Transcriptional responses to starvation of pathogenic Vibrio harveyi strain DY1*
LIU Xiaodan, GAO Xiaojian, CHEN Nan, ZHANG Yingying, LI Xixi, ZHANG Yue,ZHANG Xiaojun
College of Animal Science and Technology, Yangzhou University, Yangzhou 225009, China
Abstract Vibrio harveyi is a pathogen of various aquatic organisms that has been recently associated with massive mortality episodes in the aquaculture industry. Recurrent outbreaks of vibriosis are closely correlated with the capacity of this bacterial species to survive long-term starvation conditions. To study the regulation mechanism of gene expression at the transcriptional level in V. harveyi under starvation conditions, the transcriptomic response prof iles were determined of the Portunus trituberculatus pathogen V. harveyi strain DY1 under normal conditions and after four weeks of starvation. A total of 4 679 and 4 661 genes were expressed in the non-starved and starved cells, respectively. The signif icantly diff erentially expressed genes (DEGs) between non-starved and starved groups were identif ied, in which 255 genes were up-regulated and 411 genes were down-regulated. GO analysis and KEGG enrichment analysis were used to analyze the DEGs and revealed the involvement of these DEGs in many pathways, including ABC transporters, f lagellum assembly, and fatty acid metabolism. Several DEGs were randomly selected and their expression levels were conf irmed by quantitative real-time PCR (qRT-PCR). This is the f irst comprehensive transcriptomic analysis of starvation eff ects in V. harveyi. Our f indings will facilitate future study on stress adaptation and survival mechanisms of V. harveyi.
Keyword: Vibrio harveyi; starvation stress; transcriptome sequencing; diff erentially expressed genes;adaptation and survival mechanisms
1 INTRODUCTION
Vibrioharveyiis a facultatively anaerobic,bioluminescent, and Gram-negative rod-shaped bacterium (Johnson and Shunk, 1936). Bacterial infection causes serious vibriosis in marine f ish and invertebrates (Austin and Zhang, 2006), particularly in juvenile populations (Pujalte et al., 2003), leading to massive deaths and major economic losses in aquaculture.V.harveyihas been isolated from multiple sources including common snook(Centropomusundecimalis) in USA (Kraxberger-Beatty et al., 1990), silvery black porgy(Acanthopagruscuvieri) and cultured brown spotted grouper (Epinepelustauvina) in Kuwait (Saeed,1995), and sunf ish (Molamola) in Spain (Hispano et al., 1997).V.harveyi-associated diseases are major constraint on the production of marine invertebrates,particularly in South America and Asia.V.harveyiinfections have also been reported in common dentex(DentexdentexL.) (Company et al., 1999), salmonids(SalmosalarL.) (Zhang and Austin, 2000), seahorse(Hippocampuskuda) (Alcaide et al., 2010), farmed sole (Soleasenegalensis) (Zorrilla et al., 2003),summer f lounder (Paralichthysdentatus)(Gauger et al., 2006), and gilthead sea bream (Sparusaurata)(Haldar et al., 2010). In addition,V.harveyihas been isolated from several crustacean larvae, including black tiger prawn (Penaeusmonodon) (Lavilla-Pitogo et al., 1990), kuruma prawns (Penaeusjaponicas)(Liu et al., 1996), rock lobster (Jasusverreauxi)(Diggles et al., 2000), and oysters (Ortigosa et al.,1994). Infected animals became inappetent and developed inf lammation and necrotic subdermal cysts in various species of f ishes, such as crustaceans and mollusks (Austin, 2010).
Bacteria could enter viable but non-culturable(VBNC) or normal dormant state by a variety of environmental stresses (Wolf and Oliver, 1992;Biosca et al., 1996; Jiang and Chai, 1996). Moreover,nutrient def iciency is the most common environmental stress in natural ecosystems for microorganisms. It is reported that some species could survive for a long time in response to starvation condition such asV.harveyi,V.cholerae,V.f ischeri,V.anguillarum,V.campbellii,V.mimicus,V.proteolyticus,V.vulnif icus(Jiang and Chai, 1996; McDougald et al.,1998). Like other bacterial species,V.harveyimay be chronically exposed to nutrient-poor waters and its ability to survive in water for extended periods of time is an adaptive strategy against stress. Imaging showed that starved cells alter their morphology from rod to spherule cells (Novitsky and Morita, 1976; Sun et al., 2016). Starved cells will adjust their gene expression to adapt to environmental stress, with potential changes to transcriptional regulators, heat shock proteins, virulence regulators, and genes related to metabolism. However, little is known about gene expression prof iles under starvation stress.
Hence, we obtained whole transcriptomic prof iles ofV.harveyiDY1 strain under starvation stress by high-throughput sequencing and determined the gene expression changes compared to non-starved cells.This is the f irst comprehensive transcriptomic analysis of starvation eff ects in pathogenicV.harveyi, and the results greatly strengthen our understanding of the molecular pathogenic and transmission mechanisms ofV.harveyistrain DY1.
2 MATERIAL AND METHOD
2.1 Bacterial strain
TheV.harveyiDY1 strain was f irst isolated from cultured megalopa of swimming crab (Portunus trituberculatus) in 2011 by Zhang et al. (2014) in Jiangsu, China. DY1 strain was recovered from -80°C as glycerol stocks and incubated with LB broth(Hopebio Qingdao, China) at 28°C overnight with shaking.
2.2 Starvation stress and cell enumeration
Overnight cultures were diluted (1:100) with fresh LB broth and grown to the mid-log phase at 28°C.Cultures were collected by centrifugation at 12 000×gfor 10 min. The pellets were washed twice with sterile artif icial seawater to rinse nutrients off and suspended in the starvation regimes. Then the cells in triplicate were placed in Erlenmeyer f lasks containing 100 mL sterile artif icial seawater to give a f inal concentration of 108CFU/mL. Cells were incubated in the dark at 25°C and monitored for a period of four weeks according to the previous method. Cells in starvation regimes were sampled at 1, 7, 14, 21, and 28 days and counted by plate count method (Sun et al., 2016). The cell number was counted and converted to base-10 logarithms to f it normal distribution model.
2.3 Morphology analysis
The morphology changes ofV.harveyiDY1 strain after four weeks of starvation was observed by SEM as previously described (Arias et al., 2012). The nonstarved and starved cells were centrifuged at 10 000×gfor 10 min at 4°C. The pellets were washed with PBS,and f ixed with glutaraldehyde (0.25 g/L). A graded ethanol series was used to dehydrate the samples,then coated with gold palladium alloy in an Electron Microscopy Science (EMS 550X), and examined by Zeiss EVO 50 (Zeiss, Germany).
2.4 Library construction and Illumina sequencing
Cells starved for four weeks and the non-starved cells were subjected to total RNA isolation and cDNA preparations. Three independent trials were performed for sequencing. Total RNA was extracted from cells using the EasyPure RNA kit (TransGen Biotech,Beijing, China). rRNA was removed after total RNA was collected from prokaryocyte. Fragmentation buff er was added for splitting mRNA to short fragments. Taking these short fragments as templates,random hexamer-primer were used to synthesize the f irst-strand cDNA and the second-strand cDNA was synthesized using buff er, RNase H, dNTPs, DNA polymerase I. Short fragments were purif ied with QiaQuick PCR extraction kit and resolved with elution buff er for end reparation and adding poly (A).After that, the short fragments were connected with sequencing adapters and the UNG enzyme was used to degrade the second-strand cDNA, and the product was purif ied by MiniElute PCR purif ication kit before PCR amplif ication. At last, the library was sequencedusing Illumina HiSeq2000. Images generated by sequencers were converted by base calling into nucleotide sequences, which were called raw data or raw reads and are stored in FASTQ format. Raw reads produced from sequencing machines contain dirty reads that contain adapters, unknown or low quality bases. After removing reads with adaptors and low quality reads, clean reads were obtained. The clean reads were mapped to reference genome and genes sequences respectively using SOAP2 (Li et al., 2009).

Table 1 Primers used for the detection of DEGs by qRT-PCR
2.5 Gene annotation and diff erence analysis
After data f iltering, we compared the clean reads with the reference genome and sequence which has been deposited at DDBJ/EMBL/GenBank under the accession numbers CP009467.2 and CP009468.1.Gene coverage is the percentage of a gene covered by reads. This value equals to ratio of the number of bases in a gene covered by unique mapping reads to number of total bases in that gene. The threshold of theP-value in multiple tests was determined by setting FDR (false discovery rate) at 0.000 1. The FPKM (fragments per kb per Million reads) method was applied to calculate gene expression (Mortazavi et al., 2008). To screen the DEGs in DY1 cells potentially related to starvation stress, statistical and analysis of gene expression was applied using fold change >2 andP≤0.001 as standards.
2.6 Functional annotation and GO/KEGG enrichment analysis
For gene ontology analysis, the alignment results were parsed for assigning GO terms by using Blast2GO software. Gene sequences ofV.harveyiDY1 strain were aligned against Gene Ontology database. The calculatedP-value went through Bonferroni Correction, taking correctedP≤0.05 as a threshold. GO terms fulf illing this condition are def ined as signif icantly enriched GO terms in DEGs.
KEGG pathway enrichment analysis identif ies signif icantly enriched metabolic pathways or signal transduction pathways in DEGs comparing with the whole genome background. The calculating formula is the same as that in GO analysis.
2.7 Quantitative real-time PCR analysis
RNA were extracted using an EasyPure RNA kit(TransGen Biotech, Beijing, China) and reversetranscribed by TransScript One-Step gDNA Removal and cDNA Synthesis Supermix (TransGen Biotech,Beijing, China) according to the manufacturer’s instructions. The cDNA was synthesized using anchored oligo (dT) 18 primer and incubated for 15 min at 42°C. Real-time PCR was performed with 10 μL SYBR®SuperMix, 1 μL diluted cDNA, 0.4 μL of forward primer, 0.4 μL of reverse primer and 8.2 μL of nuclease-free water. Reactions were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s. Each sample was run in triplicate for analysis. The relative gene expression were calculated using 2-ΔΔCtmethod and16S rRNA gene was chosen as an internal control for normalization.Specif ic primers were designed according to the corresponding sequences ofV.harveyi(Table 1).
2.8 Statistics analysis
The data are presented as the mean±SD (n=3).One-Way Analysis of variance (one-way ANOVA)was used to evaluate the diff erential expression by SPSS (20.0).
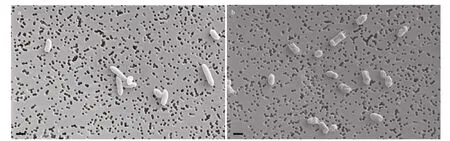
Fig.1 SEM micrographs of V. harveyi
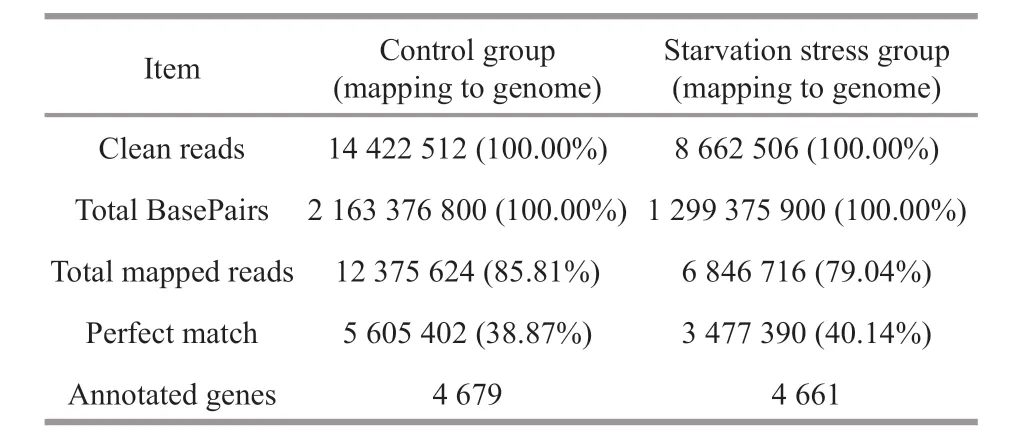
Table 2 Summary of the reads and annotated genes in nonstarved group and starved group
3 RESULT
3.1 Morphology analysis
Vibrioharveyiwas found decreased in size and changed in shape from a rod to a sphere after four weeks of starvation shown in SEM. There was also a signif icant decrease in size of cells after starvation compared to the initial population (Fig.1).
3.2 De novo assembly and gene annotation
In this study, RNA-Sequencing was used to study the adaptation ofV.harveyiunder four-week starvation using Illumina HisSeqTM2000 system.After f iltering the raw data, 14 422 512 and 8 662 506 clean reads were obtained in control and starvation stress group (Table 2). Proportions of clean reads mapped back to genome and genes can provide an overall assessment of the sequencing. The reads were aligned toV.harveyireference genome using the SOAPaligner/soap2 software and mismatches no more than 5 bases were allowed in the alignment.There were 12 375 624 (85.81%) and 6 846 716(79.04%) clean reads were mapped to reference genome in control and starvation stress group (Table 2). There were 5 605 402 (38.87%) and 3 477 390(40.14%) reads and ratio of perfect matches in total mapped reads in the control and starvation stress group (Table 2). There were 5 337 genes annotated in the transcriptome analysis.V.harveyistrain DY1 revealed 4 679 and 4 661 annotated genes before and after starvation stress, respectively.
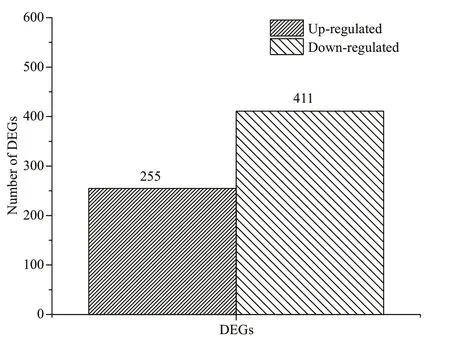
Fig.2 The number of DEGs in control and starvation stress groups
3.3 Gene expression diff erence analysis
A total of 666 diff erentially expressed genes(DEGs) were identif ied in the samples from the fourweek starved group compared to the samples from the untreated group, including 411 down-regulated genes and 255 up-regulated genes (Fig.2). Thus, about 14.2% of the total number of genes showed signif icantly altered expression levels after starvation stress for four weeks. The up-regulated DEGs included 20 genes with more than 10-fold expression change between the starved group and the untreated group. Similarly, of the down-regulated DEGs, there were four genes with more than 10-fold expression change.
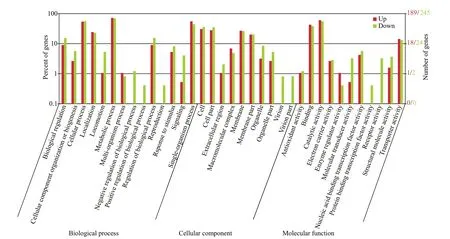
Fig.3 GO analysis of diff erential expression genes between the initial and the four-week starved V. harveyi
3.4 GO functional classif ication of DEGs
GO analyses were performed to classify gene functions ofV.harveyiunder starvation stress. GO analysis of DEGs inV.harveyiafter starvation revealed that in the biological process catalogue, the most abundant category of the diff erentially expressed genes control metabolism. There were 136 upregulated genes assigned metabolic process, such as hydroxyethylthiazole kinase and peptidase. There were 170 downregulated genes assigned metabolic process including hemolysin, oxidoreductase,6-phosphogluconate dehydrogenase, et al. In cellular component, the top three categories were cell (55 upregulated, 88 down regulated), cell part (53 upregulated, 86 down regulated) and membrane process (51 upregulated, 64 down regulated). In the molecular function catalogue, the most abundant terms of the diff erential expression gene were catalytic activity, followed by binding and transporter activity process, primarily including cell division protein ZapB, f lagellar motor switch protein FliM, sulfur transfer protein TusE and transcriptional regulator(Fig.3).
3.5 KEGG enrichment analysis of DEGs
Expression of some genes plays a signif icant role in protection against environmental stress. KEGG enrichment analysis was used to identify the most enriched pathways inV.harveyiafter starvation stress. In KEGG enrichment, the DEGs were assigned into 120 pathways. The most abundant pathways were metabolic pathways, microbial metabolism in diverse environments and Biosynthesis of secondary metabolites. Flagellar assembly pathway and Butirosin and neomycin biosynthesis pathway were the most representative pathways related to starvation stress (Fig.4). Moreover, ABC transporters pathway,f lagellar assembly pathway, fatty acid metabolism pathway, fatty acid degradation pathway, biosynthesis of antibiotics pathway and beta-Lactam resistance pathway also involved many diff erentially expressed genes.
3.6 Verif ication of the DEGs by qRT-PCR
To validate the sequencing results, eight diff erentially expressed genes (danK,f imT,hcp,impA,aphB,rnfA,fur,htpG) ofV.harveyiwere randomly selected for qRT-PCR. Among these genes,four genes (danK,f imT,impA,aphB) are up-regulated genes and the other four (hcp,rnfA,fur,htpG) are down-regulated genes. The cell samples that subjected for qRT-PCR are the same as those used for transcriptome sequencing. The expression prof ile of these DEGs are similar to those obtained in sequencing results (Fig.5).
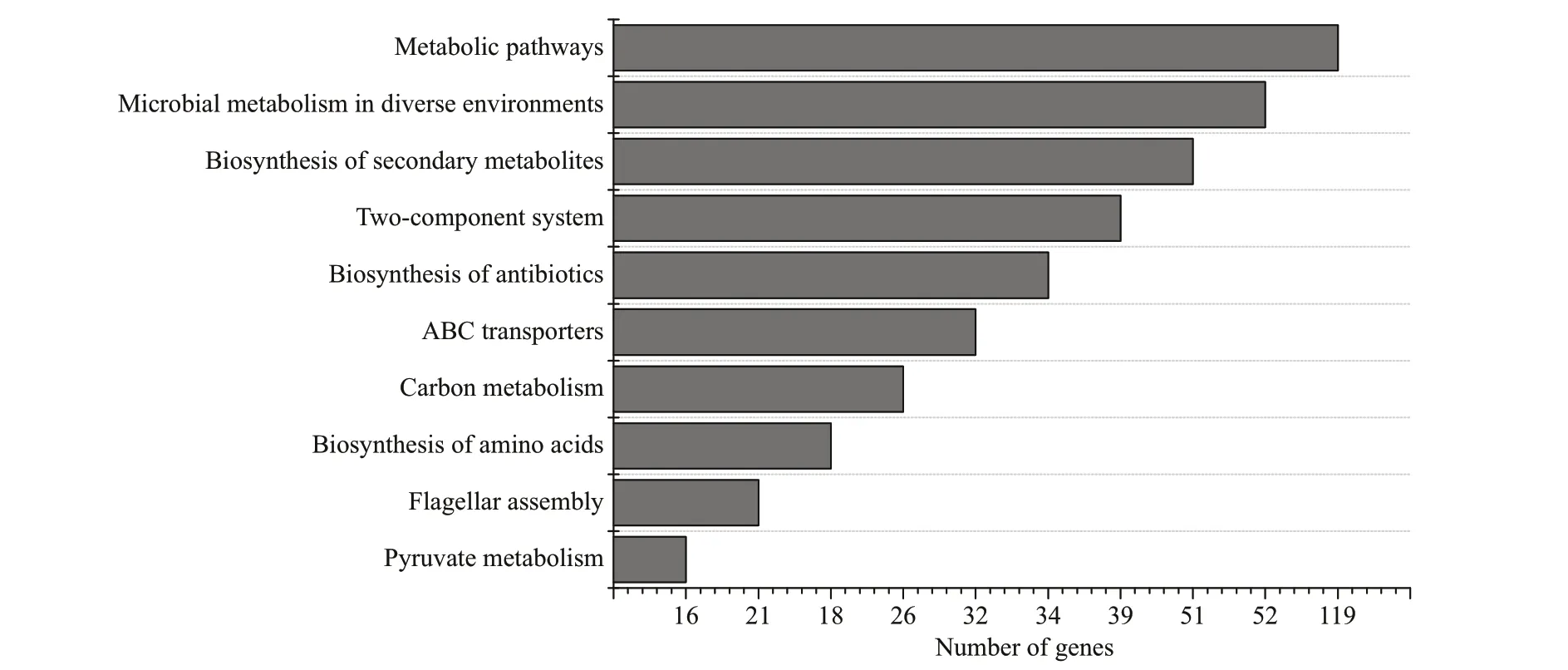
Fig.4 KEGG pathway enrichment analysis of the DEGs between the initial and the four-week starved V. harveyi
4 DISCUSSION
High mortality rates in the early stages of the marine f ish and invertebrates caused byV.harveyiis one of the most important reasons for economic losses during the production period. However, the survival strategies ofV.harveyiin the aquatic environment are not well understood. Microorganisms must adapt to environmental stress to grow and survive (Poindexter,1981; Kjellerberg et al., 1983; Kunttu et al., 2009).Previous studies showed that the most widespread environmental stress for microbes in a natural environment is a lack of nutrients (Wai et al., 1999;Vatsos et al., 2003; Suzina et al., 2004; Montánchez et al., 2014; Kaberdin et al., 2015; Parada et al., 2016;Montánchez et al., 2019). To study the physiological response and the long-term survival mechanism upon starvation stress is urgent and important.
In this study, we compared the size of the starved cells with cells from the control group, and observed that the starved cells were obviously shorter (Fig.1).This implied thatV.harveyilikely triggered specif ic regulatory mechanisms to adjust its shape and size.This result was previously reported (Sun et al., 2016).Parada et al. (2016) found that morphological change was not directly related to the entry of cells into the VBNC state, although they observed a gradual decrease in the size ofV.harveyicells during incubation. Montánchez et al. (2014) found that incubation in cold seawater for 12 h did not cause any signif icant morphological changes inV.harveyi,suggesting thatV.harveyilikely elicits specif ic adaptation mechanisms maintaining its culturability under stress conditions.
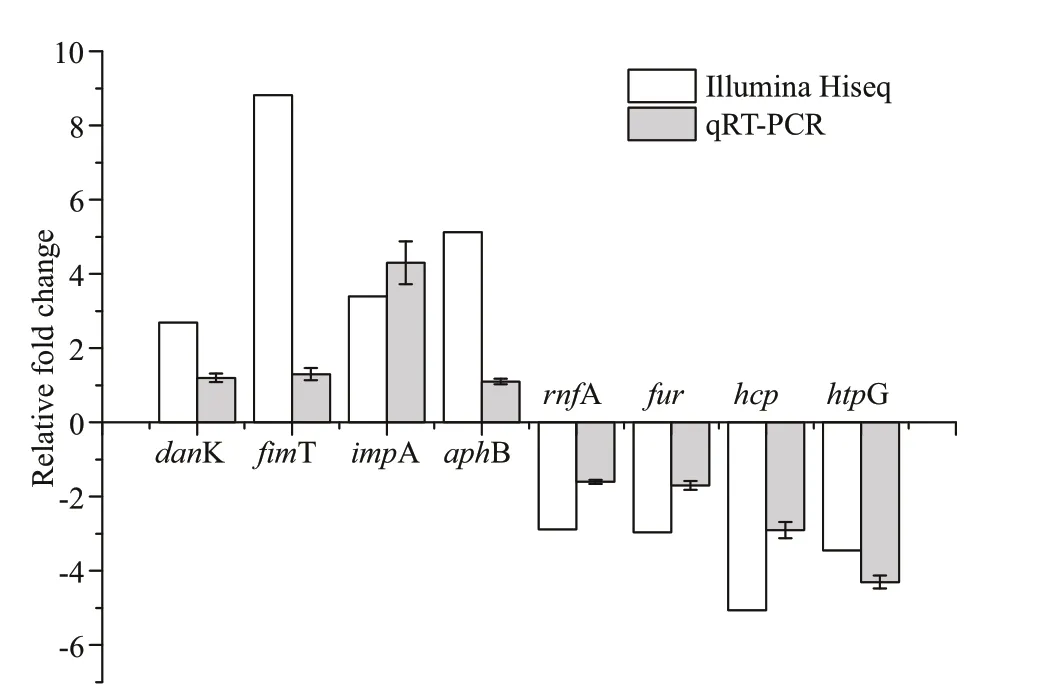
Fig.5 Verif ication of the diff erentially expressed genes of V.harveyi before and after starvation stress by RNASeq and qRT-PCR
The reads were mapped to the reference genome.The gene coverage was above 70% for both groups.The RNA-seq data can be used to assess the variation in expression of virulence-related and metabolismrelated factors inV.harveyi, which suggest the strategies used by these bacteria to survive under starvation conditions. There were 255 up-regulated genes and 411 down-regulated genes under starvation stress, and these genes very likely act in the response to starvation stress (Fig.2). The analysis shown in Fig.3 revealed 35 GO categories relevant to starvation stress ofV.harveyi. The main terms associated with starvation stress were binding and transporter activity,metabolic process, and cell and catalytic activity.Previous proteomic study revealed that the level of membrane proteins participating in cellular transport,maintenance of cell structure, and bioenergetics processes remained unchanged during starvation at low temperature, suggesting thatV.harveyimight need these proteins for long-term survival or for the resuscitation process after dormancy (Parada et al.,2016).
The analysis of KEGG enrichment may help us to understand the molecular mechanisms utilized byV.harveyifor long-term survival under starvation stress. The most abundant KEGG pathways included antibiotic biosynthesis, ABC transporters, butanoate metabolism pathway, starch and sucrose metabolism,and beta lactam resistance pathways, which are processes that have previously been related to starvation stress (Higgins, 2001; Kim et al., 2013;Svensson et al., 2014). Genes related to nutrient transport, such as phospholipids, oligopeptides, and simple sugars, were down-regulated after four weeks starvation. However, genes related to nutrient transfer,such as the phospholipid transfer protein gene and the monosaccharide transporter gene, were signif icantly up-regulated under starvation stress, representing an active metabolism pathway inV.harveyi. This change may be supplementation sparked by the compensatory response, and changes in the expression of genes controlling biosynthesis of lipids and molecular transport likely aff ect the composition and properties of theV.harveyicell envelope, an obvious adaptation to stress. We suspect thatV.harveyicomprehensively utilized multiple strategies and adaptation mechanisms to sustain key physiological functions under nutritional stress. At least in part,V.harveyicompensated for the reduced expression of biosynthetic genes by the upregulation of transporter genes controlling the uptake of amino acids (e.g. amino acid ABC transporters). Previous research found that limitation of nutrients leads to the signif icant downregulation of genes controlling central carbon metabolism,biosynthesis of lipids, amino acids, and nucleotides(Montánchez et al., 2014).
To better understand the survival strategies ofV.harveyiunder starvation stress, the fatty acid metabolism pathway was analyzed. The genes related with fatty acid synthesis were signif icantly downregulated and the expression level of fatty acid degradation related genes were signif icantly increased,producing a large number of acetyl coenzyme A for TCA cycle. The observation of the up-regulation of fatty acid degradation correlates to previous work by Kaberdin et al. (2015). Moreover, the f ine-tuning of these metabolic pathways appears to be attained through the action of small regulatory RNAs known for their essential roles in post-transcriptional control of gene expression (Kaberdin and Bläsi, 2006).
Virulence genes played a crucial role in environmental adaptation and virulence. Previous study analyzedV.harveyiadaptation in sea water microcosms at elevated temperature and found that elevated temperature also aff ected regulation ofV.harveyigenes controlling its virulence and ancillary mechanisms (i.e. production and secretion of virulence factors, biof ilm formation and motility)(Montánchez et al., 2019). The sequencing analysis revealed downregulated expression levels of virulence regulator genetoxRand f lagellin A. These virulence genes regulate a considerable number of genes involved in environmental adaptation and virulence.Other down-regulated genes include f lagellinassociated genes, such asf lgB,f lgC,f lgD,f lgF,f lgG,f lgH,f lgI,f lgK,f lgL,f liM, andf liO. Down-regulation of f lagellin-associated genes will directly reduce f lagellar synthesis ofV.harveyi, resulting in reduced motility and adhesive strength, which may be critical during the initials steps of infection. Thus, the pathogenicity may decrease under starvation stress.The decreased expression of f lagellin-associated genes suggests that under starvation conditions,V.harveyireduced energy requirements by reducing the synthesis of non-essential structures.
Under starvation stress, many genes were enriched in metabolic pathways involved in the synthesis of macrolides and ketolide antibiotics (Fig.4). The sequencing revealed up-regulated expression levels of many genes related to the synthesis of macrolides and ketolides and beta lactam resistance genes,includingTolC(outer membrane protein),acrA(membrane fusion protein, multidrug effl ux system),andopp(oligopeptide transport system). These resistance-related genes may signif icantly improve the survival rate ofV.harveyi.
5 CONCLUSION
This study presented the transcriptomic response prof iles of thePortunustrituberculatuspathogenV.harveyistrain DY1 under starvation conditions, to elucidate the changes inV.harveygene expression due to starvation in seawater.V.harveyimay survive under starvation conditions by regulating the expression of virulence and metabolism-related genes. The identif ied genes may be important highvalue drug targets, suggesting new ways to eff ectively control clinical infection ofV.harveyi. Our f indings should facilitate future study on stress adaptation and environmental survival mechanisms ofV.harveyi.
6 DATA AVAILABILITY STATEMENT
All sequence data that support the findings of this study have been deposited in the NCBI Short Read.The sequence read archive (SRA) accession number:PRJNA507871.
 Journal of Oceanology and Limnology2020年2期
Journal of Oceanology and Limnology2020年2期
- Journal of Oceanology and Limnology的其它文章
- Erratum to: Seabed domes with circular depressions in the North Yellow Sea*
- Heritability of resistance-related gene expression traits and their correlation with body size of clam Meretrix petechialis*
- Establishment and characterization of a new cell line derived from half-smooth tongue sole Cynoglossus semilaevis kidney*
- Eff ects of f lorfenicol exposure on growth, development and antioxidant capacity of f lounder Paralichthys olivaceus larvae at diff erent developmental stages*
- A new free-living marine nematode species of Rhinema from the South China Sea*
- Sabatieria sinica sp. nov. (Comesomatidae, Nematoda) from Jiaozhou Bay, China*
