Knockdown of SLC2A6 gene inhibits the proliferation of lung adenocarcinoma A549 cells by inducing G2/M arrest
TIAN Di, CHEN Chun-fa, LIU Jia, HUANG Zhao, ZHAO Ping
(Institute for Medical Biology & Hubei Provincial Key Laboratory for Protection and Application of Special Plants in the Wuling Area of China, College of Life Sciences, South-Central University for Nationalities, Hubei Medical Biology International Science and Technology Cooperation Base, Wuhan 430074, China)
Abstract SLC2A6 is a member of the facilitative glucose transporter family, it codes GLUT6 protein. SLC2A6 is up-regulated in several numerous cancers, but is not widely expressed in normal tissues. In order to elucidate the functions and underlying mechanisms for SLC2A6 controlling cancer cell proliferation and apoptosis, we screened two SLC2A6 knockdown lung adenocarcinoma A549 cell lines by CRISPR/Cas9 to illustrate the effects of SLC2A6 on biological process. SLC2A6 expression in mRNA and protein levels has been reduced in SLC2A6-knockdown A549 cells. Western Blot analysis confirmed that SLC2A6 protein expression decreased by 74% in KD1 and 83% in KD2 cells, respectively, compared with the control vehicle. In SLC2A6 mRNA expression level, there are 70.6%±2.0% and 76.9%±4.1% reduction in KD1 and KD2 cells, respectively, compared with wild-type A549 cells. The results of MTT assay indicated that the proliferation rate of KD cells was slower. The fold change of cell number on the fifth day of growth decreased from 22.1±1.0 in WT cells to 13.5±0.9 or 12.2±1.0 than in KD1 and KD2 cells (P<0.001). The cell cycle analysis via flow cytometry demonstrateed that SLC2A6 KD cells arrested at G2/M phase, as the percentage of cells in the G2 phase increased from 16.0%±1.0% of wild type (WT) cells to 23.0%±1.1% of KD1 cells or 27.4%±0.4% of KD2 cells (P<0.05). Moreover, the results of cell cycle regulators RNA expression showed that G2/M arrest was associated with the regulation of expression APC, Rad9a, CDK1, cdc25 and p21Waf1/Cip1. In conclusion, our findings revealed a novel role for SLC2A6 in regulating cancer cell growth, which suggested that SLC2A6 may represent a viable anti-cancer drug target.
Key words SLC2A6; A549 cells; CRISPR/Cas9; cell proliferation; cell cycle
Lung cancer is a highly prevalent disease that has been ranked among the leading causes of death worldwide[1]. It was reported that 1.8 million new cases of lung cancer were diagnosed in 2012. Lung cancer was the most frequently diagnosed cancer and accounted for 13% of total cancer diagnoses[2]. Current treatment strategies of lung cancer include chemotherapy, radiation therapy, targeted therapy, or a combination of treatments, depending on the disease type and stage[3]. The majority of lung cancers are Non-small cell lung cancer (NSCLC) which accounts for 85% of lung cancers[4]. Molecular dissection of abnormalities in NSCLC reveals that many pathways are disrupted and a number of oncogenes are characteristically overexpressed including K-ras, erbB2 and Bcl-2[5]. For the past few years, the first approved targeted therapy for lung cancer blocks the activity of the epidermal growth factor receptor which was overexpressed in greater than half of NSCLC. However, its clinical impact is only modest. In addition, tumor suppressor genes are frequently deleted in lung cancer, and they control cell growth and division regulated by the integration of mitogenic and inhibitory signals. It could be a good curative method if therapies are directed at these targets, such as inhibiting overexpressed oncogenes or adding tumor suppressor genes, to halt malignancy from progression.
Glucose uptake and glycolytic-lipogenic metabolism are both regulated by the Glucose transporter family (GLUTs1-14)[6]. Many of these are overexpressed in normal tissues even cancer cells and tissue[7]. For example, up-regulation ofGLUT1 (Glucose transporter 1) andGLUT6 (Glucose transporter 6) have been both reported in many malignancies[8-9].GLUT6 (originally namedGLUT9) was first discovered in 2000 when searching the human genome for sequences similar to other members of the GLUT family[10]. However, it is unclear whether it transports other substrates and its affinity (Km) remains to be determined.SLC2A6 is only found primarily in spleen, brain and leukocytes[11]. Alterations inSLC2A6 expression have been linked to numerous cancers, butSLC2A6 is not widely expressed in normal tissues. The oncomine database shows thatSLC2A6 is upregulated in cutaneous melanoma, medullary breast carcinoma, oesophageal cancer, head and neck squamous cell carcinoma and etc[12-13]. Previous studies have shown thatSLC2A6 knockdown kills endometrial cancer cells that express elevated levels of the protein[9]. However, knockdown ofSLC2A6 in healthy mouse had no adverse impact on whole-body glucose metabolism or metabolic physiology[14].
Therefore, we speculateSLC2A6 might be a potential anti-cancer drug target if its inhibition does not have deleterious impacts on healthy tissues but harm cancer cells. To elucidate the function ofSLC2A6 in cancer cells, we chose human lung adenocarcinoma A549 cells as a subject for study. In the present report, we knocked out theSLC2A6 gene in A549 cell line by using CRISPR/Cas9 technology[14]. Furthermore, we attempted to elucidate its influence on cell proliferation and reveal the downstream effectors responsible for cell proliferation and apoptosis. Lastly, we found that knockdown ofSLC2A6 inhibits the proliferation of human lung adenocarcinoma A549 cells through the induction of G2/M phase arrest.
1 Materials and methods
1.1 Chemicals
RPMI-1640 mediate and fetal bovine serum (FBS) were both purchased from Gibco (Grand Island, NY). LipofectamineTM3000 was purchased from Invitrogen (Thermo Fisher Scientific, China). β-Actin antibody was purchased from Beyotime (Jiangsu, China) andGLUT6 antibody was purchased from Omnimabs (California, USA). All other chemicals were purchased from Sinopharm Chemical Reagent Co. (Shanghai, China).
1.2 sgRNA design and plasmid construction
One pair of CRISPR-Cas9 sgRNAs was designed to targetSLC2A6 (NM_001145099.1) exon 1 by an online tool (http://crisp r.mit.edu), and sgRNA sequences are shown in Figure 1-A. The couple sgRNAs were annealing and inserted into linearized pSpCas9(BB)-2A-GFP plasmid (pX458 vector, Addgene plasmid # 48138) byBbsI (New England Biolabs, R0539L) site using T4 polynucleotide kinase (New England Biolabs, M0201S) for expressing sgRNA and Cas9. The recombined plasmid was extracted with the endo-free plasmid extraction maxi kit (CWBIO, CW2104M) and verified by DNA sequencing.
1.3 Cell culture, transfecting and screening of SLC2A6-knockdown A549 cells
Human lung adenocarcinoma A549 cell line were obtained from Procell Life Science & Technology company (Wuhan, China). A549 cell culture medium was prepared with 10% serum, 1% antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) and 89% RPMI-1640. A549 cells were cultured at 37 ℃ in a 5% CO2incubator. The cells were seeded in a 6-well cell culture plate and transfected verified positive recombined plasmid using LipofectamineTM3000. The transfected cells were cultured at 37 ℃ in a 5% CO2incubator within 48 h and observe cells by using microscope. The cells were washed with 1×phosphate buffered saline (PBS) for three times; next, cells were suspended and filtered through a 300-mesh nylon mesh into a 5 mL tube, and filtered single cell suspension was subsequently sorted by flow cytometry (Sony, model SH800) to isolate GFP protein positive cells. Gained positive single cells were seeded in each well of 96-well plates. After monoclonal cells expansion, then passage cells and genomic DNA of part cells in each well were extracted after clonal. The PCR products with sgRNA sequence ofSLC2A6 gene were amplified, purified and sequenced by Sanger sequencing to confirm the KD phenotype of the cells.
1.4 RNA isolation and quantitative real-time PCR
Total cellular RNA of A549 cells was extracted using TRIzol reagent and dissolved in diethyl pyro carbonate water. The corresponding cDNA was obtained using a reverse transcription kit. Real-time reaction was performed using the 7500 rapid real-time PCR system instrument and SYBR dye. The sequences of primers used were shown at Table 1.
1.5 Western blotting
The protein was extracted from A549 cells. The cells were washed with PBS three times and resuspended in HES buffer containing PMSF. Cells were then lysed by passing through a 22-gauge needle 30 times followed by an additional 30 passes through a 27-gauge needle. The lysate was then centrifuged at 500 g for 10 mins, and the supernatant was collected and heated at 65 ℃ for 10 min in sodium dodecyl sulfate (SDS) protein loading buffer. All samples were subjected to 8% or 10% SDS-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was later incubated with primary and HRP-conjugated secondary antibodies. Protein bands were incubated using ECL kits and then detected and quantified using the Chemi-Doc XRS system (Bio-Rad, Hercules, CA).
1.6 Cell proliferation assay
Cells were treated with MTT (Sigma, St Louis, MO, U.S.A.) to detect the effects ofSLC2A6 knockdown on cell proliferation. In brief, the cells were cultured by seeding 103cells/well into a 96-well tissue culture plate. Then, 20 μL of the MTT reagent was added to each well for 4 h at 37 ℃. Next, 200 μL DMSO was added to each well, and the optical density was measured at 490 nm after cultivating for 0, 24, 48, 72 and 96 h.
1.7 Cell cycle assay
In brief, approximately A549 cells were pelleted and fixed with 70% ethanol for 1 h at 4 ℃. Cells harvested were treated with RNase A (100 μg/mL) and propidium iodide (PI, 50 μg/mL) at 37 ℃ for 30 min in a dark chamber. The cell cycle distribution was assessed using a BD FACS caliber flow cytometer (BD Biosciences, San Jose, CA, U.S.A.) according to the manufacturer′s instructions. A bar chart of cell cycle distribution was generated with cell phase (G1, S, and G2/M) on thex-axis and the percentage (%) on they-axis.
1.8 Cell apoptosis assay
The apoptotic and necrotic cells were analyzed by Annexin V/PI apoptosis detection Kit (Yeasen) according to the manufacturer′s protocol. The cells incubated with complete medium only were set as the blank control. Next, the cells were washed with PBS and re-suspended in 200 μL of binding buffer containing 5 μL Annexin V and 10 μL PI. After incubation in dark at room temperature for 15 min, 400 μL of binding buffer was added to each sample, and the cells were immediately analyzed by BD FACS caliber flow cytometer (BD Biosciences, San Jose, CA, U.S.A.). The data analysis was performed with Flow Jo 7.6 software. Positioning of quadrants on Annexin-V/PI plots was performed to distinguish living cells (Annexin V-/PI-), early apoptotic cells (Annexin V+/PI-), late apoptotic/necrotic cells (Annexin V+/PI+).
1.9 Data analysis
To determine whether there were significant differences between groups, we performed Student′st-tests by using Origin 9.0 software (Origin Lab, Northampton, MA). Data are shown as the mean±standard error of the mean. Thenvalues represent the number of cells. Differences withP< 0.05 are considered statistically significant.
2 Results
2.1 Knockdown of SLC2A6 in lung adenocarcinoma A549 cells
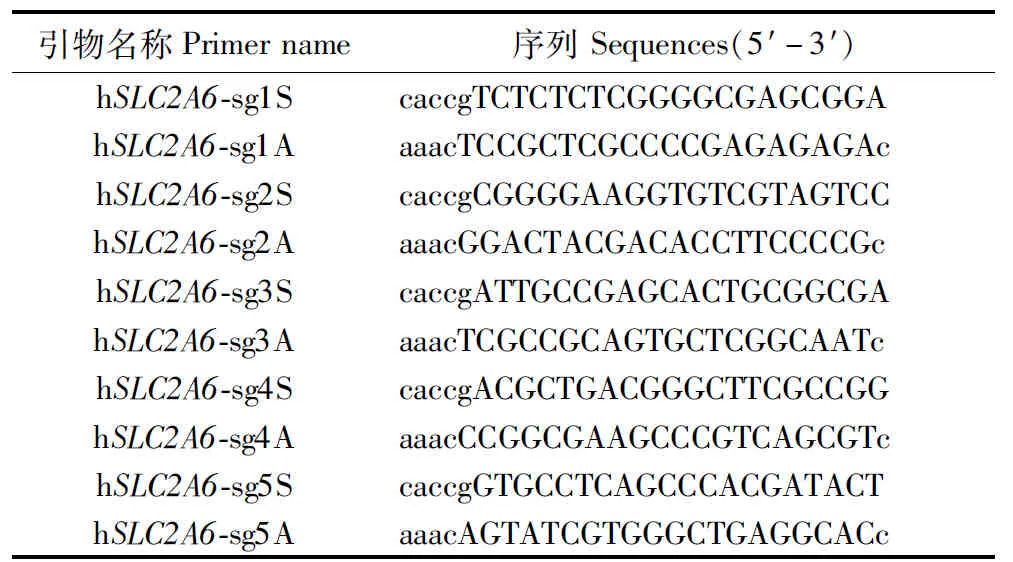
Table 1 hSLC2A6 sgRNA sequences
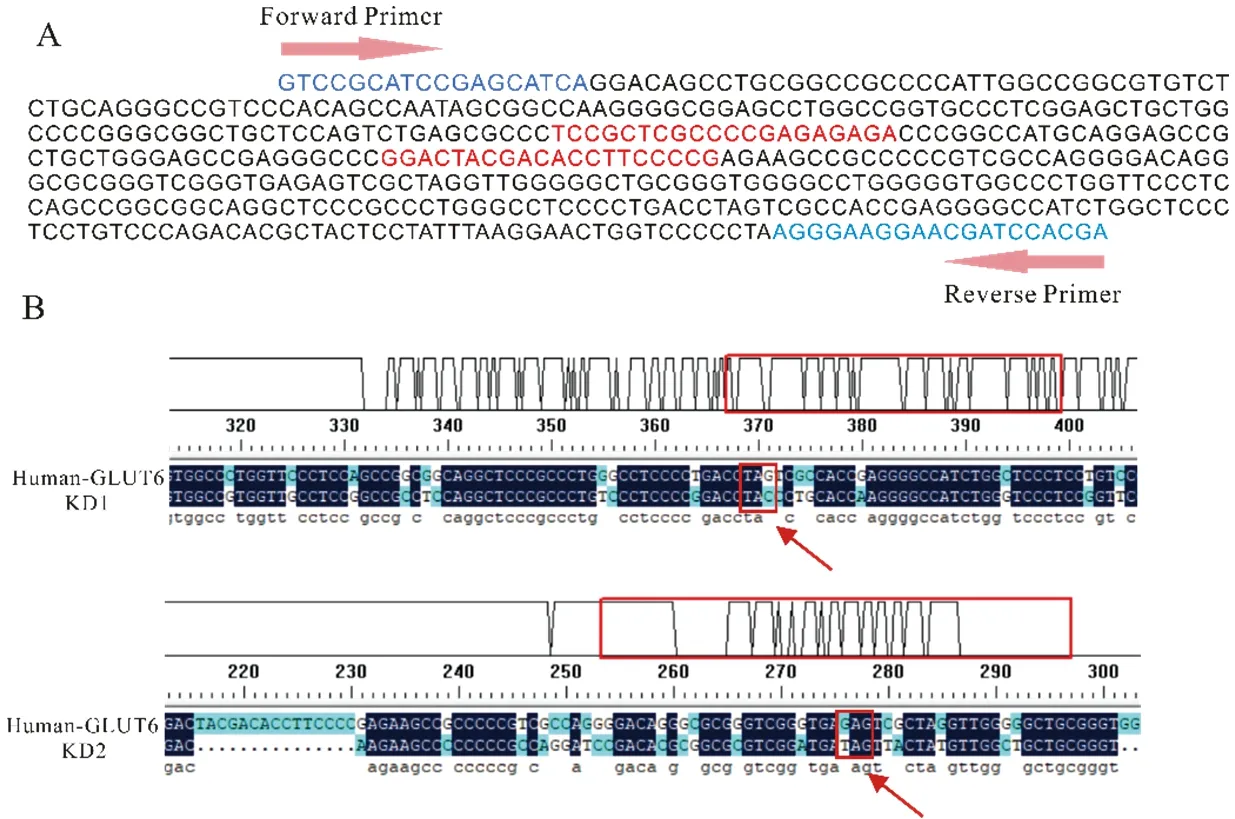
(A) sgRNAs were designed to target SLC2A6 exon 1 by an online tool. The red nucleotides indicated the sgRNA sequence. Arrows indicated the location of forward and reversed genotyping primers. The blue nucleotides indicated primers. (B) The results of DNA sequencing about KD A549 cells compared with A549 wild type by using DNAMAN software
Firstly, five pairs of sgRNAs were designed and double-strand oligo-DNA of sgRNA forSLC2A6 was successfully inserted into pX458 vector, the first four sequences were effective targets of exon 1 atSLC2A6 gene (Table 1). This two different sgRNA were located in exon1 of humanSLC2A6 gene sequence (red nucleotides) (Figure 1-A). Secondly, after using LipofectamineTM3000 Transfection Reagent to transfectSLC2A6 vector into A549 cells, cells with GFP green fluorescence were shown in Figure 2-A.
The results of sequencing indicated that numerous deletion and mutation occurred in DNA sequences of KD1 and KD2 A549 cells. In all of bases mutants, the base replacement occupies main portion of the total mutants with only a small portion of gene deletion. For KD1 cells, the key of mutation is TAG → TAC, which makingSLC2A6 gene transcript longer RNA sequences. The key of mutation was GAG → TAG transition for KD2 cells, which leading to terminateSLC2A6 gene transcription. (Figure 1-B).

(A) After transfection, the bright-field image and the dark-field image of cells are shown (10 ×), scale bar = 100 μm. (B) SLC2A6 gene protein expression in A549 cells and KD cells was detected by Western Blot. (C) RNA expression levels of SLC2A6 in KD and WT A549 cells were detected by Real-time PCR. GAPDH served as an internal control. “**”: P<0.001(n=3)
2.2 SLC2A6 expression in mRNA and protein levels had been reduced in SLC2A6-knockdown A549 cells
To investigate whetherSLC2A6 RNA and protein expression had changed in twoSLC2A6-knockdown cell lines, we implemented Western Blot analysis and Real-time PCR experiment. The results confirmed thatSLC2A6 protein expression decreased by 74% in KD1 and 83% in KD2 cells, respectively compared with the control vehicle (Figure 2-B). InSLC2A6 mRNA expression level, there were 70.6%±2.0% and 76.9%±4.1% reduction in KD1 and KD2 cells, respectively, compared with wild-type A549 cells (Figure 2-C). These data proved that two stable knocking downSLC2A6 gene cell lines had been successfully established. The twoSLC2A6-knockdown cell lines were used in all remaining experiments.
t-test was performed using Oringin 9.0. Value are the mean±SD of three determinations from separate experiments. “*”: P<0.05; “**”: P <0.01; “***”: P<0.001(n=3)
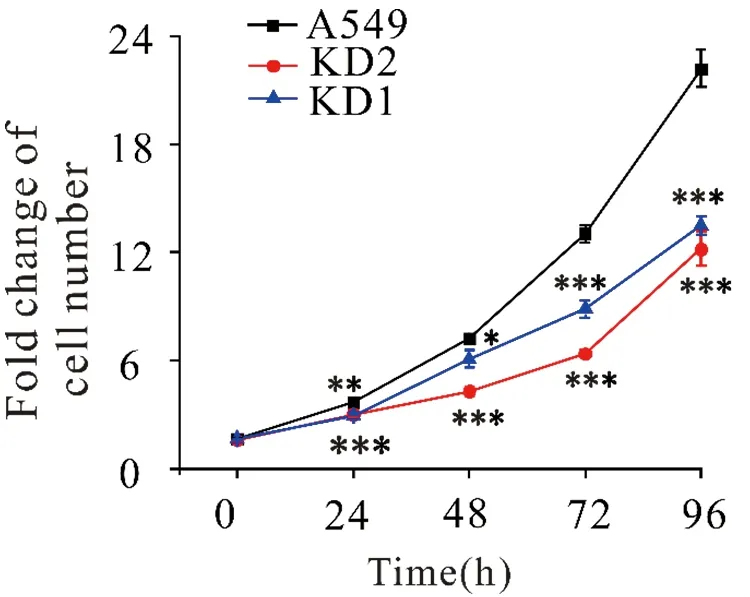
2.3 SLC2A6 knockdown inhibited cellular proliferation in A549 cells
Using these twoSLC2A6-knockdown cell lines, the function ofSLC2A6 on cellular proliferation in lung adenocarcinoma A549 cells was further explored by MTT assay. After 24, 48, 72 and 96 h of synchronous culture, we found thatSLC2A6-knockdown cells exhibited a decreased proliferative capacity compared with wild-type A549 cells (Figure 3). Especially after 96 h cell synchronous culture, the fold change of cell number decreased from 22.1±1.0 in WT cells to 13.5±0.9 or 12.2±1.0 than in KD1 and KD2 cells (***P<0.001). Wild-type A549 cells grown approximately twice as fast asSLC2A6 knockdown cells. These results indicated that knockdown ofSLC2A6 gene inhibited cell proliferation in lung adenocarcinoma A549 cells.
2.4 Knockdown of SLC2A6 gene redistributed the cell cycle in A549 cells
To determine the phase at whichSLC2A6 inhibits cell growth in the cell cycle progression of A549 cells, FACS analysis was employed to evaluate the impact ofSLC2A6 expression on cell cycle progression. Compared with WT A549 cells, knockdown ofSLC2A6 gene in KD A549 cells resulted in a higher percentage of cells at the G2/M phase and a lower percentage of cells at the G1phase, which displayed G2/M phase arrest (Figure 4-A, B and C). The statistical results of cell cycles in KD1, KD2 and WT A549 cells were shown in Figure 4-D. Percentages of cells at G1phase decreased from 62.2%±0.5% of wild type (WT) cells to 49.9%±0.9% of KD1 cells (*P<0.05) or 46.9%±1.2% of KD2 cells (**P<0.01), and as the percentage of cells in the G2phase increased from 16.0%±1.0% of wild type (WT) cells to 23.0%±1.1% of KD1 cells (*P<0.05) or 27.4%±0.4% of KD2 cells (**P<0.01).
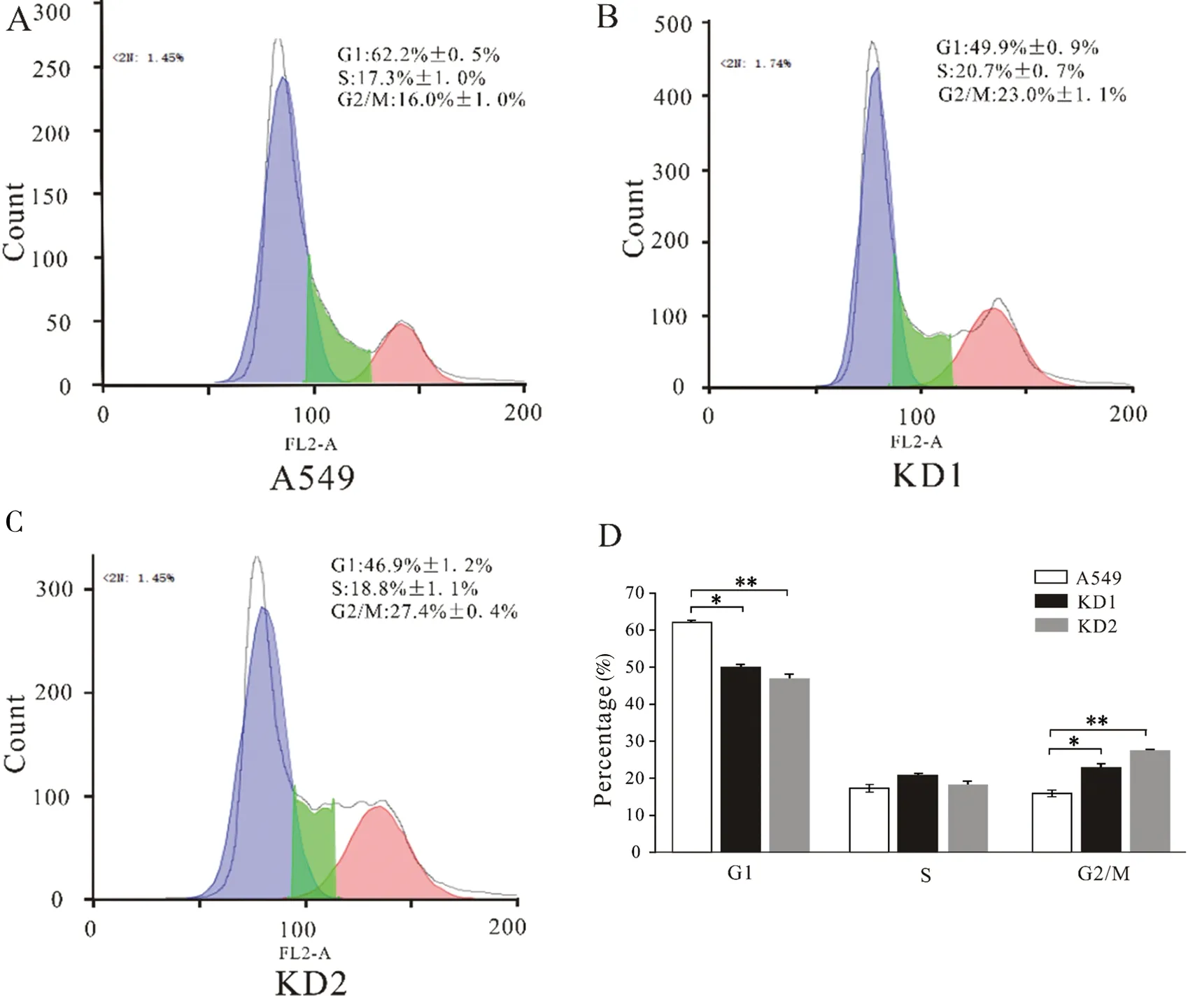
(A) The cell cycles of WT A549 cells were analyzed by FACS. The blue section indicated G1 phase, the green section indicated S phase and the red section indicated G2/M phase. Ordinate represents number of cells. (B) The cell cycles of KD1 A549 cells were analyzed by FACS. (C) The cell cycles of KD2 A549 cells were analyzed by FACS. (D) The statistical result of cell cycles in KD1, KD2 and WT A549 cells, revealed percentage of different cell phase. “*”:P<0.05; “**”:P<0.01(n=3)
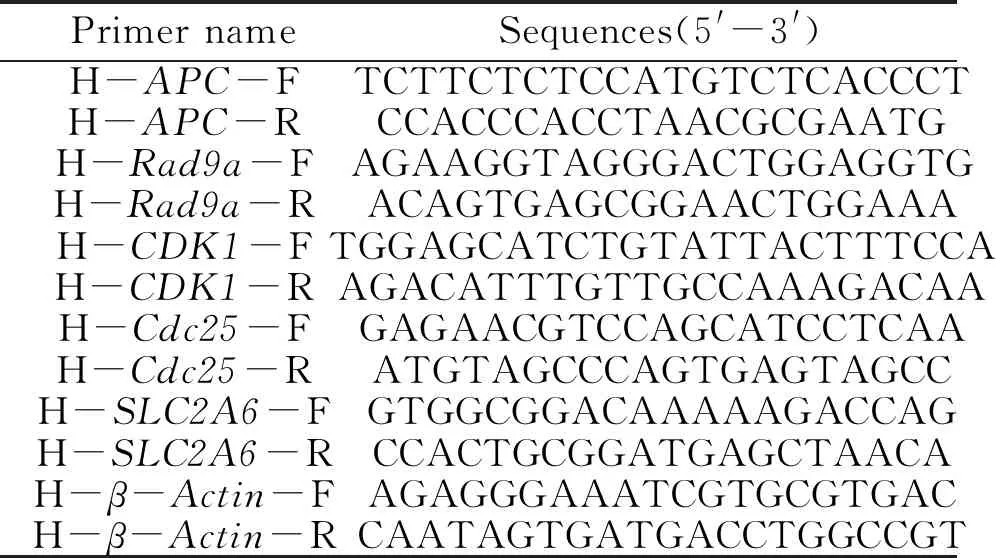
Table 2 Sequence of primers used in the present study
2.5 Cell cycle regulators induce G2/M arrest in SLC2A6 knockdown A549 cells
Subsequently, alterations of cell cycle-related markers in RNA expression were detected. Real-time PCR analysis of G2/M phase-related genes, includingAPC,Rad9a,CDK1,cdc25 andp21Waf1/Cip1. The results revealed that KD A549 cells expressed lower levels ofAPC,Rad9a,CDK1 andcdc25 compared with wild-type A549 cells. However, the analysis results showed RNA expression level ofp21Waf/Cip1was higher after knockdownSLC2A6 gene (Figure 5). These data confirmed thatSLC2A6 may affect the proliferation of A549 cells by altering the cell cycle and the expression of cell cycle-related markers in RNA levels.

Real-time PCR analysis of the RNA levels of APC, Rad9a, CDK1, cdc25 and p21Waf1/Cip1 in KD1, KD2 and WT A549 cells. β-Actin was used as a loading control. *: P<0.05; **: P<0.01 (n=3)
2.6 SLC2A6 knockdown does not impact cell apoptosis in A549 cells
To demonstrate whether the cell apoptosis impacts the cell numbers when knocking downSLC2A6 gene, FACS analysis with the Annexin V-FITC/PI dual labeling was carried out. However, there are not difference whether for early apoptotic cells or late apoptotic cells in KD A549 cells and WT A549 cells (Figure 6-A, B and C). The cells′ apoptosis rate did not display significantly difference betweenSLC2A6 knockdown and wild-type A549 cells (Table 3). The results proved that knockdown ofSLC2A6 does not impact cell apoptosis in A549 cells.
3 Discussion
Due to tumor recurrence and metastasis, lung cancer is known to be a highly treatment-refractory cancer. Especially non-small cell lung cancer, its treatment was regard as a great challenge for clinical therapy[15]. A series of tumors-enriched oncogenes play vital roles in mitogenic, material exchange and cell growth in cancer cells.SLC2A6 is known to transport glucose and was detected in multiple tumor tissues such as breast cancers and pancreatic adenocarcinomas, but not normal breast and pancreas tissues[16]. Although several studies have indicated the potential role ofSLC2A6[6-9, 11], the exact function ofSLC2A6 is still unclear. Previous studies indicated that endometrial cancer cells might rely onSLC2A6-mediated glucose transport and glycolytic-lipogenic metabolism for survival[9], but in aSLC2A6-deficient mouse, whole body metabolic physiology was not affected[14].
An extensive literature indicated that the molecular and physiological functions ofSLC2A6 in cell proliferation, cell cycle and apoptosis remain poorly understood. To further reveal the role ofSLC2A6 in cell proliferation, cell cycle and apoptosis, CRISPR/Cas9 technology was performed to knockoutSLC2A6 gene of A549 cells. Then, we studied the effects ofSLC2A6 knockdown on proliferation, cell cycle and apoptosisinvitro.
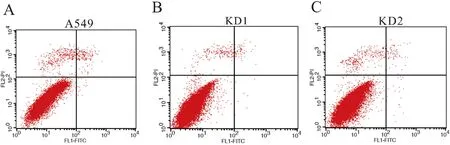
(A) Cell apoptosis in WT A549 cells was analyzed by FACS. (B) Cell apoptosis in KD1 A549 cells was analyzed by FACS. (C) Cell apoptosis in KD2 A549 cells was analyzed by FACS. Table 3 shown the results about percentage of apoptosis in KD1, KD2 and WT A549 cells
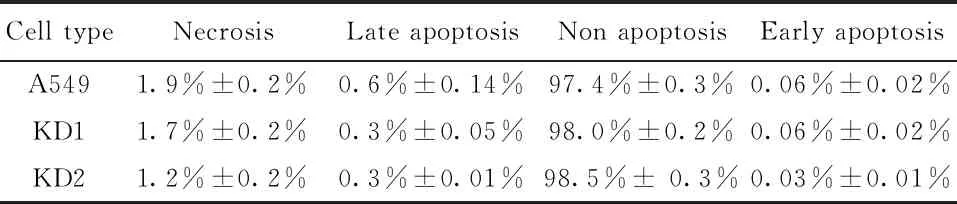
Table 3 Percentage of apoptosis
The sequencing results for two cell clones screened from CRISPR/Cas9 demonstrated that many deletions and mutations within Exon 1 ofSLC2A6 might have led to functional defects on domains (Figure 1-B). Using these two stable cell lines, it was demonstrated that RNA and protein expression ofSLC2A6 were significant inhibited (Figure 2-B and C). The results of cell proliferation showed that the proliferation ofSLC2A6 knockdown A549 cell was significantly inhibited compared with wild-type A549 cell (Figure 3), which is consistent with the already known reports in endometrial cancer cells[9].
As is well known that the inhibition of cell growth is typically accompanied by cell cycle redistribution[17-18], and the following flow cytometric analysis for cell cycle also confirmed this point. The FACS data indicated that the cell cycle ofSLC2A6 knockdown cell was more tended to be arrested in G2/M phage compared with the wild-type A549 cells (Figure 4), which further implies the roles ofSLC2A6 in inhibiting cell proliferation. During the cell cycle progression through the G2/M checkpoint, a series of cell cycle regulators are involved. Interaction of APC with microtubule-binding proteins and further association to the centrosomes and/or kinetochores during late S to G2/M can compete with β-catenin interaction, thus favoring stabilization of cytoplasmic β-catenin at late S/G2phases, and ensuring the correct progression of cells into the cell cycle[19]. Cell cycle G2/M transition is positively regulated byCDK1 (cdc2) andCyclinBcomplex[20-21].Cdc25 family of phosphatases plays a crucial role in cell cycle regulation through dephosphorylation of the inhibitory phosphorylation on cdc2 at Thr14 and Tyr15, caused by Wee1[21]. What′s more, the phosphorylation of cdc2 at Tyr15 and Thr14 is known to be a critical regulatory step that inhibits cdc2 activity and causes G2/M arrest. Previous study indicated that G2checkpoint proteinRad9Ais one of three proteins that sense damaged DNA, forming a proliferating cell nuclear antigen-like heterotrimeric ring around DNA[22]. Rad9A is constitutively phosphorylated at five C-terminal sites and ablation of these sites causes prolonged G2/M arrest[23]. Moreover, cyclin dependent kinase inhibitor (CDKI),p21Waf1/Cip1, also regulates the transitions of G2/M phase by binding and inhibiting the activity of Cdc2-cyclin B1 complex[17].
Our Real-time PCR analysis demonstrated that the knockdown ofSLC2A6 gene reducedAPCexpression and subsequently decreasedRad9aexpression levels, which inhibited thecdc2/cyclinB1 kinase activity and caused G2/M arrest. Meanwhile, the down-regulating ofcdc25 andCDK1 gene also accelerated the blocking of G2/M cell-cycle progression inSLC2A6-knockdown cells. Furthermore, the elevated RNA levels ofp21Waf1/Cip1also indicated thatSLC2A6-knockdown cells were arrested in the G2phase (Figure 5). Beyond that, we further detected whether the cell apoptosis impacted the cell numbers inSLC2A6-knockdown A549 cells. However, the result revealed that cell apoptosis was not involved in this process of cell proliferation (Figure 6 and Table 3).
Overall, our study revealed for the first timeSLC2A6 knockdown in lung adenocarcinoma A549 cell could cause a decrease in cell proliferation ability via inducing G2/M arrest. These findings provide a basis to explore further the physiological significance ofSLC2A6 in cell growth. However, the understanding ofSLC2A6 remains poor. The other important biological functions ofSLC2A6 and the molecular mechanism underlying its regulation of cellular proliferation warrant further investigation.

