Genetic variation within and among range-wide populations of three ecotypes of the Japanese grenadier anchovy Coilia nasus with implications to its conservation and management*
XUE Dongxiu , , YANG Qiaoli , , ZONG Shaobing , GAO Tianxiang , LIU Jinxian ,
1 Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
2 Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China
3 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
4 University of Chinese Academy of Sciences, Beijing 100049, China
5 School of Fisheries, Zhejiang Ocean University, Zhoushan 316022, China
Received Apr. 9, 2019; accepted in principle Jun. 17, 2019; accepted for publication Aug. 15, 2019 © Chinese Society for Oceanology and Limnology, Science Press and Springer-Verlag GmbH Germany, part of Springer Nature 2020
Abstract Studies of range-wide populations can contribute to the comprehension of the relative roles of historical events and contemporary factors that infl uence genetic variation within and among populations. Japanese grenadier anchovy, Coilia nasus, is a commercially important fi sh, which widely distributes in the Changjiang River, the coasts of China and Korea, and the Ariake Sea of Japan. This species exhibits three life-history strategies (anadromous, landlocked and freshwater resident forms). Using two fragments of nuclear DNA, genetic variation within and among 18 populations across the rivers and coast of China and Ariake Sea of Japan was examined. Patterns of genetic diversity and divergence among populations varied widely across C. nasus’ range, and indicated the difference erent efference ects of historical events and anthropogenic factors. Strong genetic divergence between freshwater resident populations and other populations suggested that historical geographical factors greatly infl uenced the genetic structure of C. nasus. Signifi cant genetic difference erentiation observed among lakes in lower Changjiang River and Huaihe River might be probably infl uenced by hydraulic facilities. The population genetic structure among the three ecotypes revealed in the present study indicated an important role for environment variation, and the factors responsible for shaping C. nasus difference erent life history strategies might also impact population structure.
Keyword: Japanese grenadier anchovy; population structure; anthropogenic factors; life-history; nuclear DNA sequences
1 INTRODUCTION
Conservation of life history variations is an important consideration for many species with tradeofference s in migratory characteristics (Nichols et al., 2016). The Japanese grenadier anchovy, Coilia nasus, is widely distributed along the coasts of China, the western coasts of the Korean Peninsula, the Ariake Sea of Japan, and the Changjiang (Yangtze) River and other major rivers in China (Whitehead et al., 1988). Based on the previous morphological and ecological studies of this species, three ecotypes of grenadier anchovy were identifi ed: anadromous, landlocked, and freshwater resident populations (Cheng, 2011). Anadromous grenadier anchovy spends adult life in the ocean and breeds in freshwater, whereas landlocked and freshwater resident grenadier anchovy inhabit only freshwater (Yuan et al., 1980; Cheng, 2011). In the past, the freshwater resident grenadier anchovy used to be named as Coilia brachygnathus, which resides Changjiang River and its adjoining lakes. The jaw length of the freshwater-resident ecotype (shortjaw tapertail anchovy) is shorter than those of the anadromous and landlocked ecotype. The landlocked grenadier anchovy used to be named as Coilia nasus taihuensis, which inhabits the lakes isolated to the Changjiang River and Huaihe River (such as Taihu Lake, Chaohu Lake, and Hongze Lake). The landlocked grenadier anchovy is a typical example of a human-induced alteration of the life history of the anadromous fi shes (Yang et al., 2006). The analysis based on the microchemistry patterns of element strontium and calcium in otoliths also supported that grenadier anchovy individuals in Taihu Lake are landlocked (Yang et al., 2006). There seems to be no evidence of large-scale geological activity that would have blocked spawning migrations or isolated inland lakes (e.g. Taihu Lake, Chaohu Lake, and Hongze Lake) (Xue et al. 2019). However, hydraulic facilities are a ubiquitous feature of Taihu Lake, Chaohu Lake, and Hongze Lake undoubtedly disrupted the migrations of anadromous fi shes, including grenadier anchovies, which migrate up the lakes outlet upon which some hydraulic facilities (e.g. Chaohu Sluice, Yuxi Sluice, Taipu Sluice, Sanhe Sluice, and Erhe Sluice) were constructed since 1950s. As a lucrative commercial fi shery, natural anadromous populations have been declining dramatically in China due to over-exploitation and deterioration of the coastal environment and spawning grounds (Zhang et al., 2005). When population sizes reduced to a level, inbreeding and loss of genetic diversity occur and may result in extinction of local populations (Reed et al., 2003). To protect this species, national aquatic germplasm resources conservation area of China of grenadier anchovy have been built in Changjiang River.
Genetic diversity and population genetic structure are fundamental to the understanding of the evolutionary and ecological process that infl uence biodiversity and provide explicit framework for conservation of species (Frankham et al., 2009). Fisheries management can benefi t from considering population genetic diversity and difference erentiation as well as how biological characteristics and environmental factors infl uence population connectivity. Most of the previous studies on C. nasus were mainly focused on the genetic relationships among the three ecotypes of C. nasus based on mitochondrial DNA markers, and only a small number of populations were analyzed (Tang et al., 2007; Cheng et al., 2008; Liu et al., 2012, 2014; Yang et al., 2017). Also, previous fi ndings on the genetic relationships among difference erent ecotypes were somewhat confl icted (Yang, 2012; Liu et al., 2014; Yang et al., 2017). Yet, little is known about a current genetic background of C. nasus across its distribution range, which may become an impediment to the ongoing fi sheries management and conservation of C. nasus.
In the present study, two anonymous fragments fl anking C. nasus (AGAT)Nmicrosatellite loci were tested for utility as ‘putatively neutral’ markers for inference of genetic variation within and among 17 populations of C. nasus from China and one population from the Ariake Sea in Japan. The aim of this study was to estimate the levels of genetic diversity within each population and to infer population structure among the 18 populations. The results would provide useful information for proper scientifi c and judicious management actions to ensure sustainability of C. nasus populations.
2 MATERIAL AND METHOD
2.1 Sample collection and DNA extraction
Coilia nasus specimens were collected from 18 sites in China and Japan (17 to 24 individuals per site) during 2006-2013 (Table 1). Genomic DNA was extracted from muscle tissue by proteinase K digestion followed by a standard phenol-chloroform method.
2.2 Nuclear DNA sequencing
PCR primers were designed to amplify 361 bp and 264 bp anonymous fragments fl anking C. nasus (AGAT)Nmicrosatellite loci (Locus H1 and Locus H4, CnasH1F: 5′-GTC CGT AGT AAC CGA AAT TGT G, CnasH1R: 5′-TCC CTC GAC TGT CAT ATC ATG T; CnasH4F: 5′-CCA GTC ATG AGT ATG ACC TC, CnasH4R: 5′-CCT CTT TGT ATT ATT GGG AA). The nuclear sequences were amplifi ed for 17-24 individuals per site (Table 1). PCRs were performed in 25 μL volumes followed by Wilson and Veraguth (2010). PCR amplifi cation included 5 min initial denaturation at 94°C, and 35 cycles of 94°C (40 s), 56°C (H1) or 53°C (H4) (50 s) and 72°C (1 min), and a fi nal extension at 72°C for 10 min. PCR products were sequenced on an ABI 3730XL automated sequencer with both forward and reverse primers (Life Technologies Biotechnology Co., China).
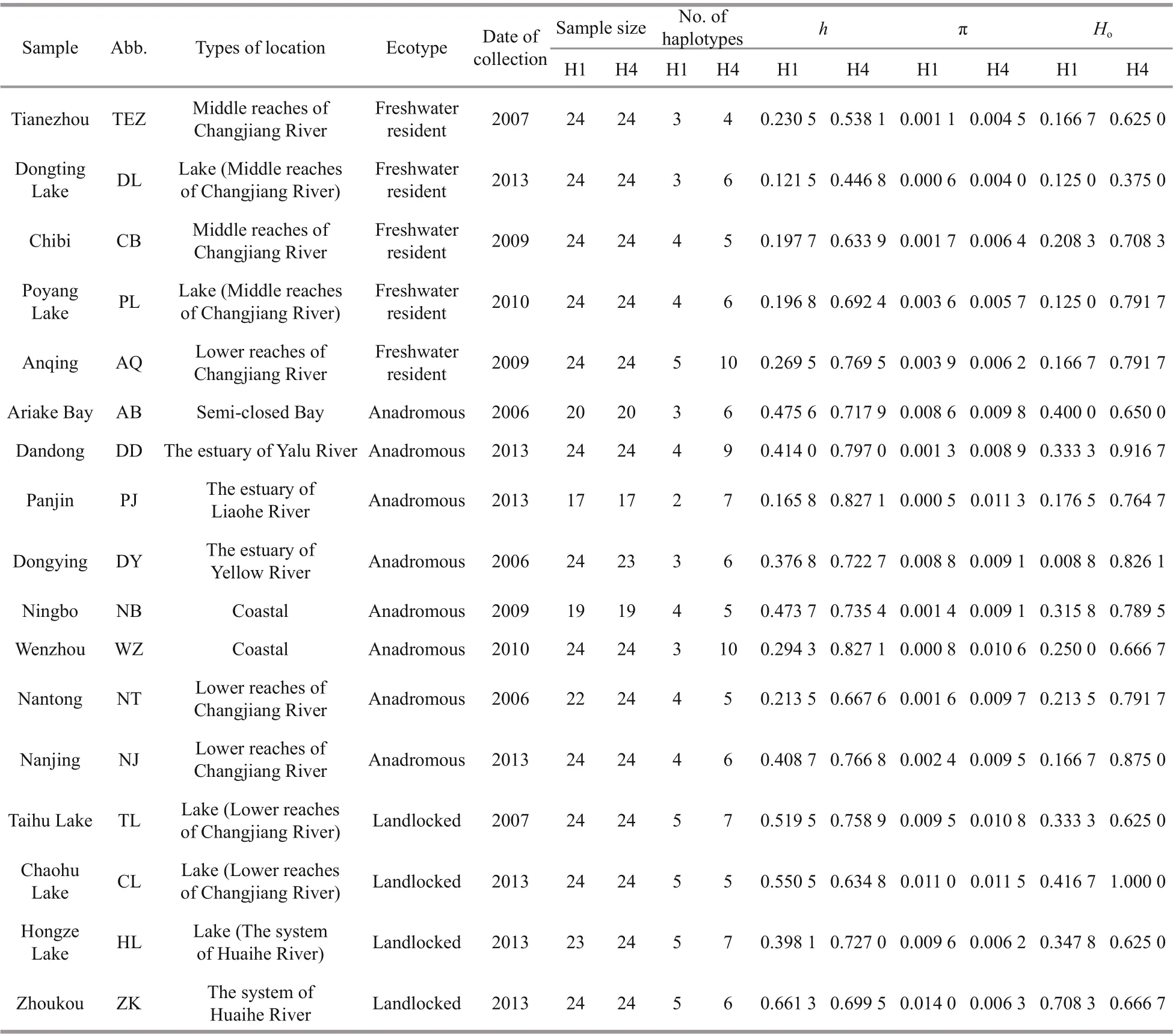
Table 1 Molecular diversity indices for the H1 and H4 nuclear fragments
Allelic sequences were inferred using a two-step procedure (Wilson and Veraguth, 2010). The fi rst step involved the statistical inference of allelic phase using the default settings of PHASE V2.1 (http://stephenslab.uchicago.edu/software.html#phase). This method provides accurate measures in statistically estimated alleles that are well-represented in a diploid data set, but may be uncertain about reconstructing allelic phase for unique genotypes (Harrigan et al., 2008; Wilson and Veraguth, 2010). Individuals for which allelic phase was uncertainty were reamplifi ed and cloned into TA cloning vectors (TaKaRa) following the manufacturer’s protocol. Three clones were picked randomly and sequenced for each individual, allowing the direct determination of allelic identify.
2.3 Statistical analysis
Observed heterozygosity of H1 and H4 alleles was calculated using Arlequin v3.5 (Excoき er and Lischer, 2010). Molecular diversity indices such as haplotype diversity ( h), nucleotide diversity (π), number of polymorphic sites and haplotypes were obtained using the program Arlequin v3.5 (Excoき er and Lischer, 2010).
Phylogenetic relationships among haplotypes were constructed using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). A MrBayes setting for the best fi t model (HKY+I+G) was selected by the hierarchical likelihood ratio tests with MrModeltest 2.3 (Nylander, 2004) in conjunction with PAUP 4.0b10 (Swofference ord, 2002). Markov chains were run for 107generations (the average SD of split frequencies<0.01) with four chains starting from a random tree. Sampling frequency was set at 100 generations and the fi rst 7 500 samples (25%) were excluded as burn-in (Shikano et al., 2010). Genealogical relationships among haplotypes for both the H1 and H4 gene were further assessed using a minimum spanning tree constructed by Arlequin V3.5 (Excoき er and Lischer, 2010).
Population structure was evaluated with AMOVA analyses that incorporate haplotype frequency and sequence divergence among haplotypes (Excoき er et al., 1992). We conducted AMOVA analyses with two groups: the freshwater resident group, anadromous and landlocked group. The signifi cance of covariance component associated with the difference erent possible levels of genetic structure was tested using 10 000 permutations. In addition, the fi xation index ΦSTwas used to compare the genetic divergence values between populations, and statistical signifi cance was evaluated by 10 000 permutations for each pairwise comparison (Excoき er et al., 1992). All the population divergence calculations were performed in Arlequin V3.5 (Excoき er and Lischer, 2010).
3 RESULT
3.1 Genetic diversity
Sequence comparison for the 361 bp of the H1 nuclear fragment revealed 19 distinct haplotypes defi ned by 27 polymorphic sites. A single-base deletion in Locus H1 was common in freshwater resident individuals, but rare in the rest individuals. For H1 fragment, the number of haplotypes ranged from 2 (Panjin) to 5 (Anqing, Taihu Lake, Chaohu Lake, Hongze Lake, and Zhoukou), haplotype diversity ( h) ranged from 0.121 5 (Dongting Lake) to 0.661 3 (Zhoukou), nucleotide diversity (π) ranged from 0.000 6 (Dongting Lake) to 0.014 0 (Zhoukou), and the observed heterozygosity ( Ho) ranged from 0.125 0 (Dongting Lake and Poyang Lake) to 0.708 3 (Zhoukou) (Table 1). For the 264 bp of the H4 nuclear fragment, 19 distinct haplotypes were defi ned by 15 polymorphic sites. The number of haplotypes ranged from 4 (Tianezhou) to 10 (Anqing and Wenzhou), the value of h ranged from 0.446 8 (Dongting Lake) to 0.827 1 (Panjin and Wenzhou), the value of π varied from 0.004 0 (Dongting Lake) to 0.011 5 (Chaohu Lake), and Horanged from 0.375 0 (Dongting Lake) to 1.000 0 (Chaohu Lake) for H4 fragment (Table 1).
3.2 Population structure
For the H1 fragment, pairwise FSTvalues ranged from -0.021 3 between Ariake Bay and Dongying to 0.980 9 between Dongting Lake and Panjin (Table 2). All of the fi ve freshwater resident populations were signifi cantly difference erent from the rest 13 populations after a Benjamini-Yekutieli correction ( P <0.008 9). However, genetic difference erences among the fi ve freshwater resident populations were non-signifi cant ( P >0.008 9). The genetic divergences were signifi cant between Ariake Bay in Japan and fi ve anadromous populations in China (Dandong, Panjin, Wenzhou, Nantong and Nanjing). The four landlocked populations (Taihu Lake, Chaohu Lake, Hongze Lake and Zhoukou) were signifi cant signifi cantly difference erent from six anadromous populations except for Dongying population. Dongying population was signifi cantly difference erent from fi ve anadromous populations (Dandong, Panjin, Wenzhou, Nantong, and Nanjing) and one landlocked population (Zhoukou).
For the H4 fragment, pairwise FSTvalues ranged from -0.020 3 between Panjin and Wenzhou to 0.543 5 between Chibi and Ningbo (Table 2). All of the fi ve freshwater resident populations were signifi cantly difference erent from the rest 13 populations after a Benjamini-Yekutieli correction ( P <0.008 9) except for Poyang Lake & Hongze Lake, Anqing & Hongze Lake, and Anqing & Zhoukou. The two landlocked populations (Hongze Lake and Anqing) of Huaihe River were signifi cantly divergent with the eight anadromous populations and the other two landlocked populations (Taihu Lake and Chaohu Lake) ( P <0.008 9).
Using the haplotypes of H1 fragment, two distinct lineages (labelled A and B) were revealed by Bayesian inference of phylogeny under the HKY+I model (Fig.1). The lineage A dominated the fi ve freshwater resident populations. This result was further supported by the minimum spanning tree for H1 fragment (Fig.2). The minimum spanning tree for H1 fragment indicated that the two lineages were separated from each other by seven mutational steps (Fig.2). There were strong geographical difference erences in haplotype frequencies of H1 fragment. Three haplotypes (HAP1, HAP4 and HAP5) were common in the fi ve freshwater resident populations. However, the HAP1 was also found in four landlocked populations (Hongze Lake, Zhoukou, Chaohu Lake, and Taihu Lake) and two anadromous populations (Dongying and Ariake Bay). Additionally, three common haplotypes (HAP2, HAP3, and HAP10) were observed in the rest 13 populations from Huaihe River, Lower Changjiang River and coastal populations. HAP2 was also existed at low frequency in two freshwater resident populations (Poyang Lake and Anqing) (Fig.3).
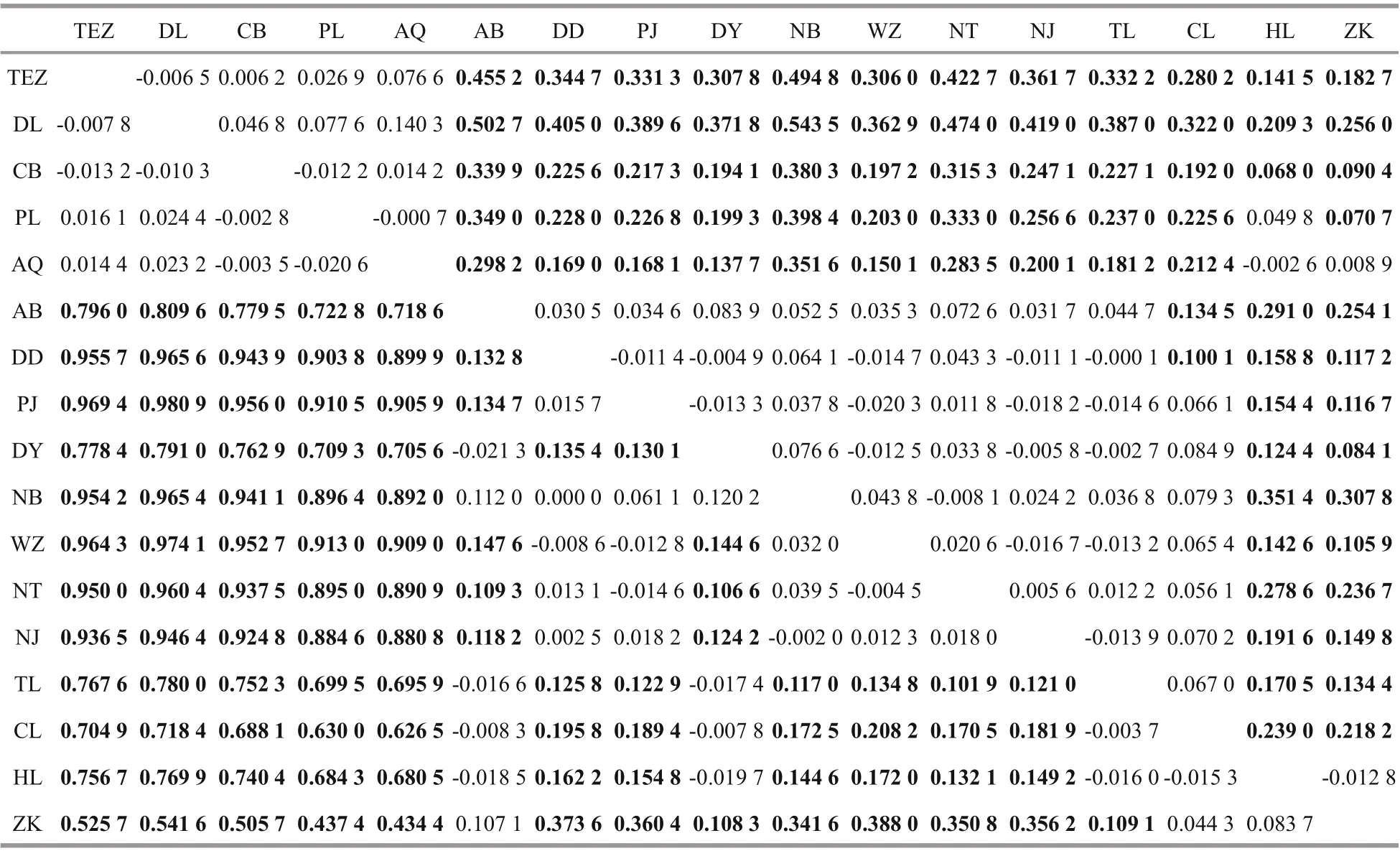
Table 2 Pairwise F ST among C. nasus populations for H1 nuclear fragment (below diagonal) and H4 nuclear fragment (above diagonal)
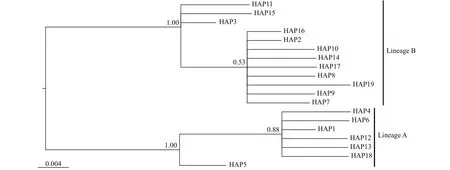
Fig.1 Bayesian tree for H1 haplotypes, as constructed under the HKY+I substitution model with MRBAYES
For the H4 fragment, the phylogenetic tree was obtained by Bayesian analysis under the F81+I model (Fig.4). The minimum spanning tree for this fragment was defi ned by fi ve common haplotypes (HAP1, 2, 4, 6, and 7, Fig.5), one of which (HAP7) was mainly restricted to the fi ve freshwater resident populations, and one landlocked population from Chaohu Lake. HAP6 was found at high frequency in the seven anadromous populations in China and two landlocked population (Taihu Lake and Chaohu Lake) from the Lower Changjiang River (Fig.6). HAP2 was found at high frequency in one freshwater resident population (Anqing), seven anadromous populations in China and three landlocked populations (Taihu Lake, and Hongze Lake, and Zhoukou) (Fig.6).
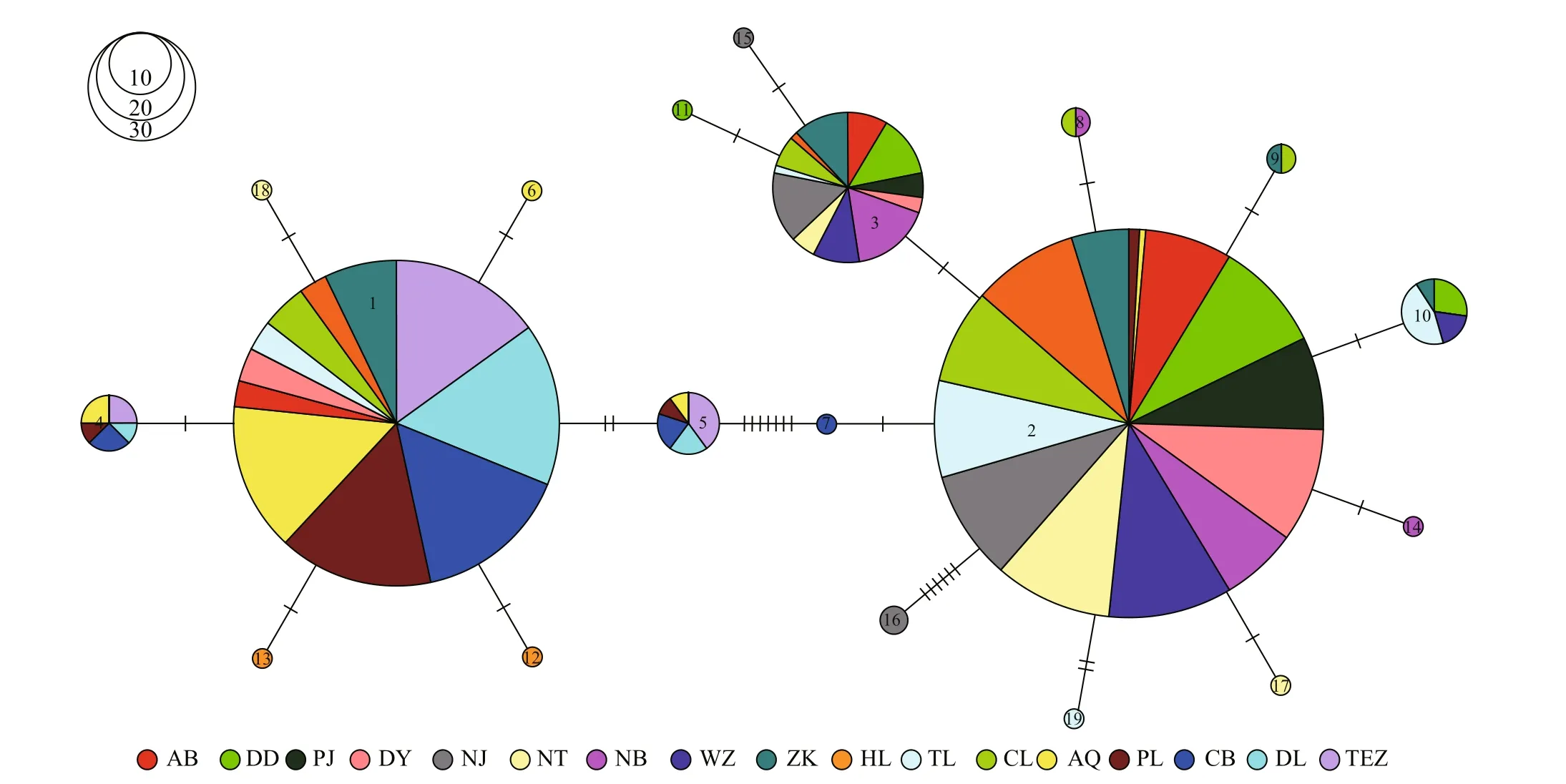
Fig.2 Minimum spanning tree showing genetic relationship among H1 haplotypes
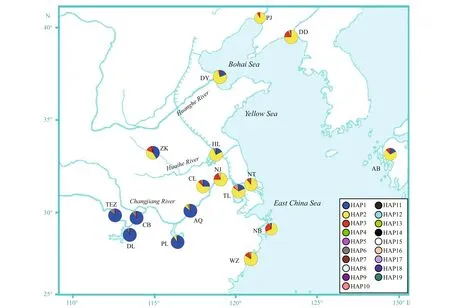
Fig.3 Pie charts show the frequencies of H1 haplotypes among populations of C. nasus
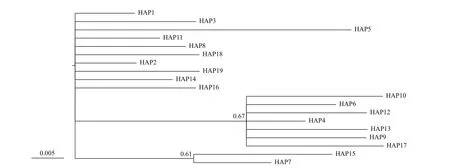
Fig.4 Bayesian tree for H4 haplotypes, as constructed under the F81+I substitution model with MRBAYES
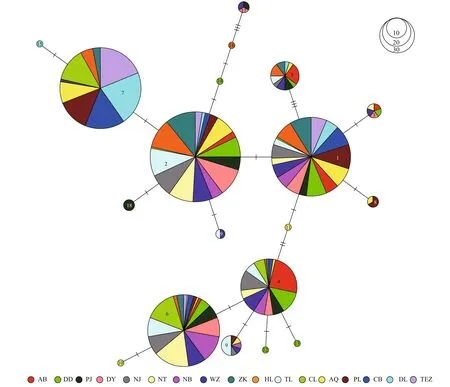
Fig.5 Minimum spanning tree showing genetic relationship among H4 haplotypes
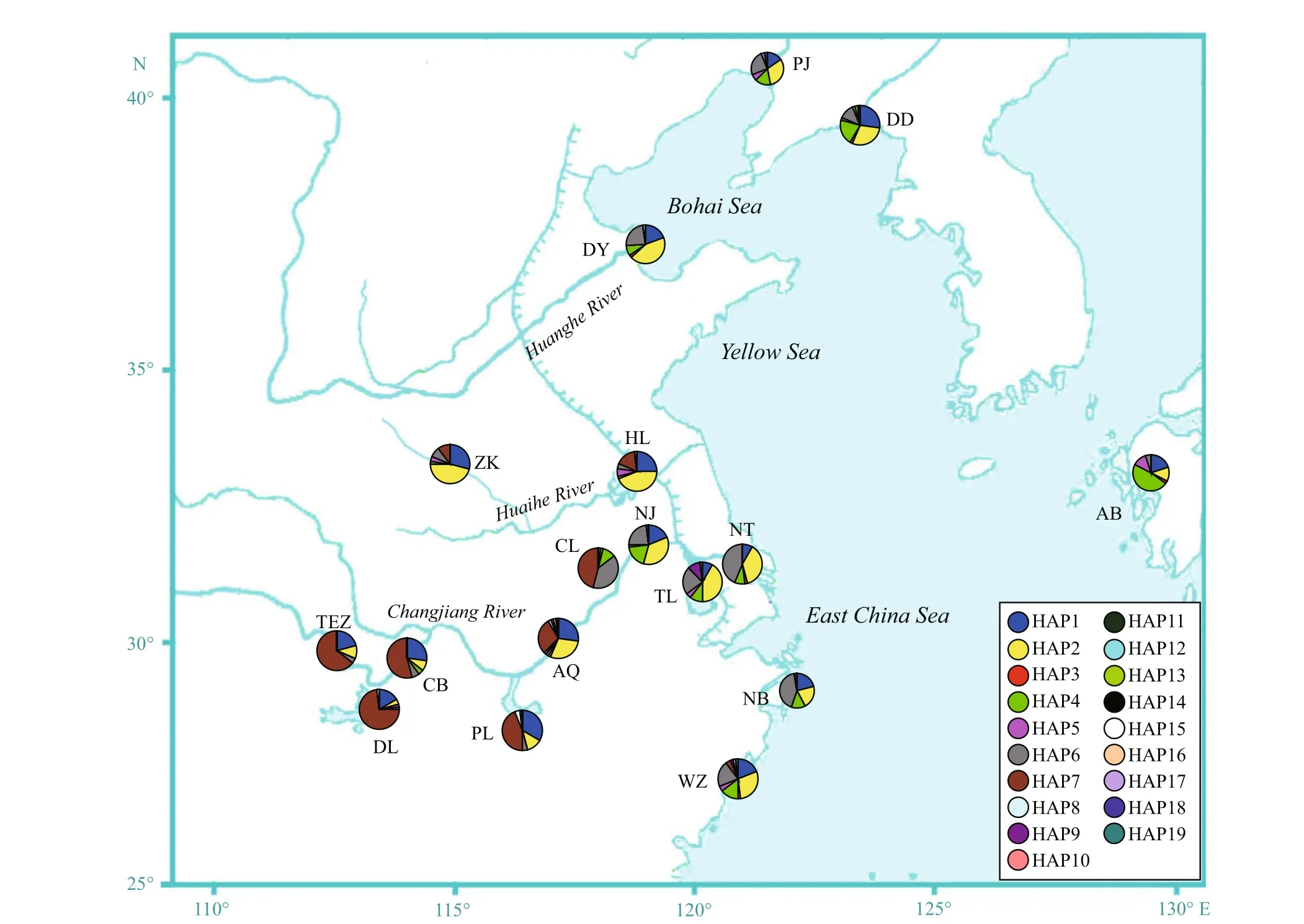
Fig.6 Pie charts show the frequencies of H4 haplotypes among populations of C. nanus
Based on the results of pairwise FSTvalues, Bayesian trees and minimum spanning trees, AMOVA analyses were conducted with two groups: freshwater resident group, and (anadromous & landlocked) group. For the H1 fragment, the AMOVA analysis revealed that most of the genetic variation was distributed between two groups (77.43%, P=0.00). Smaller but signifi cant amount of variances ( P=0.00) were also found within populations (19.77%) and among populations within groups (2.79%). For the H4 fragment, AMOVA analysis revealed that 72.98% of the genetic diversity was found within populations ( P=0.00). Smaller but signifi cant amount of variances ( P=0.00) were also found between two groups (21.03%) and among populations within groups (5.99%).
4 DISCUSSION
The level of genetic diversity of C. nasus was characterized by moderate haplotype diversity (10 of 18 populations has low values based on H1 fragment) and low nucleotide diversity. In addition, the relative low haplotype diversity, nucleotide diversity, and observed heterozygosity based on both H1 and H4 fragments were found in the fi ve freshwater resident populations from Middle Changjiang River. These results were consistent with the conclusions of a previous survey of microsatellite variation in marine, freshwater and anadromous fi shes reported by DeWoody and Avise (2000). Anadromous fi sh populations tend to display higher levels of genetic diversity than do freshwater fi shes, which probably are attributable in part to difference erences in evolutionarily efference ective population sizes and chances for admixture of typifying species inhabiting these realms (DeWoody and Avise, 2000).
The results of AMOVA, pairwise FSTvalues, the Bayesian phylogenetic tree and haplotypes networks revealed high levels of genetic divergence between freshwater resident group and (anadromous & landlocked) group. Additionally, strong allele frequency changes of two nuclear fragments and high pairwise FSTvalues indicated limited genetic exchange between freshwater resident populations and anadromous middle and lower reaches of Changjiang River populations in the absence of contemporary dispersal barriers. Similar genetic divergences between populations in the middle and lower reaches of the Changjiang River have also been reported in some other aquatic organisms (Lu et al., 1997; Feng et al., 2008; Zhao et al., 2008). These concordant patterns reveal that historical geographical factors greatly infl uenced the evolutionary genetic structure of aquatic organisms in the middle and lower reaches of the Changjiang River (Liu et al., 2007). During the last Pleistocene glaciation, the water level of the Changjiang River dropped more than 20 m with the fall of sea levels (Yang, 1986). Some geological researches have proved that the Changjiang River was an inland river and did not fl ow into East China Sea during last glacial maximum for the impacts of arid climate, reduced rainfall and desertifi cation (Xiao et al., 2003). Thus, some populations of C. nasus were probably isolated in main deep-water channel of the Changjiang River in Pleistocene glaciations and became freshwater resident populations eventually (Xue et al., 2019; Yang, 2012).
Besides historical geographical factors, contemporary anthropogenic factors also play an important role in shaping the present-day genetic structure. The landlocked ecotype of C. nasus was formally described in 1976 (Yuan et al., 1976, Yang et al., 2006). In recent years, human activities have made a drastic impact on the Changjiang River and Huaihe River, and many hydraulic facilities have been constructed in Chaohu Lake, Taihu Lake, and Huaihe River basin, including sluices and dams (Yang et al., 2006; Hu et al., 2008). Anadromous C. nasus have become the landlocked individuals when their migration waterway was blocked. Such rapid phenotypic evolution was common in fi shes, such as in guppies, rainbow trout and stickleback (Messer et al., 2016). One prominent example is the life history traits change of marine sticklebacks to landlocked ecotype, which has occurred within the last 50 years after an earthquake created multiple freshwater ponds (Lescak et al., 2015). In the present study, pairwise FSTvalues showed signifi cant genetic difference erentiation (pairwise FSTvalues) observed between the four landlocked populations and the six Chinese anadromous populations except Dongying population based on the H1 fragment. For the H4 fragment, the two landlocked populations (Hongze Lake and Anqing) of Huaihe River were signifi cantly divergent with the eight anadromous populations and the other two landlocked populations (Taihu Lake and Chaohu Lake). However, the haplotypes frequencies of nuclear H1 fragment showed HAP2 dominated the four landlocked populations and the nine anadromous populations. For the H4 fragment, HAP2 and HAP6 dominated the four landlocked populations and the nine anadromous populations. In this case, the genetic difference erentiation between landlocked and anadromous populations could refl ect the recency of the formation of all the landlocked populations and genetic bottleneck.
5 CONCLUSION
In summary, our range-wide examination of genetic variation among populations of C. nasus suggested that the relative infl uence of geographic isolation, anthropogenic factors, biological characteristics, and historical events on contemporary patterns vary spatially. The results of our study detected signifi cant genetic structure of C. nasus, which might arise from current geographic segregation, difference erent life-history strategy, and historical geographical factors. C. nasus freshwater resident populations refl ected a unique genetic cluster and relative lower genetic diversity. Patterns of genetic difference erentiation observed on the landlocked populations (Chaohu Lake, Taihu Lake, Hongze Lake, and Zhoukou) were probably the genetic efference ects of hydraulic facilities in lower Changjiang River and Huaihe River. Populations with demographic independence should be monitored and managed separately as management units (MUs). MUs are the basis for the short-term management of populations and are crucial for monitoring the efference ects of human activity on species diversity (Palsbøll et al., 2007). The most obvious conservation fi nding from our results is the identifi cation of at least three MUs: the freshwater resident populations, the landlocked populations, and the anadromous populations. The results in the present study provide complementary information for conducting genetic stock identifi cation and setting stock specifi c management plans for C. nasus in China. Our study demonstrated the utility of putatively neutral nuclear data to inform conservation in highly exploited species and improve stock-specifi c management in China.
6 DATA AVAILABILITY STATEMENT
Sequence data that support the fi ndings of this study are available from GenBank under the accessions Nos. MK777930-MK777967.
References
Cheng Q Q, Cheng H P, Wang L, Zhong Y, Lu D R. 2008. A preliminary genetic distinctness of four Coilia fi shes (Clupeiformes: Engraulidae) inferred from mitochondrial DNA sequences. Russian Journal of Genetics, 44(3): 339-343.
Cheng W X. 2011. The Research of Some Phenotypic Difference erences from Difference erent Ecotypes of Coilia nasus from Yangtze River. Shanghai Ocean University, Shanghai. (in Chinese with English abstract)
DeWoody J A, Avise J C. 2000. Microsatellite variation in marine, freshwater and anadromous fi shes compared with other animals. Journal of Fish Biology, 56(3): 461-473.
Excoき er L, Lischer H E L. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3): 564-567.
Excoき er L, Smouse P E, Quattro J M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131(2): 479-491.
Feng J B, Sun Y N, Cheng X, Li J L. 2008. Sequence analysis of mitochondrial COI gene of Macrobrachium nipponense from the fi ve largest freshwater lakes in China. Journal of Fisheries of China, 32(4): 517-525. (in Chinese with English)
Frankham R, Ballou J D, Briscoe D A. 2009. Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK. 704p.
Harrigan R J, Mazza M E, Sorenson M D. 2008. Computation vs. cloning: evaluation of two methods for haplotype determination. Molecular Ecology Resources, 8(6): 1 239-1 248.
Hu W W, Wang G X, Deng W, Li S N, 2008. The infl uence of dams on ecohydrological conditions in the Huaihe River basin, China. Ecological Engineering, 33(3-4): 233-241.
Lescak E A, Bassham S L, Catchen J, Gelmond O, Sherbick M L, von Hippel F A, Cresko W A. 2015. Evolution of stickleback in 50 years on earthquake-uplifted islands. P roceedings of the National Academy of Sciences of the United States of America, 112(52): E7 204-E7 212.
Liu D, Guo H Y, Tang W Q, Yang J Q. 2012. Comparative evolution of S7 intron 1 and ribosomal internal transcribed spacer in Coilia nasus (Clupeiformes: Engraulidae). International Journal of Molecular Sciences, 13(3): 3 085-3 100.
Liu D, Li Y Y, Tang W Q, Yang J Q, Guo H Y, Zhu G L, Li H H. 2014. Population structure of Coilia nasus in the Yangtze River revealed by insertion of short interspersed elements. Biochemical Systematics and Ecology, 54: 103-112.
Liu J X, Gao T X, Wu S F, Zhang Y P. 2007. Pleistocene isolation in the Northwestern Pacifi c marginal seas and limited dispersal in a marine fi sh, Chelon haematocheilus (Temminck & Schlegel, 1845). Molecular Ecology, 16(2): 275-288.
Lu G Q, Li S F, Bernatchez L. 1997. Mitochondrial DNA diversity, population structure, and conservation genetics of four native carps within the Yangtze River, China. Canadian Journal of Fisheries and Aquatic Sciences, 54 (1): 47-58.
Messer P W, Ellner S P, Hairston N G. 2016. Can population genetics adapt to rapid evolution? Trends in Genetics, 32(7): 408-418.
Nichols K M, Kozfkay C C, Narum S R. 2016. Genomic signatures among Oncorhynchus nerka ecotypes to inform conservation and management of endangered Sockeye Salmon. Evolutionary Applications, 9(10): 1 285-1 300.
Nylander J A A. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
Palsbøll J P, Bérubé M, Allendorf F W. 2007. Identifi cation of management units using population genetic data. Trends in Ecology & Evolution, 22(1): 11-16.
Reed D H, Lowe E H, Briscoe D A, Frankham, R. 2003. Inbreeding and extinction: Efference ects of rate of inbreeding. Conservation Genetics, 4(3): 405-410.
Ronquist F, Huelsenbeck J P. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1 572-1 574.
Shikano T, Shimada Y, Herczeg G, Merilä J. 2010. History vs. habitat type: explaining the genetic structure of European nine-spined stickleback ( Pungitius pungitius) populations. Molecular Ecology, 19(6): 1 147-1 161.
Swofference ord D L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0b10. Sinauer Associates, Sunderland, MA.
Tang W Q, Hu X L, Yang J Q. 2007. Species validities of Coilia brachygnathus and C. nasus taihuensis based on sequence variations of complete mtDNA control region. Biodiversity Science, 15(3): 224-231. (in Chinese with English)
Whitehead P J P, Nelson G J, Wongratana T. 1988. FAO species catalogue. Clupeoid fi shes of the world (Suborder Clupeoidei). An annotated and illustrated catalogue of the herrings, sardines, pilchards, sprats, shads, anchovies and wolf-herrings. Part 2: Engraulididae. FAO Fisheries Synopsis, 125(7): 305-579.
Wilson A B, Veraguth I E. 2010. The impact of Pleistocene glaciation across the range of a widespread European coastal species. Molecular Ecology, 19(20): 4 535-4 553.
Xiao S B, Huang P, Wan S M, Li A C. 2003. Entering sea history of the Yangtze River during the last pleniglacial stage. Journal of the University of Petroleum, China, 27(6): 125-130. (in Chinese with English abstract)
Xue D X, Yang Q L, Li Y L, Zong S B, Gao T X, Liu J X. 2019. Comprehensive assessment of population genetic structure of the overexploited Japanese grenadier anchovy ( Coilia nasus): implications for fi sheries management and conservation. Fisheries Research, 213: 113-120.
Yang D Y. 1986. The paleoenvironment of the mid-lower regions of Changjiang in the full-glacial period of late Pleistocene. Acta Geographica Sinica, 53(4): 302-310. (in Chinese with English abstract)
Yang J Q, Hsu K C, Zhou X D, Kuo P H, Lin H D, Liu D, Bao B L, Tang W Q. 2017. New insights on geographical/ecological populations within Coilia nasus (Clupeiformes: Engraulidae) based on mitochondrial DNA and microsatellites. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 29(1): 158-164.
Yang J, Arai, T, Liu H, Miyazaki N, Tsukamoto K. 2006. Reconstructing habitat use of Coilia mystus and Coilia ectenes of the Yangtze River estuary, and of Coilia ectenes of Taihu Lake, based on otolith strontium and calcium. Journal of Fish Biology, 69(4): 1 120-1 135.
Yang Q L. 2012. Phylogenetic Analysis of Genus Coilia in China and Molecular Phylogeography of C. nasus and C. mystus. Ocean University of China, Qingdao. (in Chinese with English abstract)
Yuan C M, Lin J B, Qin A L, Liu R H. 1976. Historical and present taxonomic status about the genus Coilia in China. Journal of Nanjing University, (2): 1-12. (in Chinese with English abstract)
Yuan C M, Qin A L, Liu R H, Lin J B. 1980. On the classifi cation of the anchovies, Coilia, from the lower Yangtze River and the southeast coast of China. Journal of Nanjing University, (3): 67-77. (in Chinese with English abstract)
Zhang M Y, Xu D P, Liu K, Shi W G. 2005. Studies on biological characteristics and change of resource of Coilia nasus Schlegel in the lower reaches of the Yangtze River. Resources and Environment in the Yangtze Basin, 14(6): 694-698. (in Chinese with English abstract)
Zhao L, Zhang J, Liu Z J, Funk S M, Wei F W, Xu M Q, Li M. 2008. Complex population genetic and demographic history of the Salangid, Neosalanx taihuensis, based on cytochrome b sequences. BMC Evolutionary Biology, 8(1): 201.
 Journal of Oceanology and Limnology2020年3期
Journal of Oceanology and Limnology2020年3期
- Journal of Oceanology and Limnology的其它文章
- List of the Most Outstanding Papers Published by CJOL/JOL in 2017-2018
- Efference ects of dietary cottonseed meal protein hydrolysate on growth, antioxidants and immunity of Chinese mitten crab Eriocheir Sinensis*
- bHLH genes polymorphisms and their association with growth traits in the Pacifi c oyster Crassostrea gigas*
- Complete mitochondrial genomes of two deep-sea pandalid shrimps, Heterocarpus ensifer and Bitias brevis: insights into the phylogenetic position of Pandalidae (Decapoda: Caridea)*
- Unraveling enhanced membrane lipid biosynthesis in Chlamydomonas reinhardtii starchless mutant sta6 by using an electrospray ionization mass spectrometry-based lipidomics method*
- Behavioral responses to ocean acidifi cation in marine invertebrates: new insights and future directions*
