Comparison of the Detection Results of Salmonellae in Samples by Two Methods
Jian LI Chengmei WANG
Abstract Salmonellae samples provided by FAPAS were detected by VIDAS and GB 4789.4-2016, respectively. The advantages and disadvantages of each detection method were discussed in the paper, so as to provide theoretical support for the detection of Salmonellae.
Key words Mini VIDAS; GB 4789.4-2016; Salmonellae
Salmonella is a group of Gram-negative sporeless bacteria parasitic in the intestines of humans or animals. It has a wide variety of bacterial types and is widely distributed. It is an important zoonotic pathogen. At present, 2 400 serotypes of Salmonella have been found in the world. According to reports, a total of 285 serotypes of Salmonella were found in China from 1911 to 1995, belonging to 37 O serogroups. Not only can it cause typhoid fever, sepsis, acute gastroenteritis, osteomyelitis, cholecystitis, arthritis, chicken diarrhea and other diseases, but in food poisoning around the world, the cases caused by salmonella occupy the first or second place. Not only does it cause huge losses to the national economy, it also seriously threatens the health of the people and the healthy development of animal husbandry. Therefore, the detection of Salmonella in humans and animals has always been a worldwide problem. In the requirements of hygienic testing, countries around the world generally stipulate that Salmonella should not be detected in food[1] . At present, the commonly used detection methods are: conventional test methods using biochemical reactions, such as national standard method (GB) and the industry standards for import and export commodity inspection (SN), VIDAS method using enzyme-linked immunoassay technology, and the use of molecular biology such as PCR method. Various methods have certain advantages and disadvantages. Shandong China Quality Inspection Co., Ltd. detected two cases of Salmonella by two different detection methods of mini VIDAS and GB 4789.4-2016 in the "Verification of Salmonella Detection Ability in Frozen Chicken" organized by Fapas in 2019.
Materials and Methods
Test samples
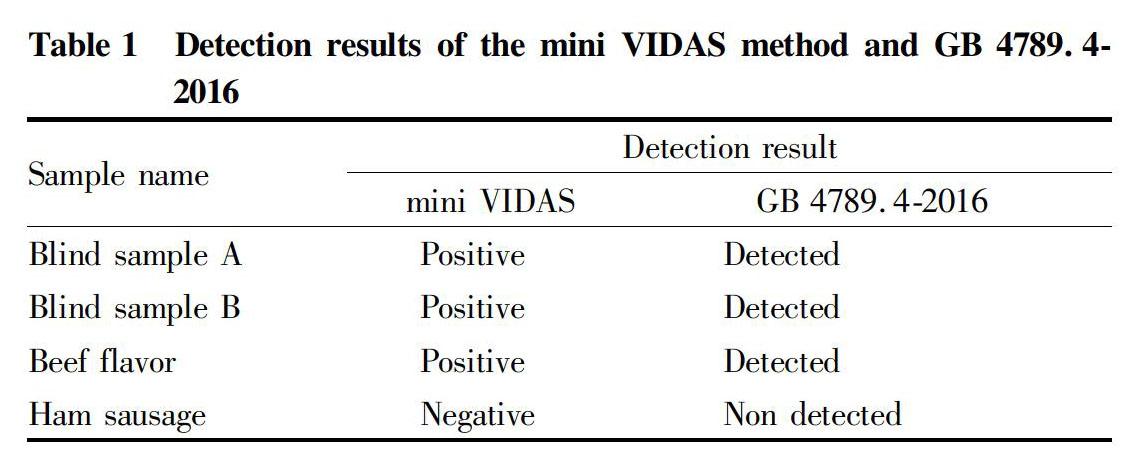
Fapas provided blind sample A, blind sample B, a laboratory-made positive quality control sample beef flavor and a negative quality control sample ham sausage.
Test strains: Salmonella standard strain and Escherichia coli standard strain were provided by ATCC (American Type Culture Collection).
Instruments
miniVIDAS automatic fluorescence immunoassay analyzer, purchased from Bio Mérieux.
Reagents
The reagents used included VIDAS Salmonella (SLM) kit, purchased from Bio Mérieux, France; M broth, purchased from Beijing Land Bridge Technology Co., Ltd.; Salmonella biochemical identification kit, purchased from Beijing Land Bridge Technology Co., Ltd.; diagnostic serum, purchased from Lanzhou Institute of Biological Products Co., Ltd. The above reagents were used within the validity period.
Media
Salmonella enrichment broths used included buffered peptone water (BPW), selenite cystine enrichment liquid (SC), TTB, RVS. The Salmonella selective agar plates used included bismuth sulfite agar (BS), xylose lysine desoxycholate (XLD) agar and Salmonella chromogenic medium. All of the above were purchased from Beijing Land Bridge Technology Co., Ltd. and used within the validity period.
Inspection basis
The inspection was carried out according to the mini VIDAS method and GB 4789.4-2016, of which the mini VIDAS method was executed according to the Chinese operating instructions provided by the manufacturer.
Detection procedures
GB 4789.4-2016 detection procedures
A 25 g of sample was weighed in 225 ml BPW, and the mixture was homogenized for 2 min and incubated at 36 ℃ for 18 h. The cultured sample mixture was gently shaken, and 1 ml was transferred to 10 ml TTB, followed by incubation at 42 ℃ for 24 h. Meanwhile, another 1 ml was transferred to 10 ml SC, and cultured at 36 ℃ for 24 h. A 3 mm loop was used to streak-inoculated one loop of each enriched broth to one BS agar plate, one XLD agar plate and one chromogenic medium plate, respectively. The inoculated bacteria were cultured at 36 ℃ for 48 and 24 h, respectively. Typical and suspicious colonies were picked for a full set of biochemical identification. Finally, serological identification was performed[2].
mini VIDAS detection procedures
A 25 g of sample was weighed in 225 ml BPW. After homogenization, the mixture was incubated at 37 ℃ for 20 h to enrich bacteria. Then, 0.1 ml of the above bacterial liquid was inoculated into 10 ml RVS and incubated at 41.5 ℃ for 8 h for secondary enrichment. Next, 1 ml of the RVS bacterial liquid was transferred into 10 ml of M broth, followed by incubation at 41 ℃ for 20 h for secondary enrichment. Inactivation was performed on 1 ml of the M broth in a 100 ℃ water bath for 15 min. Finally, 500 μl of the inactivated enriched liquid was loaded on mini VIDAS for detection according to the instructions, which was completed 45 min later[3].
Results and Analysis
Test results
GB 4789.4-2016 method: For blind sample A, blind sample B and beef flavor samples, the BS plate, XLD plate and color plate showed typical colonies; the triple sugar iron agar slant had sugar production, gas production and hydrogen sulfide production; the lysine decarboxylase test was positive; the sorbitol test was positive; and the mannitol test was positive; the urease test was positive; the indigo substrate test, potassium cyanide test and ONPG test were all negative; the physiological saline test showed no self-coagulation; the AF polyvalent "O" slide agglutination test was positive and the multivalent flagellar antigen (H) slide agglutination test was positive. The final result was that Salmonella was detected. As to the ham sausage sample, the colonies of the BS plate, XLD plate and color plate were not typical, no biochemical identification was carried out, and the final result was determined that no Salmonella was detected[4].
The output of the mini VIDAS method instrument was positive, that is, Salmonella was detected (Table 1).
The tests carried out simultaneously using standard Salmonella strain showed that both GB 4789.4-2016 method and mini VIDAS method detected Salmonella.
The tests carried out simultaneously using E. coli standard strain showed that both GB 4789.4-2016 and mini VIDAS did not detect Salmonella.
The two methods were compared as follows:
Accuracy: Both GB 4789.4-2016 and mini VIDAS detected Salmonella, but mini VIDAS had higher sensitivity and stronger specificity.
Time consumption: The national standard method took about 120 h. mini VIDAS only needed to undergo sterilization-sample processing (100 ℃ 15 min)-sample loading, and the time consumption was about 49 h.
Detection procedures: The GB method underwent pre-enrichment-selective enrichment-selective separation-biochemical test-serological test process and was cumbersome. The mini VIDAS method needed the steps of enrichment-water bath treatment (100 ℃ 15 min)-sample loading, and thus had a simple process.
Experimental analysis
At present, GB 4789.4-2016 mainly relies on biochemical and serological reactions to test and identify Salmonella, which not only has a long testing period, but also has complicated biochemical test procedures and needs many reagents. mini VIDAS uses ELISA (enzyme linked fluorescence immunoassay) technology, in which the detection of antigens (bacteria, viruses) of the test substance uses a sandwich technology (i.e., antibody-antigen-antibody), and the solid-phase container SPR is a disposable device similar to a sampling head, used as a solid phase and a sampler. SPR has been coated with antibodies during manufacture, and the reagents required for the test are sealed on the reagent strip. Each reagent strip has 10 wells, of which the 1st well is a specimen well, and the 2nd to 9th wells are reagent wells, among which well 6 contains antibody-alkaline phosphatase and well 10 contains fluorescent substrate (4-methyl-coumarin-phosphate) which is an optical colorimetric ring used for measurement. The remaining well contains washing liquid. During the operation, 0.5 ml of the inactivated sample inoculum was added to the 1st well, which will be circulated regularly in the SPR. The antigen in the sample binds to the monoclonal antibody coated on the inner wall of the SPR, and the unbound sample is washed away in wells 2 to 5. The antibody-alkaline phosphatase complex in well 6 of the reagent strip circulates inside and outside the SPR and binds to the antigen captured by the inner wall of the SPR, and the unbound alkaline phosphatase complex is washed away in wells 7-9. The fluorescent substrate 4-methyl-coumarin-phosphate is added to the SPR, and the enzyme remaining on the SPR wall decomposes the catalytic substrate into 4-methyl-umbelliferone. The light scanner of VIDAS automatically measures the fluorescence intensity at 450 nm. At the end of the test, the computer automatically analyzes the results, obtains the test values, and prints out the result report for each sample. The fluorescence value measured by the instrument is proportional to the antigen in the sample, and when the ratio of the sample fluorescence to the standard fluorescence is less than the standard value, the result is negative, otherwise it is positive. Some detection methods of mini VIDAS have been approved by AOAC, AFNOR and other organizations in the world. In September 2000, E. coli O157:H7 in this method was compiled into the industry standard SN/T 0973-2000 in China. mini VIDAS provides us with a quick and convenient detection method. The tested samples do not need to be separated and purified after being enriched, and can be directly loaded on the machine after inactivation. The test can be finished within 45 or 80 min after being loaded on the machine, and finally the test results are automatically printed. Its advantages are high detection sensitivity, strong specificity, fast speed and high efficiency. This instrument in our laboratory has two test chambers with a total of 12 test positions. Different items can be tested at once, and multiple samples can be tested simultaneously.
Conclusions and Discussion
The above two Salmonella detection methods, the mini VIDAS method has the simplest operation steps, the shortest time consumption, and the highest efficiency, and does not require pure culture and is not prone to miss detection, but the equipment and consumables are relatively expensive. Furthermore, according to the AOAC report of the United States, the detection of Salmonella by mini VIDAS has a certain false positive rate, and the positive result generally needs to be confirmed. The GB 4789.4-2016 method has the most complicated operation steps, the longest time consumption, and the lowest efficiency. However, it is a traditional method for identification of Salmonella, which is widely recognized internationally and is the benchmark method for identifying Salmonella. In case of controversy, this method should be used as the benchmark method. The interfering bacteria added to the blind sample of the ability verification, the colony morphology on the selective plate is almost the same as the target bacteria, which brings us great difficulty in picking the colonies for biochemical identification. How can the target bacteria be selected correctly for biochemical identification? The only answer is to pick more suspicious colonies, rather than missing one. In the GB 4789.4-2016 method, as long as there is a suspicious colony on the selective plate, whether it is Salmonella or not, we have to pick it for identification. When the output of mini VIDAS is negative, we can confidently determine that there is no Salmonella in the sample, and there is no need to do subsequent isolation culture, biochemical identification, and serological identification. When the output of mini VIDAS is positive, the subsequent confirmation test will be targeted. GB 4789.4-2016 has complicated operation steps, longest time consumption and lowest efficiency, while mini VIDAS has simple operation steps, short time consumption and high efficiency. When preparing testing samples for ability verification, it is recommended that GB 4789.4-2016 and mini VIDAS can be used together to ensure success. By such, we can quickly screen a large number of negative samples in a short time to improve work efficiency in daily testing.
References
[1] CHEN YC, NIU CL, LI J, et al. Common bacteria and practical inspection techniques[M]. Beijing: China Science and Technology Press, 2004. (in Chinese)
[2] GB 4789.4-2016 Food microbiology inspection: Salmonella inspection[S]. Beijing: China Standard Press. (in Chinese)
[3] LU Y, CUI SH, YANG MC, et al. Technical specifications for food inspection operations (microbiological inspection)[M]. Beijing: China Medical Science and Technology Press, 2019. (in Chinese)
[4] LI J, LIU Z, HE YL. Graphic manual for quality control of microbial dehydrated medium[M]. Beijing: China Science and Technology Press, 2007. (in Chinese)
- 农业生物技术(英文版)的其它文章
- Investigation on Agronomic Characters of Dwarf Mutant 778 in Broomcorn Millet (Panicum miliaceum L.) and Analysis of Its Sensitivity to GA
- Construction of Technology System on Development and Repropagation in Vitro of Several Cultivars in Pear
- Construction of Camellia oleifera Cultivation Standardization System
- Nutrients Determination in Nuts from Different Torreya grandis Cultivars
- Photosynthetic Physiological Response to Drought Stress of Populus euphratica at Different Ages in Minqin
- Effects of UV-B Radiation on the Activity of Antioxidants in Flue-cured Tobacco Leaves

