Heritability of resistance-related gene expression traits and their correlation with body size of clam Meretrix petechialis*
JIANG Fengjuan , YUE Xin ZHANG Shujing , YU Jiajia , WANG Rui ,LIU Baozhong , WANG Hongxia
1 Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
2 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266000, China
3 University of Chinese Academy of Sciences, Beijing 100049, China
Abstract Gene expression variation can be considered as a phenotype, and it plays an important role in both acclimation and adaption. However, genetic variation of gene expression received much less attention than traditional commercial traits in aquaculture. To estimate the genetic variation and heritability of gene transcription in clam Meretrix petechialis, f ive Vibrio resistance-related genes were selected for gene expression analysis in the digestive gland, and an animal linear model was used to analyze data from quantitative real-time PCR (qRT-PCR). Among the f ive genes, BIRC7 showed signif icant additive genetic variations, the heritability of this gene of 12-month- and 15-month-old clams were 0.84±0.32 and 0.91±0.34,respectively. The heritability of other four genes ( Bax, NFIL3, Big- Def, and CTL9) expression were low-tomoderate but not signif icantly expressed. Additionally, no signif icant phenotypic and genetic correlations between the BIRC7 transcription trait and body size were detected. This study highlights that certain gene expression variation is heritable and provides a reference for indirect selection of M. petechialis with high Vibrio resistance.
Keyword: Meretrix petechialis; transcription trait; body size; heritability; genetic correlation
1 INTRODUCTION
Meretrixpetechialisis one of the most important commercial clam species farmed in China (Liu et al.,2006). Despite continuous progress and improvement in artif icial reproductive technologies and aquaculture techniques, clam diseases remain a major limiting factor in the clam culture industry. Outbreaks of vibriosis in clams often aff ect the prof itability and sustainability of aquaculture operations. From a sustainability perspective, selective breeding for disease resistant clams is an attractive strategy for disease prevention (Liang et al., 2017). However, in contrast to many other economically important traits(e.g., growth rate, feed conversion effi ciency), genetic improvement of disease resistance has been hindered by the diffi culty of measuring appropriate phenotypes accurately (Bishop and Woolliams, 2014). Therefore,markers accurately ref lecting disease resistance and having a signif icant genetic component are necessary in resistance selection.
To date, some association analyses between molecular markers andVibrioresistance traits have been reported inM.petechialis(Nie et al., 2013,2015; Zou and Liu, 2016). However, the eff ectiveness of these markers is minor, accounting for only a small fraction of the genetic variation (Yang et al., 2010).Thus, more attempts are needed to develop markers that are closely linked toVibrioresistance. In previous studies, the use of gene expression prof iles as indirect tests for improved disease resistance has been proposed for selective breeding in Atlantic salmon(Robinson and Hayes, 2008). Research onM.petechialishas demonstrated that hundreds of genes are involved in the physiological and immunological response to bacterial infection (Jiang et al., 2017). Although numerous studies have demonstrated diff erences in gene transcription among genetic groups or lines in aquatic species (Langevin et al., 2012; Marancik et al., 2014; Reyes-López et al.,2015; Robledo et al., 2016), few have partitioned genetic variation and estimated heritability for gene transcription (Roberge et al., 2007; Normandeau et al., 2009; Aykanat et al., 2012; He et al., 2017).
Gene expression variation can be considered as a phenotype (Houle et al., 2010), and as with any traditional phenotypic trait, gene expression variation is inf luenced by various genetic and environmental factors (Leder et al., 2015). It is clear that gene expression variation is widespread among individuals and taxa and can be heritable (Whitehead and Crawford, 2006; Dixon et al., 2007; Powell et al.,2013). Additive genetic eff ects of gene expression represent evolutionary potential, which allows us to predict the magnitude of its response to selective pressure (Leder et al., 2015; He et al., 2017). Nonadditive eff ects also contribute signif icantly to phenotypic variation but are not heritable (Evans and Neff , 2009). All these components of genetic parameters are important contributors to phenotypic variation. Therefore, it is fundamentally important to disentangle the contributions of additive genetic and non-additive genetic variance to overall gene expression variance. This information of genetic parameter is critical for evaluating the potential for gene-expression performance markers to be used in selection programs targeting resistant clam broodstocks.
Recent studies have shown that the basal expression levels of one set ofVibrioresistance-related genes(Big-Def,BIRC7,NFIL3,CTL9, andBax) signif icantly correlated with survival in challenge tests in clamM.petechialis(Jiang et al., 2018). Therefore, it seemed that selection decisions could be made based on the expression level of resistance-related genes that are genetically correlated with disease resistance. The indirect selection by gene expression trait would simplify the data collection, avoid the challenge testing, and reducing costs and time consumption.Theoretically, higher additive genetic eff ects ref lect higher adaptive potential in response to selection pressures; thus, the gene transcription traits closely linked to phenotype and with higher additive genetic component are better candidates for an indirect selection criterion in practice. However, the genetic variance components for the transcription of resistancerelated genes and their genetic correlations with body size have not been evaluated inM.petechialis.Therefore, in this study, an animal model was used to estimate the genetic variation component forVibrioresistance-related genes, and estimate the genetic correlations betweenVibrioresistance-related genes and body sizes. The results could provide some key parameters for understanding of the potential resistant selection response of gene transcription for resistancerelated genes in non-model organisms.
2 MATERIAL AND METHOD
2.1 Experimental families
Experimental families were produced in a hatchery at the Zhejiang Mariculture Research Institute(Wenzhou, China). In July 2014, 11 two-year-old males and 11 two-year-old females were randomly selected from the clam population and used to produce 23 full-sibling families according to a slightly modif ied Berg and Henryon design (Berg and Henryon, 1999).For example, each sire was mated with 2-3 dams, and each dam was mated with 2-3 sires. Thus 11 dam halfsib families and 11 sire half-sib families were produced, the detailed breeding design for these fullsibling families was presented in Fig.1. Detailed rearing conditions for larvae have been described by Wang et al. (2011). The full-sibling families of juvenile clams were randomly distributed in pond at the Zhejiang Mariculture Research Institute. In the farming process, diff erent families were reared separately by mesh in the same pond and exposed to similar environmental conditions to minimize the eff ects of uncontrolled environmental factors. In detail, the stocking density of each family was the same, and the average survival rate of families was up to 92.4%±1.9% at 15-month-old, which means that no accidental mass mortalities occurred in the cultivation.

Table 1 The primers used for qRT-PCR in this study
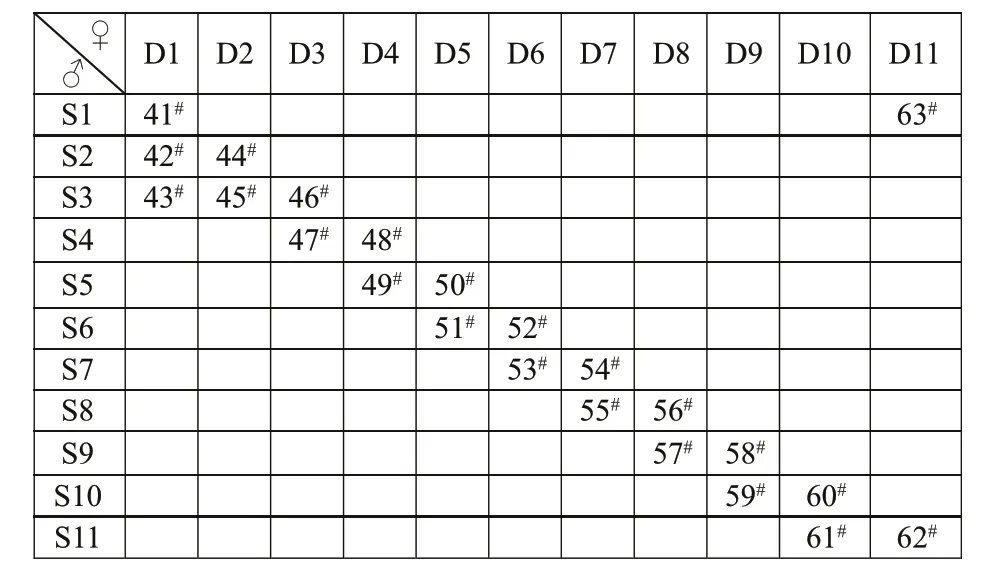
Fig.1 The schematic diagram of clam M. petechialis 23 fullsibling families
2.2 Sample collection
Before the estimation of genetic parameters, six tissues (digestive gland, mantle, gill, foot, adductor muscle, and hemocytes) were collected from four healthy clams for analysis of the expression of the detected genes between tissues. Hemolymph(~500 μL) was withdrawn from each adult clam with a 1-mL disposable syringe, and was then centrifuged at 3 000×gfor 10 min at 4°C. The pellet was used in total RNA extraction. Another f ive tissues from each clam were taken, dissected, immediately frozen in liquid nitrogen, and stored at -80°C until tissue distribution analysis could be conducted.
The 12-month- and 15-month-old clams from two batches were used to estimate genetic parameters respectively. We randomly selected f ive individuals from each family from each sampling batch for the genetic analysis. The shell width (SW) and total weight (TW) of 12-month-old clams were measured using a digital Vernier caliper and a digital balance respectively. After measuring the SW and TW, the digestive gland from each clam was dissected,immediately frozen in liquid nitrogen, and stored at-80°C until further processing.
2.3 RNA extraction and cDNA synthesis
Total RNA was extracted from samples using an EZNA®Total RNA Kit II (Omega Bio-Teck, USA)according to the manufacturer’s instructions. RNA degradation and contamination were monitored on 1% agarose gels and then quantif ied using a NanoDrop-1000 spectrophotometer (Thermo Scientif ic, USA). cDNA was synthesized from total RNA (1 μg) using a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, Japan) according to the manufacturer’s protocol.
2.4 Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed to determine the expression levels ofVibrioresistancerelated genes in diff erent tissues from adult clams. The elongation factor 1α gene (EF1α) was used as the internal reference gene in semi-quantitative RT-PCR analysis (Jiang et al., 2017). Primer sequences are listed in Table 1. PCRs were performed in a 25 μL reaction volume containing 1 μL cDNA, 0.2 mmol/L of each primer, and 12.5 μL Premix Taq™ (TaKaRa,Japan). The appropriate number of cycles of these six genes were selected, 18 cycles forEF1α, 26 cycles forBax,NFIL3andBig-Defand 28 cycles forBIRC7andCTL9. These cycling parameters could ensure that the amplif ication product is clearly visible and can be quantif ied. That is the amplif ication is in the exponential range and has not reached a plateau yet.The cycling protocol was one cycle of 94°C for 4 min;18, 26 or 28 cycles of 94°C for 15 s; 55°C for 15 s; and 72°C for 15 s, and the f inal extension time was 7 min.PCR was conducted using a Takara thermocycler.PCR products were separated via electrophoresis in 2% agarose gels that were stained with 4S Red Plus Nucleic Acid Stain (Sangon, China) and visualized under ultraviolet light. The quantif ication of bands was performed by Bio-Rad Quantity One.
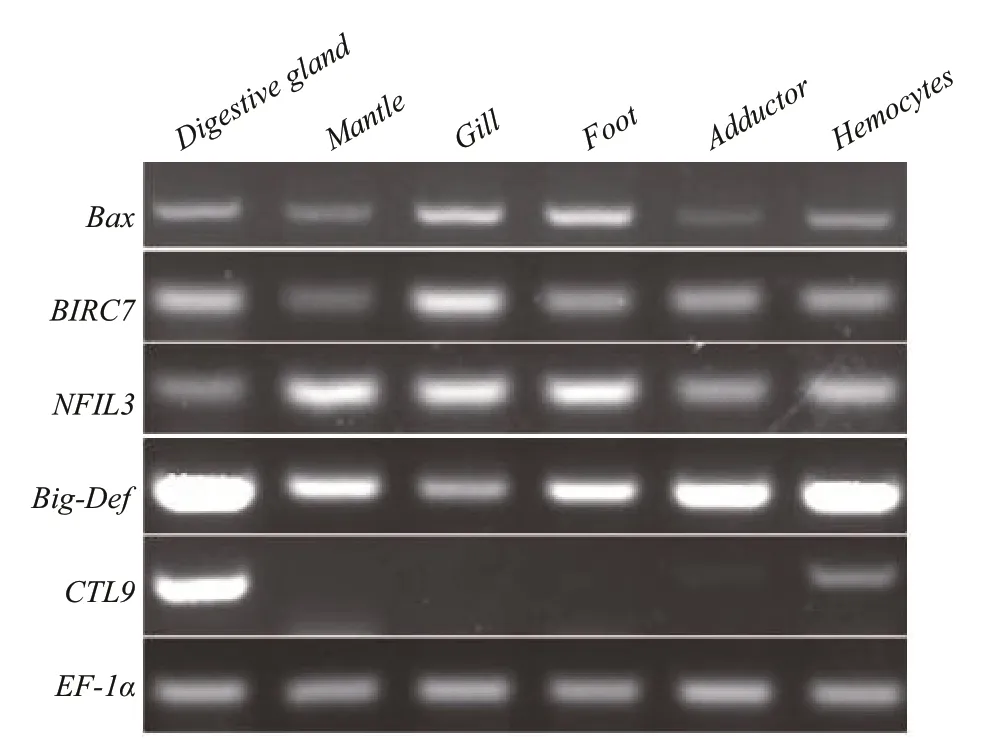
Fig.2 Expression of f ive Vibrio resistance-related genes mRNA in adult tissues
2.5 Quantitative real-time PCR
Based on a previous study (Jiang et al., 2018),5Vibrioresistance-related genes were selected for detection of the genetic parameters of gene expression levels using clams randomly selected from the 23 full sibling families by quantitative real-time PCR (qRTPCR). Theβ-actinandEF1αgenes were used as internal references to normalize the relative expression levels between samples (Jiang et al., 2017). The primers for qRT-PCR are the same used for semiquantitative RT-PCR. qRT-PCR was performed in QuantStudio 6 Flex (Applied Biosystems, USA)machine using a QuantiNova SYBR Green PCR Kit(Qiagen, Germany). Each reaction was performed in triplicate under the following conditions: 95°C for 2 min, 40 cycles of 95°C for 5 s, and 60°C for 20 s,followed by melting curve determination. The ΔCt value was used to quantify the relative gene expression value, the greater the ΔCt value, the lower the gene relative expression level (Pfaffl , 2001).
2.6 Estimation of genetic parameters
The phenotypic variance of 12-month- and 15-month-old clams were estimated using the following animal linear model:
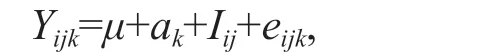
whereYijkis the phenotypic value ofkthoff spring ofithsire andjthdam;μis the mean phenotype of the sample;akis random additive genetic eff ect of clamk;Iijis the no additive eff ect due to the interaction ofithsire andjthdam; andeijkis the residual of thekthindividual ofithsire andjthdam.
For the animal linear model, heritability was calculated as follows:
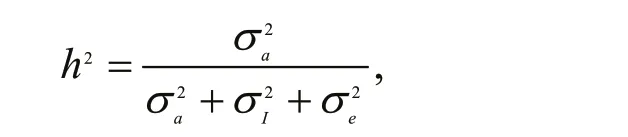
Gene transcription variance was partitioned into additive genetic variance (and the interaction between sire and damseparately. Wilcoxon signed-rank test was conducted to compare the genetic parameters of gene transcription trait between 12-month- and 15-month-old clams.
2.7 Estimation of phenotypic and genetic correlations
The phenotypic variance SW and TW were partitioned using the following animal linear model:Yijk=μ+ak+Iij+eijk, and the parameters were as described above. A bivariate linear animal model was used to estimate the variance and co-variance components for the f ive transcription traits and SW and TW. The phenotypic or genetic correlation between each paired trait was calculated as:, whereσ12is the phenotypic or genetic covariance between two traits,andare the additive phenotypic or genetic variances of trait 1 and trait 2, respectively. All the data were analyzed using the ASReml 3.0 software package in R (Gilmour et al., 2009).
3 RESULT
3.1 Tissue expression prof ile analysis
The expression levels of the f ive genes varied among diff erent tissues (Fig.2). For example,CTL9had the highest expression level in the digestive gland, relatively low expressive abundance in hemocytes and adductor muscles, and no expression in the mantle, gill, and foot. The other four genes were expressed in all six tissues, but expression levels varied among tissues. For example, the apoptosisrelated genesBaxandBIRC7both had the highest expression levels in the gills but were only moderately high in the digestive gland. The transcription factor of inf lammatory responseNFIL3had high expression level in the mantle, gill, and foot and relatively low expression in the other three tissues.Big-Defhad low expression in the gills and high expression levels in other f ive tissues, with the highest in the digestive gland. These results revealed that expression of all f ive genes could be detected in the digestive glandand hemocytes inM.petechialis. Considering that few hemocytes could be extracted from the clams, the digestive gland was selected as the major source for the following gene expression analysis.
3.2 Characteristic of the gene transcription traits
The number of observations, means, standard deviations, coeffi cients of variation (CV) for the f ive gene transcription traits (Bax,BIRC7,Big-Def,CTL9,andNFIL3) in two growth stages are shown in Table 2. The expression level diff ered among the f ive genes in digestive gland.Big-Defhad the highest relative expression level and the highest CV in 12-and 15-month-old clam group, whereas the relative expression level ofBIRC7was the lowest.Furthermore, the gene expression levels of 12- and 15-month-old clam were compared. With the exception ofBIRC7, other genes showed lower expression in 15-month-old clam group. The gene expression diff erence for f ive genes between 12- and 15-month-old clam group was 8%-23%. Compared to other genes, the expression ofBIRC7andNFIL3for 12-month-old was more similar to that of 15-month-old clam group. The result oft-test suggests that expression level of the 5 genes between 12- and 15-month old clam were statistically and signif icantly diff erent (P<0.05).
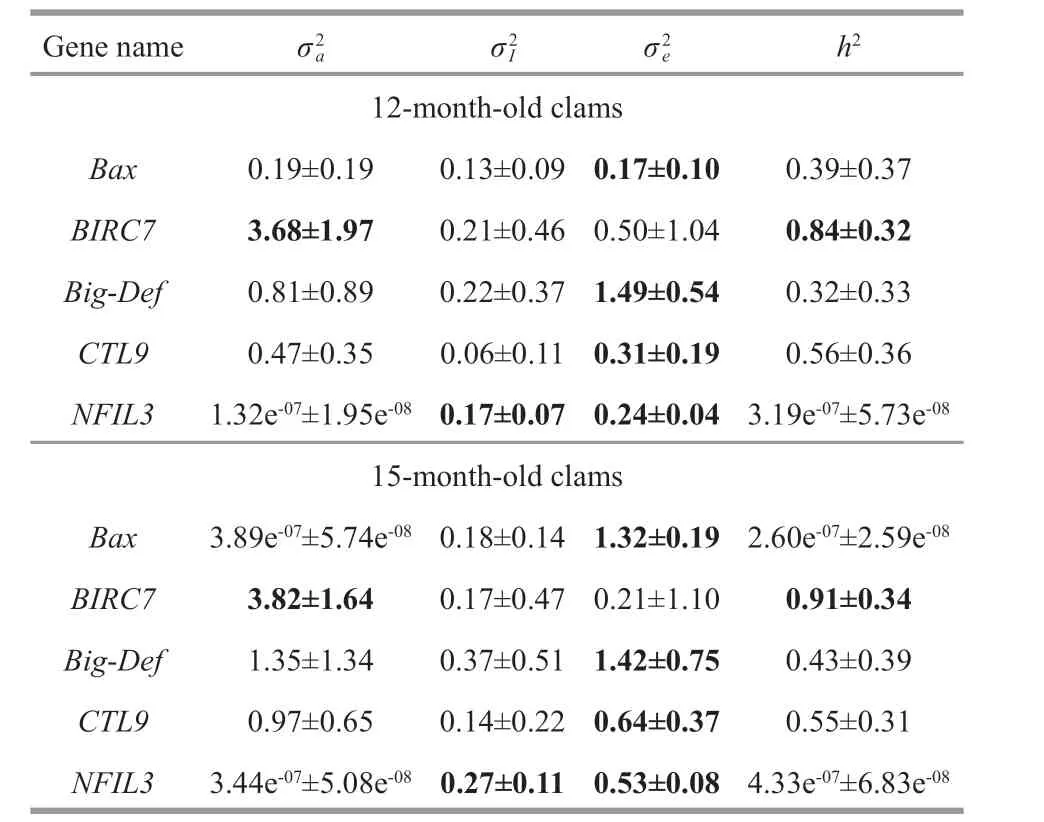
Table 3 Variance components and heritabilities of the f ive resistance-related gene expression traits(mean±standard error)
3.3 Genetic estimates of the transcription traits
Estimation of the variance components and heritability of gene transcription trait in 12-monthand 15-month-old clams are presented in Table 3. The gene transcription trait of two diff erent growth stages exhibited substantial similarities in genetic variance components. Across all f ive genes, the gene transcription traits of two stages were no signif icant diff erent in additive genetic eff ects (Wilcoxon signedrank test,P=0.31) and sire-by-dam interaction eff ects(no additive genetic eff ects) (P=0.13). In detail, theBIRC7gene showed signif icant additive genetic variations in the 12-month and 15-month-old clams,correspondingly the high heritabilities 0.84±0.32 and 0.91±0.34, respectively. Whereas, the heritabilities of other four genes (Big-Def,BAX,NFIL3, andCTL9)were low-to-moderate but not signif icantly. TheNFIL3gene showed signif icant sire-by-dam interaction eff ects.
3.4 Genetic correlation for transcription trait and body size
The phenotypic and genetic correlations were analyzed based on the body size SW, TW and expression ofBIRC7gene in the stage of 12-monthold clams. The phenotypic and genetic correlation coeffi cients between SW andBIRC7gene were 0.05±0.15, -0.04±0.44, the phenotypic and genetic correlation coeffi cients between TW andBIRC7gene were -0.18±0.15 and -0.97±0.98 respectively. No signif icant phenotypic and genetic correlations were detected betweenBIRC7gene transcription trait and body size in the 23 full sibling families.
4 DISCUSSION
The basal gene expression level could vary markedly in diff erent organs of the same animal(Engwerda and Kaye, 2000). Therefore, the choice of research organ is crucial for gene expression studies.In our experiment, all the f ive resistance-related genes were highly expressed in hemocytes and digestive gland inM.petechialis. It has been shown that hemocytes play a key role in animal immune response(Lavine and Strand, 2002). However, inM.petechialis,due to few hemocytes extracted from the clams’bodies, it is not a preferred organ at the individual level or for batch analyses. In mollusks, the digestive gland is another important tissue of the immune system because their epithelial cells can produce major immune molecules (Röszer, 2014). In addition,the digestive gland has been used as a preferred research organ inM.petechialis(Gao et al., 2012;Zou and Liu, 2016). In this study, the digestive gland is the preferred tissue for gene expression analysis considering that all f ive genes expressed in digestive gland and it could be collected conveniently.
Gene expression variation can underlie complex phenotype variations (Ayroles et al., 2009) and play important roles in the response to environmental change and stress (Crawford and Powers, 1992). It has long been thought that a large part of gene expression variation is highly heritable with an underlying polygenic architecture (Cheung and Spielman, 2002; Gilad et al., 2008). In recent years,the genetic parameter of gene transcription has been estimated in some aquatic species (Aykanat et al.,2012; Brokordt et al., 2015; Leder et al., 2015; He et al., 2017). Additionally, the heritability of diff erent gene transcription was diverse. A potential explanation is that diff erent phenotypic or transcription traits undergo diff erent strength of selection throughout an animal’s life. For example, as those closely related with the biological f itness, i.e. for survival and reproduction etc., undergo more strong selective pressures than other traits, and usually show lower heritabilities. Herein, we examined the genetic parameters of gene transcription for f iveVibrioresistance-related genes in full sibling clam families.The results showed that the heritability ofBIRC7was 0.91 and the heritability of other four genes was not diff erent from zero, which showed that diff erent gene transcription traits could exhibit dramatic variation in heritability. In previous studies, Aykanat et al. (2012)found that the heritability of four cytokine genes in Chinook salmon varied from 0.05 to 0.44, and Tedeschi et al. (Tedeschi et al., 2016) reported a higher heritability of three heat shock protein genes in sea turtles (meanh2=0.58). This presence of heritability suggests thatVibrioresistance-related gene expression variation ofBIRC7gene are heritable.Thus,BIRC7gene transcription may provide reliable genetically based markers for use in selective breeding and the performance of gene transcription can be improved by successive selection.
Genetic analysis is heavily dependent on data, a larger data set will provide a more accurate estimation of genetic parameters, but the costs used in getting the data will also be increased. No research yet has provided the evidence that appropriate sample size is required for genetic analysis. Due to the measuring procedure is complicated for transcription traits and the high cost, the sample size used in genetic analysis is usually not large. For example, Aykanat et al.(2012) used a data set including 48 families and 192 individuals to partition the transcription variation of four cytokine genes into additive genetic, nonadditive genetic, and maternal components in Chinook salmonOncorhynchustshawytscha. In our study, we used 23 full sibling families to estimate the genetic parameters of 12-month- and 15-month-old clams’transcription traits. The results showed that there was no signif icant diff erences in additive genetic eff ects and sire-by-dam interaction eff ects between two growth stages (Wilcoxon signed-rank testP=0.31,P=0.13), which indicated that the genetic variance components of gene expression traits were relatively stable during short-term growth. However, the estimation of heritability in our study was accompanied by high standard error, which are likely associated with low sample sizes, and possibly resulted in our failure to detect signif icant low-to-moderate heritabilities (Table 3). Nevertheless, we observed signif icant additive eff ects inBIRC7, which might be valuable in the genetic improvement ofVibrioresistance in artif icial selection approaches.
Animals are frequently faced with trade-off s between growth and mortality rates. Enhancement of disease resistance may become with compromise in growth traits. For instance, Yáñez et al. (2016) has reported a signif icant negative genetic correlation between resistance againstPiscirickettsiasalmonisand harvest weight in coho salmon. Furthermore, a slightly negative genetic correlation between resistance to vibriosis and harvest weight was found in Atlantic cod (Bangera et al., 2011). These results suggest that selective breeding for faster growth may have a negative eff ect on disease resistance in these species. Conversely, a signif icant positive genetic correlation between resistance to f ish pasteurellosis and body length was reported in the gilthead sea bream (Antonello et al., 2009), which may be due to resistant individuals experiencing superior growth.Additionally, genetic correlations that are not signif icantly diff erent from zero have also been found between disease resistance and harvest weight in some aquatic species (Silverstein et al., 2009; Flores-Mara et al., 2017). Results of these studies suggest that genetic correlations between disease resistance and growth of diff erent species are not uniform.Therefore, genetic correlation must be evaluated case by case in practical breeding. In this study, we found that there is no signif icant genetic correlation betweenVibrioresistance-related geneBIRC7and body sizes.Our results indicate that variation in gene transcription could help the genetic improvement ofVibrioresistance and will not inf luence body size inM.petechialis.
5 CONCLUSION
In conclusion, our results f irst demonstrate the variation in the expression of theVibrioresistancerelatedBIRC7gene inM.petechialisis heritable,indicating the feasibility of improving this trait by artif icial selection. Moreover, no signif icant genetic correlation is detected between theVibrioresistancerelatedBIRC7gene and body sizes, suggesting that artif icial selection forVibrio-resistance will not inf luence body size inM.petechialis.
6 DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study. All data generated and/or analyzed during this study are included in this published article.
 Journal of Oceanology and Limnology2020年2期
Journal of Oceanology and Limnology2020年2期
- Journal of Oceanology and Limnology的其它文章
- Erratum to: Seabed domes with circular depressions in the North Yellow Sea*
- Transcriptional responses to starvation of pathogenic Vibrio harveyi strain DY1*
- Establishment and characterization of a new cell line derived from half-smooth tongue sole Cynoglossus semilaevis kidney*
- Eff ects of f lorfenicol exposure on growth, development and antioxidant capacity of f lounder Paralichthys olivaceus larvae at diff erent developmental stages*
- A new free-living marine nematode species of Rhinema from the South China Sea*
- Sabatieria sinica sp. nov. (Comesomatidae, Nematoda) from Jiaozhou Bay, China*
