叶绿体基因组在药用植物鉴定及系统进化研究中的应用
姜汶君 郭梦月 庞晓慧
摘要 叶绿体是细胞内具有自主遗传信息的重要细胞器,近年来叶绿体基因组已发展成为近缘物种分子标记、揭示植物进化和系统发育关系的有力工具。药用植物是人类赖以生存和发展的自然资源,叶绿体基因组在药用植物研究中的应用价值受到广泛关注。在总结叶绿体基因组结构研究的基础上,现就叶绿体基因组在药用植物物种鉴定、系统进化等方面的应用进行综述,以期为药用植物种质资源的开发和利用提供参考。
关键词 药用植物;叶绿体基因组;物种鉴定;DNA条形码;高变异区;系统进化;分歧时间;基因工程
Application of Chloroplast Genome in Identification and Phylogenetic Analysis of Medicinal Plants
JIANG Wenjun, GUO Mengyue, PANG Xiaohui
(Key Lab of Chinese Medicine Resources Conservation, State Administration of Traditional Chinese Medicine of the People′s Republic of China, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100193, China; Engineering Research Center of Chinese Medicine Resources, Ministry of Education, Beijing 100193, China)
Abstract Chloroplast is an important organelle with autonomous genetic information in the cell.In recent years, the chloroplast genome has been used as a powerful tool for molecular markers of related species and uncovering plant evolution and phylogeny.Medicinal plants are natural resources that human beings depend on for survival and development, and the application value of chloroplast genomes in the research of medicinal plants has been widely concerned.The paper reviewed the application of the chloroplast genome in species identification and phylogenetic evolution of medicinal plants based on summarizing the research of chloroplast genome structure, providing a reference for the development and utilization of germplasm resources of medicinal plants.
Keywords Medicinal plants; Chloroplast genome; Highly divergent regions; Species identification; DNA barcode; Phylogenetic analysis; Divergence times; Genetic engineering
中圖分类号:R284文献标识码:Adoi:10.3969/j.issn.1673-7202.2020.05.008
叶绿体(Chloroplast)被普遍认为起源于内共生的蓝藻[1],不仅是细胞内具有自主遗传信息的半自主细胞器,还是植物进行光合作用和生物合成的关键场所。近年来随着新一代测序技术的迅速发展和测序成本的下降,已发表的叶绿体基因组数量急剧增加。截至2019年12月,GenBank共收录超过3 300条叶绿体基因组序列,距离烟草和地钱叶绿体基因组(Chloroplast Genome)的首次成功测序仅有短短30余年[2-3]。叶绿体基因组包含丰富的遗传信息,以结构、大小和基因种类均较保守而著称[4],被广泛应用于物种鉴定及系统进化研究,对于筛选药用植物密切相关物种的分子标记以及破译亲缘关系较近植物类群间的系统发育关系具有重要贡献。本文对近年来叶绿体基因组的结构研究以及其在药用植物鉴定及系统进化研究中的应用进行了综述,分析了研究中存在的问题,同时对叶绿体基因组研究的发展方向提出展望。
1 叶绿体基因组结构研究
得益于高通量测序技术和生物信息技术的发展,越来越多物种的叶绿体基因组被成功测序和组装,研究表明大多数陆生植物的叶绿体基因组基本上是被2个反向重复序列(Inverted Repeat Sequence,IR)分成大单拷贝区(Large Single Copy,LSC)和小单拷贝区(Small Single Copy,SSC)的环状四分体结构[4]。伴随着IR区的扩展和收缩,叶绿体基因组大小通常为120~160 kb[4-5]。叶绿体基因组共编码120~130个基因,主要参与光合作用的光反应、转录和翻译等相关功能[5-6]。尽管短片段倒位(Small Inversion)、插入/缺失(Insertion/Deletion or Indel)、基因组结构重排、基因转移与丢失等叶绿体结构变异事件已被报道[7],但叶绿体基因组编码基因种类、排列顺序和含量通常被认为是保守的。
蛋白编码基因和tRNA的数量非常相似,基因组大小的差异主要来源于IR区基因数量的不同[5]。Guo等发现与其他被子植物相比,五味子ycf15基因丢失,IR区明显收缩10 kb,但IR区收缩机制尚不明确,收缩区的GC含量明显低于IR区,推测IR区收缩可能与GC含量相關[8];与2种其他伞形科药用植物相比,滨海前胡的IR区边界明显扩增,并且具有一个625 bp的含有trnD-GUC、trnY-GUA、trnE-UUC基因的短片段倒位[9];此外也有部分物种丢失了IR区,例如豆科植物黄耆只有一个反向重复序列[10],属于蝶形花亚科中的反向重复序列缺失分支(Inverted Repeat-Lacking Clade)。大多数寄生植物的叶绿体基因组中存在基因丢失现象[11],这与其光合作用能力的部分或全部退化、从寄主上吸收养分和水分的生活方式有关。例如广寄生和桑寄生,与烟草相比其叶绿体基因组丢失了ndh基因、infA基因、3个rpl基因(rpl32、rps15和rps16)、7个tRNA基因(trnA-UGC、trnG-UCC、trnH-GUG、trnL-GAU、trnK-UUU、trnL-UAA和trnV-UAC)和4个ycf基因(ycf1、ycf5、ycf9和ycf10)[12],研究表明ndh基因在从自养型向异养型转化过程中最先丢失[13]。其他自养型物种在进化过程中也存在基因丢失、短片段倒位等结构变异现象,其中infA基因的丢失在被子植物中最为普遍[14]。黄耆叶绿体基因组中存在3个基因(infA、rps16、rpl22)丢失,其在rps16基因和rbcL基因之间有一个50 kb的倒位,这是豆科植物中广泛存在的现象[15],此外还发现其ndhF基因和ycf1基因之间有一个20 kb的倒位[10]。黄花蒿具有一个完全倒置的SSC区(18 267 bp),这一现象在其他菊科蒿属植物中也曾被报道,此外rps19基因在LSC区而非IR区,rps19假基因在IR区缺失[16]。菊科橐吾属药用植物叶绿体基因组的LSC区具有一个约3.4 kb的小倒位嵌套在23 kb的大倒位中,这也是菊科植物的一个独特特征[17]。He等报道忍冬叶绿体基因组在trnI-CAU和trnN-GUU之间有一个独特的重排现象,ycf1假基因丢失,rps19、rpl2和rpl23基因从IR区移动到LSC区,并首次发现ycf2和rps18基因中含有内含子[18]。
2 药用植物叶绿体基因组的应用
2.1 物种鉴定
近年来,DNA条形码(DNA Barcoding)已发展成为物种鉴定的强有力工具。在植物物种鉴定中,常用的DNA片段包括叶绿体基因组序列rbcL、matK、psbA-trnH等与核基因组的ITS和ITS2[19]。其中,鉴于ITS2序列较强的鉴定能力,已被建议作为药用植物的通用DNA条形码[20]。然而,对于一些特殊药用植物类群,普通DNA条形码不具备足够的变异信息以实现物种鉴定,例如淫羊藿属和贝母属[21-22]。与普通的DNA条形码短片段相比,叶绿体全基因组包含更丰富的变异位点,鉴定效率更高。叶绿体全基因组一方面被直接用作物种鉴定的超级条形码(Super Barcode),另一方面则基于其筛选出高变区作为鉴定物种的潜在分子标记[23-25]。
2.1.1 超级条形码 Cui等比对分析了砂仁、缩砂仁、海南砂仁等32种豆蔻属叶绿体基因组,结果表明叶绿体全基因组可准确鉴定豆蔻属物种[26]。以往研究表明ITS、matK、psbA-trnH和rbcL对橐吾属的物种鉴定效率不理想[27],Chen等报道了6种橐吾属植物的叶绿体基因组并提出以全基因组作为超级条形码可有效鉴定橐吾属植物[17]。钟志敏发现基于叶绿体全基因组不仅可以有效鉴定石斛属41个物种,还可区分来自3个不同产地的铁皮石斛[28]。Xia等的研究表明了基于叶绿体全基因组可将野菊和近缘物种准确区分开,可作为鉴定这些物种的超级条形码[29]。
2.1.2 高变区筛选 李雪佩等比较分析了黄草乌和展毛黄草乌的叶绿体全基因组,发现trnR-atpA和atpF-atpH基因间隔区上的缺失可作为分子标记鉴定2个物种[30]。叶绿体InDel分子标记也被证明是鉴定白首乌[31]、6种忍冬属植物[32]和人参属[33]的有效工具。马双姣基于测定的薯蓣和叉蕊薯蓣以及从GenBank下载的4条薯蓣属植物叶绿体基因组比对分析,筛选出了5个蛋白编码基因和5个基因间隔区作为潜在分子标记[34]。Chen等通过对川贝母及其近缘物种的8条叶绿体全基因组进行比对分析,筛选出6个高变区作为鉴定贝母属植物的潜在DNA条形码,并提出叶绿体全基因组作为超级条形码可能更适合该属的物种鉴定和系统发育分析[35]。杨俏俏等通过比对分析6种葱属植物的叶绿体基因组,共筛选到7个变异较大的候选DNA条形码序列用于鉴定葱属植物[36]。李晓娟等发现华重楼叶绿体基因组中cemA基因是假基因,该基因编码序列中的终止密码子位置差异可作为区分华重楼和北重楼的依据[37]。Do等基于黄瓜假还阳参的叶绿体基因组开发了新的来自于rbcL和matK基因的SNP分子标记,可将其与相关物种进行有效区分[38]。Chen等基于3个麻黄属药用物种的叶绿体基因组筛选出11个高变区,可作为鉴定麻黄属植物的潜在DNA条形码[39]。Zhou等基于掌叶大黄、药用大黄、唐古特大黄的叶绿体基因组开发了包含鉴定位点的潜在DNA条形码并成功验证,为超级DNA条形码在大黄中的应用奠定了基础[40]。
2.2 系统进化
叶绿体基因组在大多数被子植物中为母系遗传,重组率低、核苷酸置换率适中[41],具有良好的系统发育重建潜力。叶绿体基因组的遗传信息在解决系统发育位置和不同物种间的亲缘关系方面做出了重要贡献,同时也被广泛用于揭示物种起源和估计各谱系间的分歧时间。
2.2.1 系统发育位置 魏似婕等基于藏药“巴莴色保”的基原植物獐牙菜属轮叶獐牙菜、川西獐牙菜及6种龙胆属植物叶绿体全基因组构建ML树,结果支持经典分类学观点对2个属物种的分类[42]。沈立群基于实验获得的薄荷、白毛夏枯草和活血丹3种唇形科药用植物的葉绿体基因组及从NCBI下载的唇形科序列构建了系统发育树,3种植物在进化树中的位置均被支持且与传统分类学相符[43]。张慧等基于ML系统进化分析确定了益母草在野芝麻亚科中的进化位置,发现益母草属和水苏属的亲缘关系较近[44]。在Chen等的研究中,基于对川贝母及其密切相关物种的系统发育分析显示,8个贝母属植物聚成3支且均有较强的支持率,川贝母与甘肃贝母、中华贝母密切相关[35]。Chen等报道基于叶绿体基因组构建的ML和MP系统发育树可以鉴定3种麻黄属植物,同时发现麻黄属植物与买麻藤属植物、百岁兰具有更近的系统发育关系[39]。Cui等基于已发表姜目物种的叶绿体基因组序列,探讨了姜在姜目中的系统发育位置,结果表明姜属为山柰属的姊妹分支[45]。Gao等分析了益智的叶绿体基因组并构建了姜科的系统发育树,结果强烈支持姜科植物喜阴和喜阳的特征分类[46]。Gichira等对非洲一种隶属于蔷薇科的药用植物Hagenia abyssinica的叶绿体基因组进行了测定并构建了系统发育树,结果支持将该属分在蔷薇亚科下[47]。Guo等首次报道了五味子的叶绿体基因组序列,通过构建ML和BI系统发育树确定了五味子属与八角属形成姊妹群[8]。Gu等测定了药用植物黄薇的叶绿体基因组,基于12种桃金娘目物种共有的68个CDS编码基因构建了系统发育树,结果表明其与紫薇属物种的亲缘关系更近[48]。Li等基于广寄生、桑寄生等10个物种的54个蛋白编码基因和matK基因构建的NJ和ML系统发育树拓扑结构基本一致,结果有力支持桑寄生科和槲寄生科为独立的2个单系类群[12]。
2.2.2 分歧时间估计 Lee等以Daucus carata和人参为标定点,基于14个伞形科物种的76个叶绿体基因组共有蛋白编码基因构建贝叶斯进化树,结果表明防风和滨海前胡的分歧时间大约在2.3~3.4百万年前(Million Years Ago,Mya),而珊瑚菜与紫花前胡、大叶山芹的分歧时间大约在1.6~2.8 Mya[9]。He等基于8种唇形科植物及外类群的叶绿体基因组构建系统发育树,采用分子钟模型估算广藿香分歧时间,结果表明其大约在29.45 Mya从黄芩亚科分支分化[49]。采用最大似自然法对21个毛茛目物种和2个木兰科物种的叶绿体基因组进行了系统发育推断分析,He等发现黄连与其他毛茛科物种的分歧时间约为81 Mya,而毛茛科和小檗科的分歧时间约为111 Mya,在侏罗纪时期(大约153 Mya)两者有一个共同的祖先[50]。Kim等基于叶绿体基因组比较分析了五加科的系统发育关系,树参属与五加属的分歧时间大约在4.48~5.60 Mya,人参属与楤木属的分歧时间大约在2.58~3.20 Mya,人参和西洋参的分歧时间大约在0.72~0.87 Mya[51]。Fan等基于叶绿体基因组的蛋白编码序列计算推断了14个川续断目物种的分歧时间,五福花科与忍冬科的分歧时间大约为81.14 Mya,荚蒾属与华福花属、五福花属、四福花属3个属的分歧时间大约为47.16 Mya,华福花属、五福花属、四福花属的分歧时间大约在15~16 Mya[52]。
3 小结与展望
新一代测序技术的出现和进步是叶绿体基因组学和遗传学领域快速发展的重要因素之一,众多植物叶绿体全基因组序列的研究和报道,不仅增强了我们对于叶绿体结构功能、系统进化的认知,也为叶绿体转化提供了重要信息。叶绿体基因组编码植物参与光合作用和其他必要代谢过程的关键蛋白,以应对干旱、盐、热、光等环境压力及防御入侵病原体[53]。叶绿体转化具有蛋白表达水平高、从多顺反子mRNA中表达多种蛋白的可行性、避免花粉传播造成的基因污染等优点[54]。叶绿体转基因工程在增强药用植物的优良农艺性状或生产高价值的生物医学产品等方面具有巨大潜力。
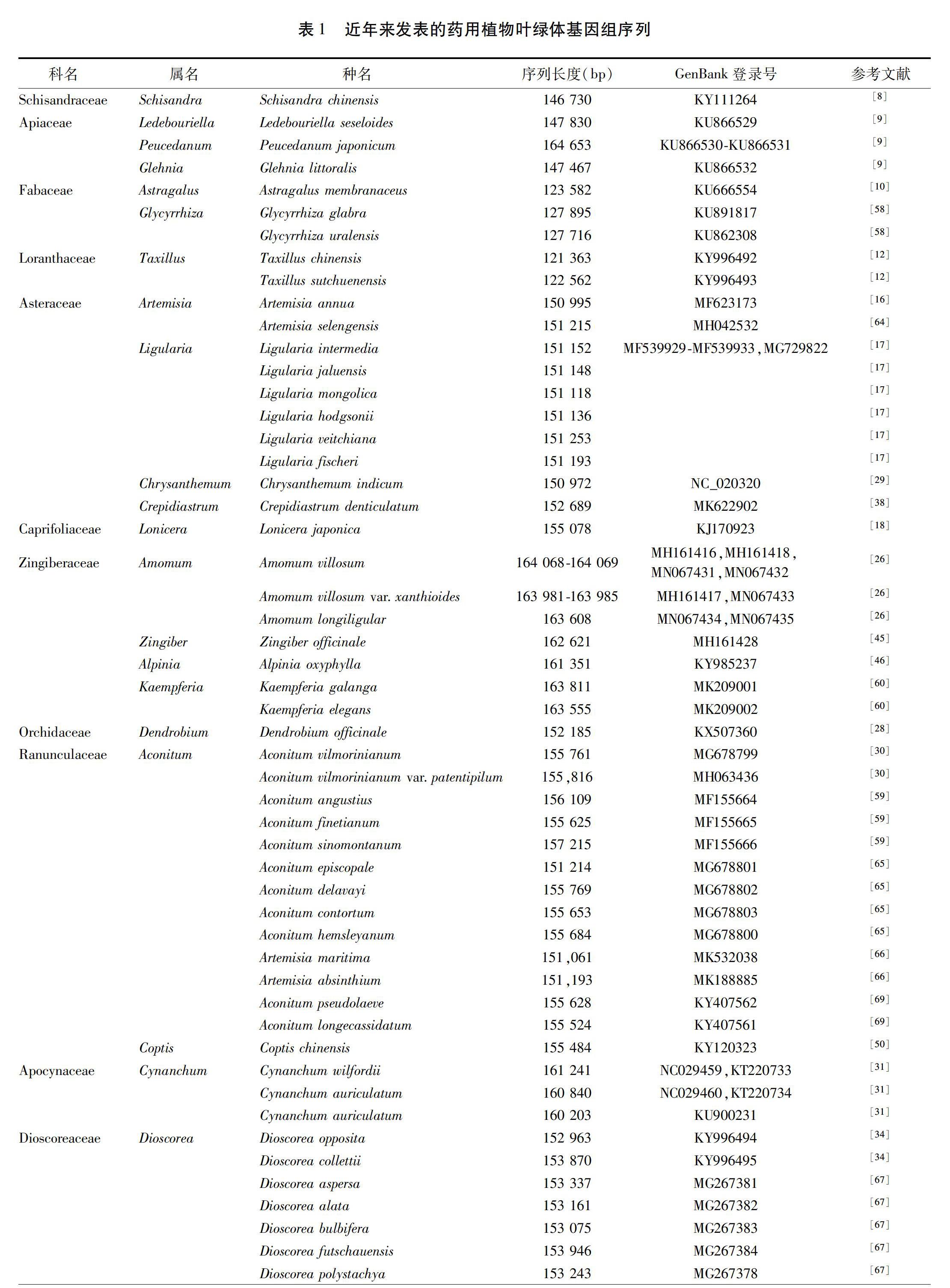
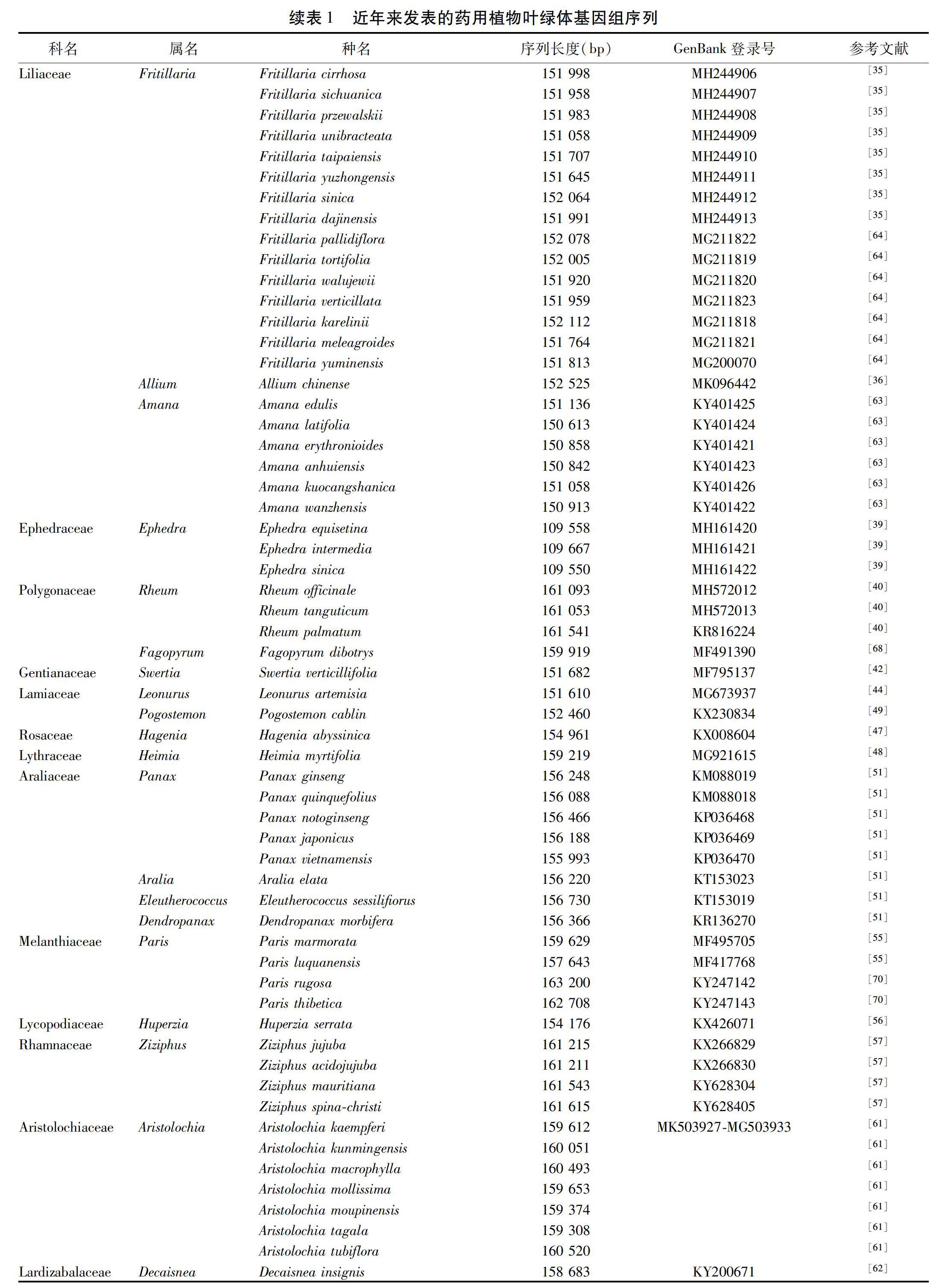
由于叶绿体自身的复制机制和相对独立的进化,现已广泛应用于药用植物的物种鉴定和系统进化研究中。见表1。物种鉴定是生物多样性研究的重要前提和基础,DNA条形码不依赖于传统的形态学鉴定经验,此概念一经提出便受到国内外学者的广泛关注和逐步认可,但目前通用的DNA条形码不足以准确鉴别所有的药用植物物种。随着新一代测序技术和生物信息技术的进步,叶绿体全基因组作为超级条形码或基于叶绿体测序分析筛选候选DNA条形码逐步发展起来,植物叶绿体基因组的数量也在逐年增加。就药用植物而言,目前已发表的叶绿体基因组序列的数量还存在不足。叶绿体基因组目前已显示出了巨大的物种识别潜力,尤其是在亲缘关系较近的类群之间,因此有必要对更多的药用植物进行叶绿体基因组测序,深入研究其鉴定与系统进化问题。
参考文献
[1]Dyall S D,Brown M T,Johnson P J.Ancient invasions:from endosymbionts to organelles[J].Science,2004,304(5668):253-257.
[2]Shinozaki K,Ohme M,Tanaka M,et al.The complete nucleotide sequence of the tobacco chloroplast genome:its gene organization and expression[J].Embo J,1986,5(9):2043-2049.
[3]Ohyama K,Fukuzawa H,Kohchi T,et al.Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA[J].Nature,1986,322(6079):572-574.
[4]Wicke S,Schneeweiss G M,Depamphilis C W,et al.The evolution of the plastid chromosome in land plants:gene content,gene order,gene function[J].Plant Mol Biol,2011,76(3-5):273-297.
[5]Green B R.Chloroplast genomes of photosynthetic eukaryotes[J].Plant J,2011,66(1):34-44.
[6]Tonti-Filippini J,Nevill P G,Dixon K,et al.What can we do with 1000 plastid genomes?[J].Plant J,2017,90(4):808-818.
[7]王玲,董文攀,周世良.被子植物葉绿体基因组的结构变异研究进展[J].西北植物学报,2012,32(6):1282-1288.
[8]Guo H,Liu J,Luo L,et al.Complete chloroplast genome sequences of Schisandra chinensis:genome structure,comparative analysis,and phylogenetic relationship of basal angiosperms[J].Sci China Life Sci,2017,60:1286-1290.
[9]Lee H O,Joh H J,Kim K,et al.Dynamic Chloroplast Genome Rearrangement and DNA Barcoding for Three Apiaceae Species Known as the Medicinal Herb “Bang-Poong”[J].Int J Mol Sci,2019,20(9):2196.
[10]Lei W,Ni D,Wang Y,et al.Intraspecific and heteroplasmic variations,gene losses and inversions in the chloroplast genome of Astragalus membranaceus[J].Sci Rep,2016,6:21669.
[11]Wicke S,Müller K F,de Pamphilis C W,et al.Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family[J].Plant Cell,2013,25(3711):113373.
[12]Li Y,Zhou J,Chen X,et al.Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species[J].Sci Rep,2017,7(1):12834.
[13]Martín M,Sabater B.Plastid ndh genes in plant evolution[J].Plant Physiol Bioch,2010,48(8):636-645.
[14]Millen R S,Olmstead R G,Adams K L,et al.Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus[J].Plant Cell,2001,13(3):645-658.
[15]Doyle J J,Doyle J L,Ballenger J A,et al.The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae[J].Mol Phylogenet Evol,1996,5(2):429-438.
[16]Shen X,Wu M,Liao B,et al.Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua[J].Molecules,2017,22(8):1330.
[17]Chen X,Zhou J,Cui Y,et al.Identification of Ligularia Herbs using the complete chloroplast genome as a super-barcode[J].Front Pharmacol,2018,9:695.
[18]He L,Qian J,Li X,et al.Complete chloroplast genome of medicinal plant Lonicera japonica:Genome rearrangement,intron gain and loss,and implications for phylogenetic studies[J].Molecules,2017,22(2):249.
[19]Hollingsworth P M,Li D Z,van der Bank M,et al.Telling plant species apart with DNA:from barcodes to genomes[J].Philos T R Soc B,2016,371(1702):20150338.
[20]Chen S,Yao H,Han J,et al.Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species[J].PloS one,2010,5(1):e8613.
[21]Guo M,Xu Y,Ren L,et al.A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L.(Berberidaceae)[J].Genes,2018,9(12):637.
[22]罗焜,马培,姚辉,等.基于ITS2序列鉴定川贝母及其混伪品基原植物[J].世界科学技术-中医药现代化,2012,14(1):1153-1158.
[23]Nock C J,Waters D L E,Edwards M A,et al.Chloroplast genome sequences from total DNA for plant identification[J].Plant Biotechnol J,2011,9(3):328-333.
[24]Kane N,Sveinsson S,Dempewolf H,et al.Ultra-barcoding in cacao(Theobroma spp.;Malvaceae)using whole chloroplast genomes and nuclear ribosomal DNA[J].Am J Bot,2012,99(2):320-329.
[25]Li X,Yang Y,Henry R J,et al.Plant DNA barcoding:from gene to genome[J].Biol Rev,2015,90(1):157-166.
[26]Cui Y,Chen X,Nie L,et al.Comparison and Phylogenetic Analysis of Chloroplast Genomes of Three Medicinal and Edible Amomum Species[J].Int J Mol Sci,2019,20(16):4040.
[27]He W Y,Pan Y Z.Study on the DNA barcoding of genus Ligularia Cass.Asteraceae)[J].Plant Divers,2015,37:693-703.
[28]钟志敏.石斛DNA条形码鉴定及系统分类研究[D].广州:广州中医药大学,2018.
[29]Xia Y,Hu Z,Li X,et al.The complete chloroplast genome sequence of Chrysanthemum indicum[J].Mitochondrial DNA A,2016,27(6):4668-4669.
[30]李雪佩,孟静,张琳娜,等.黄草乌与展毛黄草乌叶绿体全基因组结构的比较分析[J].中药材,2018,41(8):1812-1820.
[31]Kim Y,Choi H,Shin J,et al.Molecular Discrimination of Cynanchum wilfordii and Cynanchum auriculatum by InDel Markers of Chloroplast DNA[J].Molecules,2018,23(6):1337.
[32]Kang K B,Kang S J,Kim M S,et al.Chemical and genomic diversity of six Lonicera species occurring in Korea[J].Phytochemistry,2018,155:126-135.
[33]Nguyen V B,Park H S,Lee S C,et al.Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences[J].J Agr Food Chem,2017,65(30):6298-6306.
[34]马双姣.草药分子鉴定:从条形码到基因组[D].北京:北京協和医学院,2017.
[35]Chen Q,Wu X,Zhang D.Phylogenetic analysis of Fritillaria cirrhosa D.Don and its closely related species based on complete chloroplast genomes[J].PeerJ,2019,7:e7480.
[36]杨俏俏,姜梅,王立强,等.药食两用藠头叶绿体基因组解析、比较基因组学及系统发育研究[J].药学学报,2019,54(1):173-181.
[37]李晓娟,杨振艳,黄玉玲,等.药用植物华重楼(黑药花科)叶绿体全基因组研究[J].热带亚热带植物学报,2015,23(6):601-613.
[38]Do H D K,Jung J,Hyun J Y,et al.The newly developed single nucleotide polymorphism(SNP)markers for a potentially medicinal plant,Crepidiastrum denticulatum(Asteraceae),inferred from complete chloroplast genome data[J].Mol Biol Rep,2019,46(3):3287-3297.
[39]Chen X,Cui Y,Nie L,et al.Identification and Phylogenetic Analysis of the Complete Chloroplast Genomes of Three Ephedra Herbs Containing Ephedrine[J].Biomed Res Int,2019,2019.
[40]Zhou Y,Nie J,Xiao L,et al.Comparative Chloroplast Genome Analysis of Rhubarb Botanical Origins and the Development of Specific Identification Markers[J].Molecules,2018,23(11):2811.
[41]Drouin G,Daoud H,Xia J.Relative rates of synonymous substitutions in the mitochondrial,chloroplast and nuclear genomes of seed plants[J].Mol Phylogenetics Evol,2008,49(3):827-831.
[42]魏似婕,趙志礼,倪梁红,等.藏药“巴莴色保”基原鉴定及其叶绿体全基因组序列分析[J].药学学报,2018,53(6):1009-1015.
[43]沈立群.唇形科三种药用植物叶绿体全基因组及科内的比较与进化分析[D].杭州:浙江大学,2018.
[44]张慧,何帅兵,孔繁德,等.益母草叶绿体基因组序列与系统进化位置分析[J].中医药信息,2018,35(4):21-27.
[45]Cui Y,Nie L,Sun W,et al.Comparative and Phylogenetic Analyses of Ginger(Zingiber officinale)in the Family Zingiberaceae Based on the Complete Chloroplast Genome[J].Plants,2019,8(8):283.
[46]Gao B,Yuan L,Tang T,et al.The complete chloroplast genome sequence of Alpinia oxyphylla Miq.and comparison analysis within the Zingiberaceae family[J].PloS one,2019,14(6):e0218817.
[47]Gichira A W,Li Z,Saina J K,et al.The complete chloroplast genome sequence of an endemic monotypic genus Hagenia(Rosaceae):Structural comparative analysis,gene content and microsatellite detection[J].PeerJ,2017,5:e2846.
[48]Gu C,Dong B,Xu L,et al.The complete chloroplast genome of Heimia myrtifolia and comparative analysis within Myrtales[J].Molecules,2018,23(4):846.
[49]He Y,Xiao H,Deng C,et al.The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin[J].Int J Mol Sci,2016,17(6):820.
[50]He Y,Xiao H,Deng C,et al.Complete chloroplast genome sequence of Coptis chinensis Franch.and its evolutionary history[J].Biomed Res Int,2017,2017.
[51]Kim K,Nguyen V B,Dong J,et al.Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species[J].Sci Rep,2017,7(1):4917.
[52]Fan W B,Wu Y,Yang J,et al.Comparative chloroplast genomics of Dipsacales species:insights into sequence variation,adaptive evolution,and phylogenetic relationships[J].Front Plant Sci,2018,9:689.
[53]Liu X,Li Y,Yang H,et al.Chloroplast genome of the folk medicine and vegetable plant Talinum paniculatum(Jacq.)Gaertn.:Gene organization,comparative and phylogenetic analysis[J].Molecules,2018,23(4):857.
[54]Wang H H,Yin W B,Hu Z M.Advances in chloroplast engineering[J].J Genet Genomics,2009,36(7):387-398.
[55]Gao X,Zhang X,Meng H,et al.Comparative chloroplast genomes of Paris Sect.Marmorata:insights into repeat regions and evolutionary implications[J].BMC genomics,2018,19(10):878.
[56]Guo Z Y,Zhang H R,Shrestha N,et al.Complete chloroplast genome of a valuable medicinal plant,Huperzia serrata(Lycopodiaceae),and comparison with its congener[J].Appl Plant Sci,2016,4(11):1600071.
[57]Huang J,Chen R,Li X.Comparative analysis of the complete chloroplast genome of four known Ziziphus species[J].Genes,2017,8(12):340.
[58]Kang S H,Lee J H,Lee H O,et al.Complete chloroplast genome and 45S nrDNA sequences of the medicinal plant species Glycyrrhiza glabra and Glycyrrhiza uralensis[J].Genes Genet Syst,2017:17-00002.
[59]Kong H,Liu W,Yao G,et al.A comparison of chloroplast genome sequences in Aconitum(Ranunculaceae):A traditional herbal medicinal genus[J].PeerJ,2017,5:e4018.
[60]Li D M,Zhao C Y,Liu X F.Complete Chloroplast Genome Sequences of Kaempferia Galanga and Kaempferia Elegans:Molecular Structures and Comparative Analysis[J].Molecules,2019,24(3):474.
[61]Li X,Zuo Y,Zhu X,et al.Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia(Aristolochiaceae)medicinal species[J].Int J Mol Sci,2019,20(5):1045.
[62]Li B,Lin F,Huang P,et al.Complete chloroplast genome sequence of Decaisnea insignis:Genome organization,genomic resources and comparative analysis[J].Sci Rep,2017,7(1):10073.
[63]Li P,Lu R S,Xu W Q,et al.Comparative genomics and phylogenomics of East Asian tulips(Amana,Liliaceae)[J].Front Plant Sci,2017,8:451.
[64]Li Y,Zhang Z,Yang J,et al.Complete chloroplast genome of seven Fritillaria species,variable DNA markers identification and phylogenetic relationships within the genus[J].PloS one,2018,13(3):e0194613.
[64]Meng D,Xiaomei Z,Wenzhen K,et al.Detecting useful genetic markers and reconstructing the phylogeny of an important medicinal resource plant,Artemisia selengensis,based on chloroplast genomics[J].PloS one,2019,14(2):e0211340.
[65]Meng J,Li X,Li H,et al.Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species[J].Molecules,2018,23(5):1015.
[66]Shahzadi I,Mehmood F,Ali Z,et al.Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium:Comparative analyses,mutational hotspots in genus Artemisia and phylogeny in family Asteraceae[J].Genomics,2019,112(2):1454-1463.
[67]Zhao Z,Wang X,Yu Y,et al.Complete chloroplast genome sequences of Dioscorea:Characterization,genomic resources,and phylogenetic analyses[J].PeerJ,2018,6:e6032.
[68]Wang X,Zhou T,Bai G,et al.Complete chloroplast genome sequence of Fagopyrum dibotrys:genome features,comparative analysis and phylogenetic relationships[J].Sci Rep,2018,8(1):12379.
[69]Park I,Yang S,Choi G,et al.The complete chloroplast genome sequences of Aconitum pseudolaeve and Aconitum longecassidatum,and development of molecular markers for distinguishing species in the Aconitum subgenus Lycoctonum[J].Molecules,2017,22(11):2012.
[70]Song Y,Wang S,Ding Y,et al.Chloroplast genomic resource of Paris for species discrimination[J].Sci Rep,2017,7(1):3427.
(2020-02-10收稿 責任编辑:徐颖)

