in a MBW independent manner, to change vacuolar acidification and regulate anthocyanin synthesis. At the same time, light signaling factors also affected some factors in the photosynthetic electron transport chain through MBW dependent or MBW independent pathways, then affected anthocyanin synthesis in plants. How these pathways were coordinated and which pathway was preferentially responded by light environments (light intensity, light quality, light duration)? This paper provided a basis to further investigate the molecular mechanism regulating anthocyanin biosynthesis by light signalings. The study explored the effective ways and target molecules for light regulation of anthocyanin accumulation, andcreated opportunities for the development of anthocyanin-rich horticultural crops through genetic and metabolic engineering, and light environmental management.
light; anthocyanins; transcription factor; transcriptional regulation; horticultural crops

10.3864/j.issn.0578-1752.2020.23.015
2020-04-06;
2020-06-30
国家自然科学基金(31801904)、辽宁省“兴辽英才计划”(XLYC1807020)、辽宁省高等学校创新人才支持计划(LR2018027)、辽宁省博士启动基金(20180540094)、沈阳市中青年科技创新人才支持计划(RC200449)、国家重点研发计划(2018YFD1000800,2019YFD1000300)、国家现代农业产业技术体系建设专项(CARS-23-C01)、辽宁省“百千万人才工程”(LNBQW2018W0483)、沈阳农业大学科研启动基金(880418039)
通信作者王峰,E-mail:fengwang@syau.edu.cn。通信作者李天来,E-mail:tianlaili@126.com
(责任编辑 赵伶俐)
——记嘉荫县红光乡燎原村党支部

