Development and characterization of novel polymorphic microsatellite markers for the Korean freshwater snail Semisulcospira coreana and cross-species amplif ication using next-generation sequencing*
Yeon Jung PARK , Mi Nan LEE , Eun-Mi KIM , Jung Youn PARK , Jae Koo NOH ,Tae-Jin CHOI , Jung-Ha KANG ,
1 Biotechnology Research Division, National Institute of Fisheries Science, Busan 46083, Republic of Korea
2 Department of Microbiology, Pukyoung National University, Busan 48513, Republic of Korea
Abstract Korean freshwater snails of the genus Semisulcospira are widely distributed across East Asia.It has been a very popular nutritional food in Korea, and is an ecologically important water quality indicator because it lives only in clean water. However, no microsatellite markers have been generated to study the population genetic diversity of this genus. In the present study, we developed and characterized 18 novel microsatellite loci from Semisulcospira coreana genomic DNA. The microsatellites were isolated using 454 GS-FLX titanium sequencing and 18 markers were used for genotyping in S. coreana. In addition, we also tested the cross-species transferability of the microsatellite markers in four additional Semisulcospira spp. We identif ied 18 polymorphic loci and the number of alleles per loci, and their polymorphism information content values ranged from 2 to 17 and 0.203 to 0.902, respectively. The observed and expected heterozygosities of the loci ranged from 0.063 to 0.924 and 0.226 to 0.924, respectively. According to the analysis of the cross-species transferability of these markers, four species, S. forticosta, S. gottschei, S.tegulata, and S. libertina, showed a very high transferability (80%-85%). These results show that this set of nuclear markers could be useful for population genetics studies of this species and closely related species.
Keyword: Semisulcospira; next-generation sequencing (NGS); microsatellite markers; cross-species transferability
1 INTRODUCTION
Korean freshwater snails of the genusSemisulcospiraare a prominent freshwater gastropod resource across East Asia, including in Korea, Japan,southeastern China, and Taiwan, China (Davis, 1969).Freshwater snails have been used as healthy food sources in Korea and play important roles as biological indicators in the environmental monitoring of freshwater systems (Lee and Lim, 2005). Currently,the snail population is rapidly declining in Korea due to increasing human consumption, overharvesting,habitat degradation, and water pollution caused by insecticides and the restructuring of riverbeds (Kim et al., 2010). In order to recover the decreasing resources,artif icial seed production was being promoted. In addition, a large number of foreign snail species have been imported to recover the declining population(Moon et al., 2015). Many species belonging to this genus, includingS.coreana,S.tegulata,S.libertina,S.forticosta, andS.gottschei, are produced in diff erent regions. Especially, the Korean endemic species isS.coreana, which is distributed in rivers of the Midwest region, such as the Geum River, Seomjin River, and Yeongsan River in Korea (Kim et al.,2012). Of the genus,S.coreanaandS.libertinathat recognized as the major economic resource on the inland waters are principal culture species. Despite of its ecological importance, there have been a few phylogenetic, phenotypic, and genetic analyses(Urabe, 2000; Zeng et al., 2015); no population-level study using microsatellite markers has been carried out to clarify genetic diversity and population structure in the genusSemisulcospira.
Among the currently available DNA-based techniques, microsatellite DNA markers, also named simple sequence repeats (SSRs), are useful tools to assess the genetic variance and structure of populations because of their several desirable features such as variability, codominance, and high mutation rates(Féral, 2002). They have become the most widely useful DNA technology in many f ields of biology,including genome mapping, determining pedigree,population genetics, biological resource conservation,and forensic studies (Knapik et al., 1998; Luikart et al., 2003). The traditional cloning methods used to develop microsatellites involved signif icant trial and error, required knowledge of the f lanking region DNA sequence, and were time-consuming, cost prohibitive,and low throughput (Queller et al., 1993). At present,these problems have been partly resolved with the advent of next-generation sequencing (NGS)technologies that are both cost- and time-eff ective, as they can manufacture millions of base pairs of short fragment reads in a single run (Moges et al., 2016).Furthermore, when primers newly developed for one species can be used for broad taxonomic groups is increased the cost- and time-eff ectiveness of microsatellites. The cross-species amplification of microsatellites has successfully been applied to several marine species (Greenley et al., 2012).
In the present study, we developed the f irst 18 novel polymorphic microsatellite markers forS.coreanaand tested cross-species amplif ication in four additionalSemisulcospiraspp. These loci will assist in obtaining genetic information for resource management of the genusSemisulcospirathroughout the Korean drainage area.
2 MATERIAL AND METHOD
2.1 Sample collection and DNA extraction
Thirty-seven individuals ofS.coreanawere obtained from the Inland Fisheries Research Institute,National Institute of Fisheries Science (NIFS) (Table 1). Foot muscles from the samples were maintained in 100% ethanol before being transported to the laboratory. Genomic DNA extraction procedure was conducted using the MagExtractor with an MFX-6100 automated DNA extraction system (Toyobo,Osaka, Japan) under the manufacturer’s instructions.The concentration and quality of extracted genomic DNA was estimated using a spectrophotometer NanoDrop ND-1000 (Thermo Fisher Scientif ic,Barrington, IL, USA). For the cross-species transferability test to otherSemisulcospiraspecies includingS.tegulata,S.libertina,S.forticosta, andS.gottschei, DNA extraction was performed by the same method from ethanol-f ixed tissue from four relatedSemisulcospiraspp. obtained from NIFS in Ulgin, Korea.
2.2 454 GS-FLX Titanium sequencing and SSR identif ication
The NGS library was generated from ~10 μg genomic DNA and sequenced on a GS-FLX-454 pyrosequencing system (454 Life Sciences, Branford,CT, USA) at the NICEM (National Instrumentation Center for Environmental Management of Seoul National University). The obtained sequence reads were trimmed to 96% minimum overlap identity using Newbler 2.6 (Roche Diagnostics, Mannheim,Germany). To search for SSRs in the genomic sequence, the dinucleotide and trinucleotide repeats of more than seven iterations were screened using the Perl program SSR_f inder.pl (Tóth et al., 2000). Primer pairs complementary to the sequence f lanking the repeat element were designed using Primer 3 software(Untergasser et al., 2012). The optimal size for primer was set to a range as 18-26 bases and the optimal annealing temperature was set at 58°C. The optimal product size was set to 130-400 bp and the remaining parameters were kept at default settings.
2.3 DNA amplif ication and genotyping
The performances of the newly developed primer sets were tested for optimum concentration for PCR amplif ication using DNA from eight individuals ofS.coreana. The electrophoresis of PCR products were operated on a 1.5% agarose gel and 40 primer sets produced PCR products of 100-300 bp in length.Thirty of these primer sets were labeled and used to amplify DNA from the eight individuals. The forward primer was labeled at 5′ end with the f luorescent dyes 6-FAM, TAMRA, and HEX (Applied Biosystems,Foster City, CA, USA). PCR amplif ications were performed in 10-μL total volumes that included 0.4 μmol/L of the forward and reverse primer,0.2 mmol/L dNTPs, 1× PCR buff er, 0.25 U Ex Taq DNA polymerase (TaKaRa Biomedical, Inc., Shiga,Japan), and approximately 100 ng template DNA under the following conditions: pre-denaturation at 95°C for 10 min, followed by 35 cycles of 45 s at 94°C, 45 s of 58°C, and 45 s at 72°C with a f inishing of 5 min at 72°C using an ABI 2720 Thermocycler(Applied Biosystems, USA). The PCR products were detected on a 1.5% agarose gel. Fragment analysis was run on a 3730xl DNA Analyzer (Applied Biosystems, USA) using GeneScan 400HD ROX dye as the internal size standard (Life Technologies,Carlsbad, CA, USA) and analyzed using GeneMapper 5.0 (Applied Biosystems). Primer sets that generated numerous signal peaks or amplif ied monomorphic microsatellite loci were excluded.

Table 1 Information of freshwater snail species used in this study
2.4 Transferability analysis
To test the cross-species transferability of the SSR loci, we used all 18 polymorphic sites ofS.coreanaon samples of four related taxa. Transferability tests were performed using the abovementioned PCR conditions. Amplif ication products were monitored on 1.5% agarose gels, and qualitatively graded on the presence or absence of the band.
2.5 Data analysis
Numbers of alleles, eff ective numbers of alleles,expected and observed heterozygosities were assessed using Arlequin 3.0 (Excoffi er and Lischer, 2010). The signif icance level (P-value) for Hardy-Weinberg equilibrium (HWE) was evaluated using Genepop 4.0(Rousset, 2008). Polymorphic information content(PIC) was assessed statistically using Cervus 3.0 program (Kalinowski et al., 2007). The presence of null alleles and potential genotyping errors was calculated statistically using MicroChecker 2.2.3(Van Oosterhout et al., 2004).
3 RESULT AND DISCUSSION
NGS generated 17 719 000 reads of a total of 8 894 938 000 bp forS.coreana. The total number of contigs was 132 872 and the number of singletons was 1 754 529. The total number of reads after trimming low-quality sequences was 9 967 563. A total of 3 565 825 918 regions were found to contain from nucleotide repeats for more than four iterations.The amplicon sizes varied from 90 to 320 bp. Of them 41%, 27%, and 32% were di-, tri-, and tetranucleotides, respectively. CA repeats were the most common dinucleotide repeats, TTG repeats were the most common trinucleotide repeats, and TGTG repeats were the most common tetra-nucleotide repeats.
All of 78 SSRs with high copy numbers of all repeat motifs for the amplif ication and assessment of polymorphisms were synthesized to measure amplif ication effi ciency and extent of polymorphism.Among them, a total of 18 primer pairs (23%) showing clear amplif ication of polymorphic loci were applied for PCR analysis and genotyping. The primer details and f luorescent labeling information are shown in Table 2.
In the present, microsatellite developed in most marine resource, was mostly consisted as dinucleotide repeats. However, tri- and tetra-nucleotides motif have the advantages like highly polymorphic and stable than di-nucleotide repeats (Lindqvist et al.,1996). We developed 18 novel polymorphic microsatellite markers consisted of all of di-, tri-, and tetranucleotide motifs. The allele number per locus resulted from 2 to 17, with a mean of 6.9. The PIC for each locus ranged from 0.203 to 0.902, with a mean of 0.626. Botstein et al. (1980) described that a PIC value above 0.5 has suffi cient discrimination within a population. In addition, comparisons of PIC values can present an estimate of the power of the marker.Fifteen of the 18 loci were supposed to be highly informative (PIC>0.5). Another two (Sc-27 and Sc-60) were indicated reasonably informative(0.25<PIC<0.5), and one (Sc-76) was only slightly informative (PIC<0.25). This suggests that the set of microsatellites generated in the present study shows considerable potential for analyzing genetic polymorphisms.
The observed and expected heterozygosities of theloci resulted from 0.063 to 0.924 and 0.226 to 0.924,respectively. The inbreeding coeffi cient (Fis) ranged from -0.750 to 0.844. Null alleles were detected at most of the loci, except for Sc06, Sc53, and Sc60,which showed deviation from HWE after Bonferroni correction (P<0.002 7) with expected heterozygosity(He) values greater than observed heterozygosity (Ho)values. The signif icant def icits of heterozygosity are associated with the existence of null alleles (Selkoe and Toonen, 2006), which are common in freshwatermollusca (Gu at al., 2012). This is also demonstrated by the signif icant positiveFisvalues at most of the loci, except for Sc-60, indicating that the excess of homozygotes was likely due to the existence of null alleles or stuttering at these loci, as suggested by the Micro-Checker results. This is also caused by inbreeding within a population in invertebrates due to the poor mobility of freshwater snails (Gu et al.,2015).
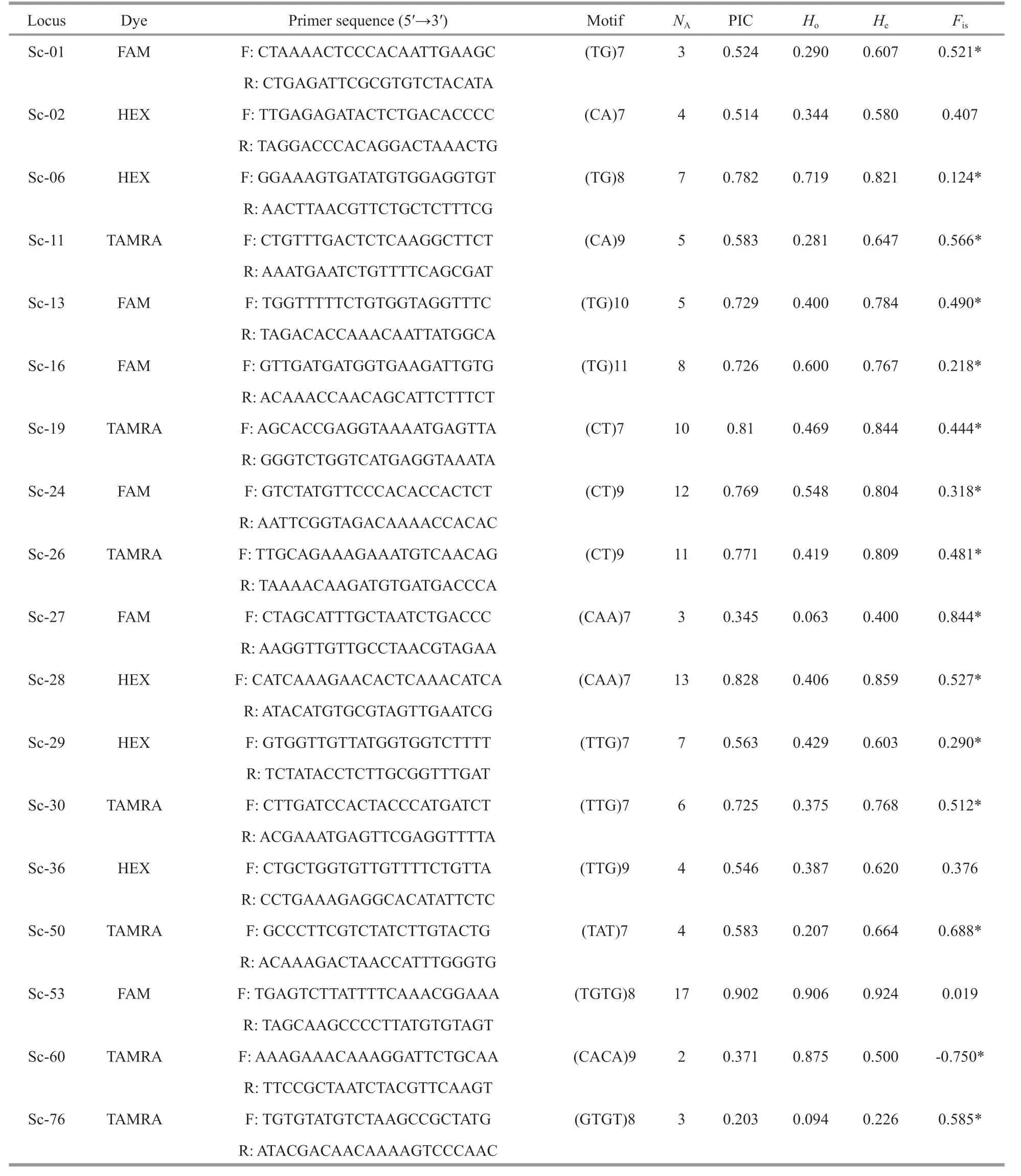
Table 2 Characteristics of Semisulcospira coreana microsatellite loci
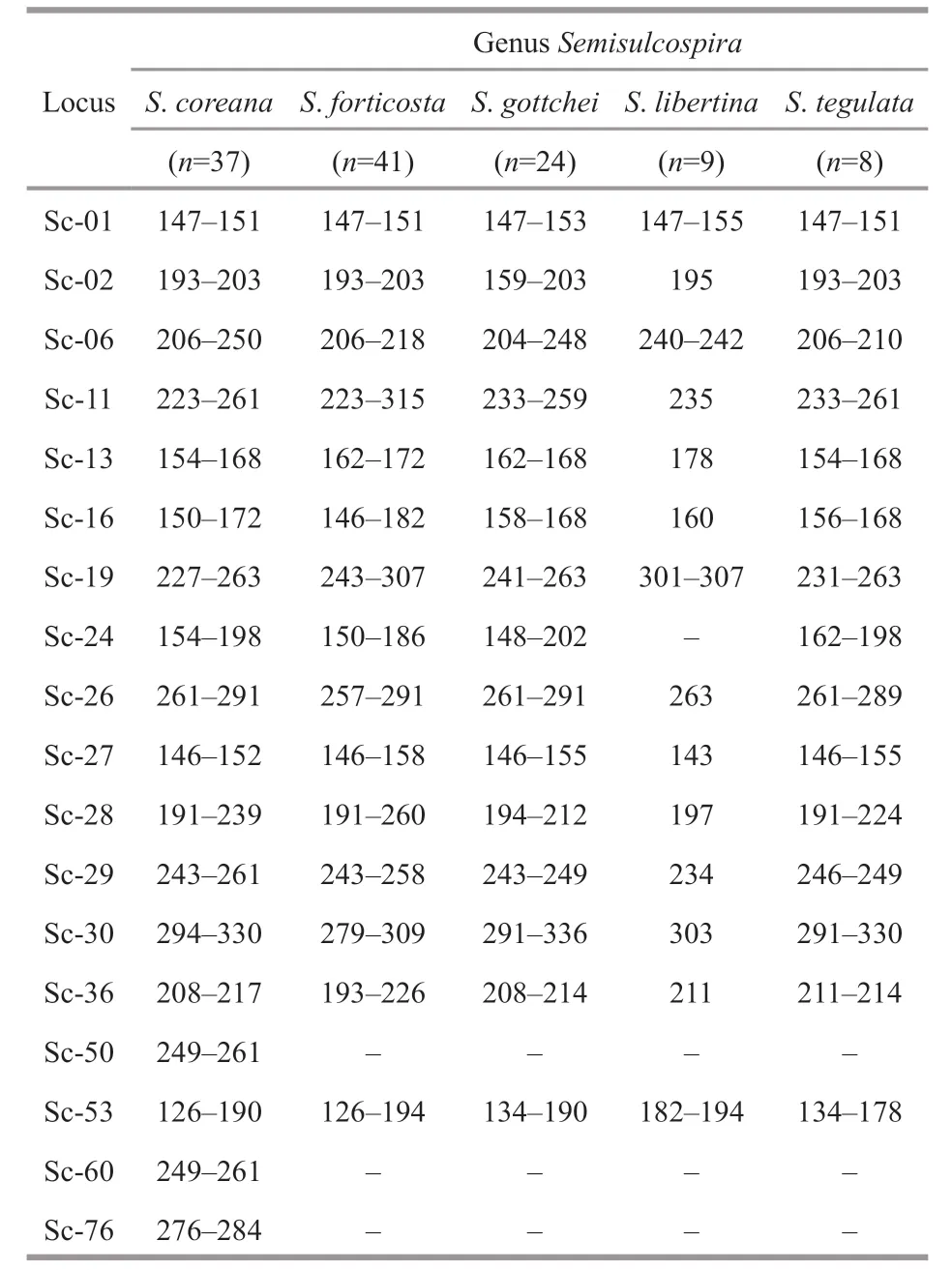
Table 3 Cross-species amplif ication in related Semisulcospira spp. using S. coreana microsatellite loci
Cross-species amplif ication of the 18 polymorphic loci was conducted within four species ofSemisulcospira,S.forticosta,S.gottschei,S.libertina,andS.tegulata(Table 3). As shown in Table 3, three polymorphic loci (Sc-50, Sc-60, and Sc-76) were not amplif ied by any species tested. These three loci could be used as identif ication ofS.coreana. Loci Sc-24 showed polymorphic amplif ication in all species tested exceptS.libertina. The transferability test of these microsatellite markers for the four species showed a very high transferability (80%-85%). The transferability of SSR markers in diff erent snail species is aff ected by the nucleotide sequence similarity in the SSR marker primer sites among related species (Barbará et al., 2007). The crossspecies transferability of SSR primers in the snails was high, indicating that there is high conservation of markers betweenSemisulcospiraspp.
We developed 18 new microsatellite markers forS.coreanausing 454 GS-FLX NGS. This is the f irst set of polymorphic microsatellite markers designed forS.coreanaand related species. These microsatellite markers will potentially be useful for genetic diversity and population genetic structure analyses of snails throughout the Korean drainage area.
4 CONCLUSION
Korean freshwater snails of the genusSemisulcospiraare economically and ecologically important inland resource. To study population genetic diversity of this genus, the development of microsatellite marker is necessary. The 18 novel polymorphic microsatellite loci fromSemisulcospira coreanawere isolated using 454 GS-FLX titanium sequencing. In addition, the transferability of all of the loci was checked onS.forticosta,S.gottschei,S.tegulata, andS.libertina. These novel microsatellite markers will aid freshwater snail resource investigations and population genetic conservation.
5 DATA AVAILABILITY STATEMENT
All data generated and/or analyzed during this study are included in this article, or are available upon the request from the readers.
6 COMPLIANCE WITH ETHICAL STANDARD
The authors declare that they have no conf lict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
 Journal of Oceanology and Limnology2020年2期
Journal of Oceanology and Limnology2020年2期
- Journal of Oceanology and Limnology的其它文章
- Erratum to: Seabed domes with circular depressions in the North Yellow Sea*
- Transcriptional responses to starvation of pathogenic Vibrio harveyi strain DY1*
- Heritability of resistance-related gene expression traits and their correlation with body size of clam Meretrix petechialis*
- Establishment and characterization of a new cell line derived from half-smooth tongue sole Cynoglossus semilaevis kidney*
- Eff ects of f lorfenicol exposure on growth, development and antioxidant capacity of f lounder Paralichthys olivaceus larvae at diff erent developmental stages*
- A new free-living marine nematode species of Rhinema from the South China Sea*
