Comparing the effects of enhancing anaplerotic pathways on L-isoleucine production performance by Corynebacterium glutamicum YI
ZHU Fuzhou, LU Nan, LI Yuhong, LIN Beibei, ZHENG Yingnan,WANG Zishen, CHEN Ning, ZHANG Chenglin, 2*
1(College of Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, China)2(Linghua Group Ltd, Jining 272073, China)
ABSTRACT To clarify and compare the effects of enhancing the anaplerotic pathways on L-isoleucine production performance by Corynebacterium glutamicum. The genes of pyc and ppc were overexpressed by replacing the native promoter with Ptuf or by inserting the genes to plasmid in a L-isoleucine producer, C. glutamicum YI.The pyc overexpressed via genome-integration in ILE01 and via plasmid in ILE02 resulted in 17.3% (6.1 g/L) and 9.6% (5.7 g/L) increase in production of L-isoleucine by batch fermentation; as well as 11.7% (24.8 g/L) and 8.1% (24.0 g/L) by fed-batch fermentation. By comparison, the ppc overexpressed via the two strategies lead to 30.8% (6.8 g/L) and 13.5% (5.9 g/L), 15.8% (25.7 g/L) and 9.5% (24.3 g/L) increase by the two fermentation processes. Meanwhile, overexpression of pyc and ppc resulted in improvement of L-isoleucine yield. However, overexpression of pyc and ppc in ILE02 and ILE04 resulted in cell growth decreased. Overexpression of pyc and ppc both resulted in remarkably promotion of L-isoleucine production performance and the genome-integration overexpressing strategy was more profitable. This study first reports the effects of enhancing anaplerotic pathways on L-isoleucine production by C. glutamicum. The results would supply the reference for metabolic engineering of C. glutamicum .
Key words Corynebacterium glutamicum; L-isoleucine; anaplerotic pathways; metabolic engineering
Amino acids are commercially important as they are widely used in food, pharmaceutical, cosmetics and animal feed industries[1].L-isoleucine, belonging to theL-aspartate family amino acids (AFAAs), has been widely used in such fields as pharmaceuticals, food additives, and precursors of herbicides[2].So far,Corynebacteriumglutamicumhas been used as the main producer ofL-isoleucine[3]. As shown in Fig. 1,C.glutamicumsynthesizesL-isoleucine in a split pathway from oxaloacetate viaL-aspartate,L-threonine, 2-ketobutyrate and 2-aceto-2-hydroxybutyrate as the main intermediates. The oxaloacetate supply has been considered as a bottleneck forL-isoleucine production[4]. InC.glutamicum, oxaloacetate is mainly replenished by the anaplerotic pathways[5]. In contrast to many other organisms which have only one anaplerotic pathway,C.glutamicumpossesses both: one is catalyzed by phosphoenolpyruvate carboxylase (PEPC, encoded byppc) converting phosphoenolpyruvate to oxaloacetate, and the other is by pyruvate carboxylase (PC, encoded bypyc) converting pyruvate to oxaloacetate with biotin as a coenzyme[6].
It seems that PC and PEPC play distinct roles in amino acid synthesis byC.glutamicum. PC was predominant during the temperature triggered glutamate fermentation, whereas PEPC was necessary for glutamate production induced under biotin-limited conditions and overexpression ofppceffectively enhanced glutamate production in the similar conditions[7-8]. We previously reported thatpycoverexpression did not result in promotion of glutamate production, but in decreased pyruvate-related byproducts accumulation[9]. Overexpression ofpycinC.glutamicumincreased lysine accumulation by 50%[10],while overexpressingppcshowed only marginal effect. The amplification of PEPC activity allowed an increase in threonine production by 12%[10-11].
In summary, the roles ofpycandppcoverexpression were focused onL-glutamate,L-threonine andL-lysine so far. Nevertheless, those inL-isoleucine synthesis were unclear. In this study, the effects ofpycandppcoverexpression on synthesis ofL-isoleucine were compared.
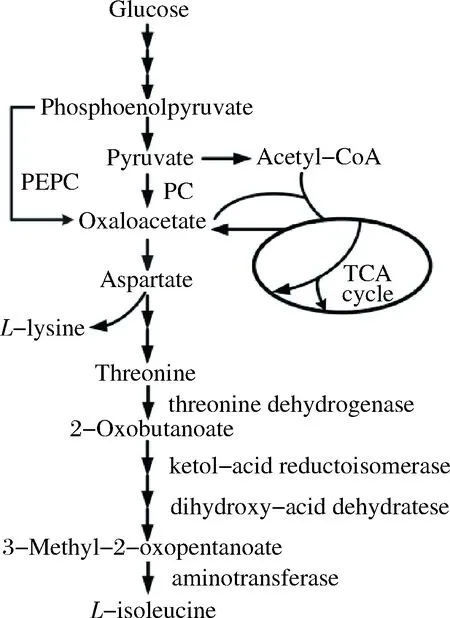
Fig.1 Biosynthesis pathway of L-isoleucine
1 Materials and methods
1.1 Materials and reagents
1.1.1 Strains, plasmids, and culture conditions
The strains and plasmids were listed in Table 1.EscherichiacoliDH5α was used for vector construction and was cultured in lysogeny broth (LB) medium.C.glutamicumstrains were cultured in LB medium supplemented with 5 g/L glucose (LBG). All the strains were grown at 32℃ in a shaker at 200 r/min. When necessary, the media were supplemented with kanamycin (25 μg/mL).

Table 1 Bacterial strains and plasmids
1.1.2 Primers
Primers used in this study were listed in Table 2.

Table 2 Primers used in research
*: Recognition sites are underlined and restriction enzymes are shown in parentheses.
1.1.3 Media
Seed medium: 50 g/L glucose, 12 g/L (NH4)2SO4, 10 mg/L FeSO4·7H2O, 1.5 g/L KH2PO4·3H2O, 0.6 g/L MgSO4·7H2O, 10 mg/L MnSO4·7H2O, 0.1 mg/L biotin, 0.4 mg/L vitamin B1.
Fermentation medium for batch fermentation: 60 g/L glucose, 15 g/L (NH4)2SO4, 10 mg/L FeSO4·7H2O, 2 g/L KH2PO4·3H2O, 0.6 g/L MgSO4·7H2O, 12 mg/L MnSO4·7H2O, 0.1 mg/L biotin, 0.5 mg/L vitamin B1.
Fermentation medium for fed-batch fermentation: 80 g/L glucose, 4 g/L (NH4)2SO4, 15 mg/L FeSO4·7H2O, 0.5 g/L MgSO4·7H2O, 15 mg/L MnSO4·H2O, 1.5 g/L KH2PO4·3H2O, 3 g/L K2HPO4·3H2O, 0.1 mg/L biotin, 5 mg/L vitamin B1, 20 mL/L soybean hydrolysate and 15 mL/L corn steep liquor.
1.1.4 Reagents
Chemicals: Sinopharm Chemical Reagent Co., Ltd. Beijing; Restriction endonucleases: Primer STAR HS DNA polymerase, RNAiso Plus kit and T4 DNA ligase, Takara Bio Inc., Dalian; Ultra-SYBR Two-Step RT-qPCR Kit: CWBIO Co., Ltd., Beijing; ClonExpressTM Ⅱ One-Step Cloning Kit:Vazyme Biotech Co., Ltd., Nanjing; Primers: GENEWIZ Bio Inc., Suzhou.
1.2 Instruments
Bio-Rad MyCycler: Bio-Rad, USA; High performance liquid chromatograph (HPLC): U3000 Thermo Fisher Scientific, USA; Stepone Real-Time PCR instrument: Applied Biosystems ABI, USA; PD-10 desalting column: GE Healthcare UK Ltd., UK; Spectrophotometer UV2600: Shimadzu, Japan; SBA biosensor analyzer SBA-40D: Institute of Biology, Shandong Province Academy of Sciences, Jinan; 5-L bioreactors: Shanghai Baoxing Bio-engineering Equipment Co., Ltd., Shanghai; ZORBAX Eclipse AAA column: Agilent, USA.
1.3 Methods
1.3.1 Construction of plasmids and recombinant strains
The primers for fragments amplification are listed in Table 2. The digestion and ligation of DNA were operated according to standard protocols[12]. The transformation ofE.coliDH5α andC.glutamicumwere by heat shock and electroporation, respectively[13].

Thepycandppcwas individually amplified fromC.glutamicumYI genomic DNA usingpyc-6,pyc-7 andppc-6,ppc-7 and was cloned into theHindⅢ site of pXT01 to construct pXT-pycand pXT-ppc; the plasmids were transformed intoC.glutamicumYI to obtain ILE02 and ILE04, respectively.
1.3.2 Real-time quantitative PCR
Total RNA was isolated from cells ofC.glutamicumstrains growing for 12 h in LBG medium according to the manufacturer’s instructions of RNAiso Plus kit and then the cDNA was synthesized. The transcription levels ofpyc(with primers ofpyc-F andpyc-R) andppc(with primers ofppc-F andppc-R) were measured and data were analyzed by the 2-ΔΔCTmethod[15]. Primers for real-time quantitative PCR were listed in Table 2.
1.3.3 Detection of enzyme activity
Enzyme activities were measured using crude cell extracts as the enzyme source.C.glutamicumYI, ILE01, ILE02, ILE03 and ILE04 cells grown in LBG medium (OD600=9-12) were collected, washed and resuspended in 10 mL of Tris-HCl (0.1 mol/L, pH 7.0) buffer. Then, the cells were disrupted by sonication for 20 min. The cell suspensions were centrifuged at 4 ℃ and 12 000gfor 20 min to remove cell debris. The supernatant was applied to a desalting column to remove salts and proteins with low molecular weight. For detection of PEPC activity, the reaction mixture includes 10 mmol/L KHCO3, 5 mmol/L MnSO4, 0.1 mmol/L acetyl-CoA, 0.15 mmol/L NADH, 2 U malate dehydrogenase, and 2 mmol/L phosphoenolpyruvate in 0.1 mol/L Tris-HCl (pH 7.8)[16]. And for PC activity, the reaction mixture contained 5 mmol/L sodium pyruvate, 5 mmol/L MgCl2, 15 mmol/L NaHCO3, 2 mmol/L ATP, 10 mmol/L KCl, 0.15 mmol/L NADH, and 2 U malate dehydrogenase in 0.1 mol/L Tris-HCl (pH 7.8)[17]. The changes in NADH concentration were detected at 340 nm. The specific activity of PEPC and PC was expressed as nmol/(min(mg protein). Concentration of total protein was determined with the Bradford method.
1.3.4 Fermentation assay
For batch fermentation, CaCO3(25 g/L) was added to the seed medium. After culturing 12 h in 500 mL shake flasks at 200 r/min and 32℃ (OD600=12-16), 3 mL of the seed culture was inoculated into 27 mL fermentation medium in 500 mL shake flasks.
The seed culture for fed-batch fermentations was the same as that for batch fermentation. 30 mL of the seed culture was inoculated into 3 L of fermentation medium. Concentration of residual glucose was kept at 20 to 25 g/L. The pH was controlled at 7.0 to 7.5 by adding 25% NH4OH. The level of dissolved oxygen was maintained at 20%-30%.
1.3.5 Analytical methods
Cell growth was monitored by measuring OD600and was converted to the dry cell weight (1 g DCW/L=0.25 OD600). Concentration of residual glucose was determined with the SBA biosensor analyzer. Amino acids in culture were quantified by HPLC using precolumn derivatisation, ZORBAX Eclipse AAA column, acetonitrile/water mixture (50∶50,V/V) with 50 mmol/L ammonium acetate as the mobile phase at the flow rate of 1 mL/min, detection wavelength of 360 nm.
1.4 Statistical analysis
Experiments were conducted in triplicates and repeated for three times. The data were averaged and presented as mean±standard deviation (SD). Significant differences were determined with one-way analysis of variance followed by Dunnett’s multiple comparison tests and statistical significance was defined atP≤0.05.
2 Results and analysis
2.1 Identification of the recombinant strains ILE01, ILE02, ILE03 and ILE04
To compare the effects of overexpressingpycandppcgene on production ofL-isoleucine, ILE01, ILE02, ILE03 and ILE04 were constructed. In ILE01 and ILE03,pycandppcwas overexpressed by replacing the native promoters with the strong and constitutive promoter Ptuf. In ILE02 and ILE04,pycandppcwas overexpressed by the plasmid driven by Ptuf. Successful construction of strains ILE01, ILE02, ILE03 and ILE04 was validated by PCR analysis (data not shown). To certify the enhanced expression ofpycandppcunder Ptufin genome or plasmids, real-time quantitative PCR together with enzyme activity assays were employed. The transcription levels ofpycin ILE01 and ILE02 were about 21- and 49-fold of that inC.glutamicumYI, respectively; and the specific activities of PC were increased by 122% and 395%. The transcription levels ofppcin ILE03 and ILE04 were approximately 20- and 52-fold of that inC.glutamicumYI, respectively, resulting in 243% and 410% improvement in the specific activities of PEPC (Table 3). Obviously, overexpression ofpycandppcin plasmids leads to higher specific activities than that in genome.
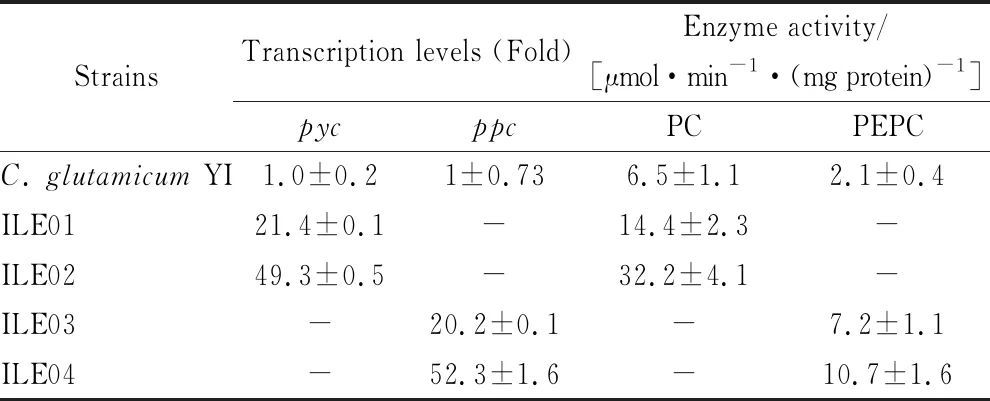
Table 3 Transcription levels of pyc and ppc and specific activities of PC and PEPC in the strains
*: -, Never detected.
2.2 Effects of pyc or ppc gene overexpression on production performance of L-isoleucine in the batch fermentations
To detect the effects ofpycorppcoverexpression on production performance ofL-isoleucine, batch fermentations were performed withC.glutamicumYI, ILE01, ILE02, ILE03 and ILE04. In comparison with the L-isoleucine production and yield byC.glutamicumYI (5.2 g/L and 0.075 g/g), the production ofL-isoleucine by ILE01 and ILE02 was increased by 17.3% and 9.6%, reaching 6.1 g/L and 5.7 g/L, respectively (Fig. 2); the yield ofL-isoleucine by the two strains was enhanced by 12.0% and 6.7%, reaching 0.084 g/g and 0.080 g/g, respectively (Fig. 2). The production ofL-isoleucine by ILE03 and ILE04 was increased by 30.8% and 13.5%, reaching 6.8 g/L and 5.9 g/L, respectively (Fig. 2); the yield ofL-isoleucine by the two strains was increased by 13.3% and 6.7%, reaching 0.085 g/g and 0.080 g/g, respectively (Fig. 2). There were no differences in the biomass of ILE01, ILE03 andC.glutamicumYI. However, biomass of ILE02 and ILE04 was decreased by 26.7% and 28.4% comparing withC.glutamicumYI, respectively.
The final concentrations of byproducts in the fermentation culture,L-alanine,L-valine,L-leucine andL-lysine were measured. Accumulations ofL-alanine by ILE01, ILE02, ILE03 and ILE04 were decreased by 33.3%, 27.8%, 50.0% and 30.6%, respectively, compared withC.glutamicumYI. Meanwhile, those ofL-valine were decreased by 24.4%, 31.7%, 73.2% and 39.0%; and those ofL-leucine were decreased by 22.7%, 40.9%, 72.7% and 50.0%, respectively. However, those ofL-lysine were increased by 28.1%, 9.4%, 43.8% and 18.8%, respectively (Fig. 3).

Fig.2 The L-isoleucine production and yield as well as biomass of strains in the batch fermentations.

Fig.3 The final concentrations of byproducts by the constructed strains in the batch fermentations
2.3 Effects of pyc and ppc overexpression on L-isoleucine synthesis in the fed-batch fermentations
To further evaluate and compare the effects of enhancingpycandppcoverexpression onL-isoleucine production, fed-batch fermentations of ILE01, ILE02, IL03, ILE04 andC.glutamicumYI (as control) were conducted in a 5-L bioreactor. Accumulation ofL-isoleucine was observed at 4 h in all the strains. The highestL-isoleucine production and yield was achieved by ILE03 (reaching 25.7 g/L and 0.162 g/g, Fig. 4), 15.8% and 24.6% higher than that byC.glutamicumYI (22.2 g/L and 0.130 g/g), respectively. Compared with that byC.glutamicumYI, production ofL-isoleucine by ILE 01, ILE02 and ILE04 was improved by 11.7%, 8.1% and 9.5%, respectively (Fig. 4). In addition, accumulation ofL-alaine,L-valine andL-leucine were all remarkably declined and that ofL-lysine was significantly increased (Fig. 5). Glucose consumption in ILE 01 and ILE03 was markedly lower than that inC.glutamicumYI (Table 4). ILE03 exhibited the lowest glucose consumption and the highest yield ofL-isoleucine (0.162 g/g). Biomass of ILE02 and ILE04 was significantly lower than that of ILE01, ILE03 andC.glutamicumYI (Fig. 6).However, the two strains exhibited the highest specific production ofL-isoleucine (1.6 g/g DCW).

Fig.4 The production of L-isoleucine by the constructed strains in the fed-batch fermentations
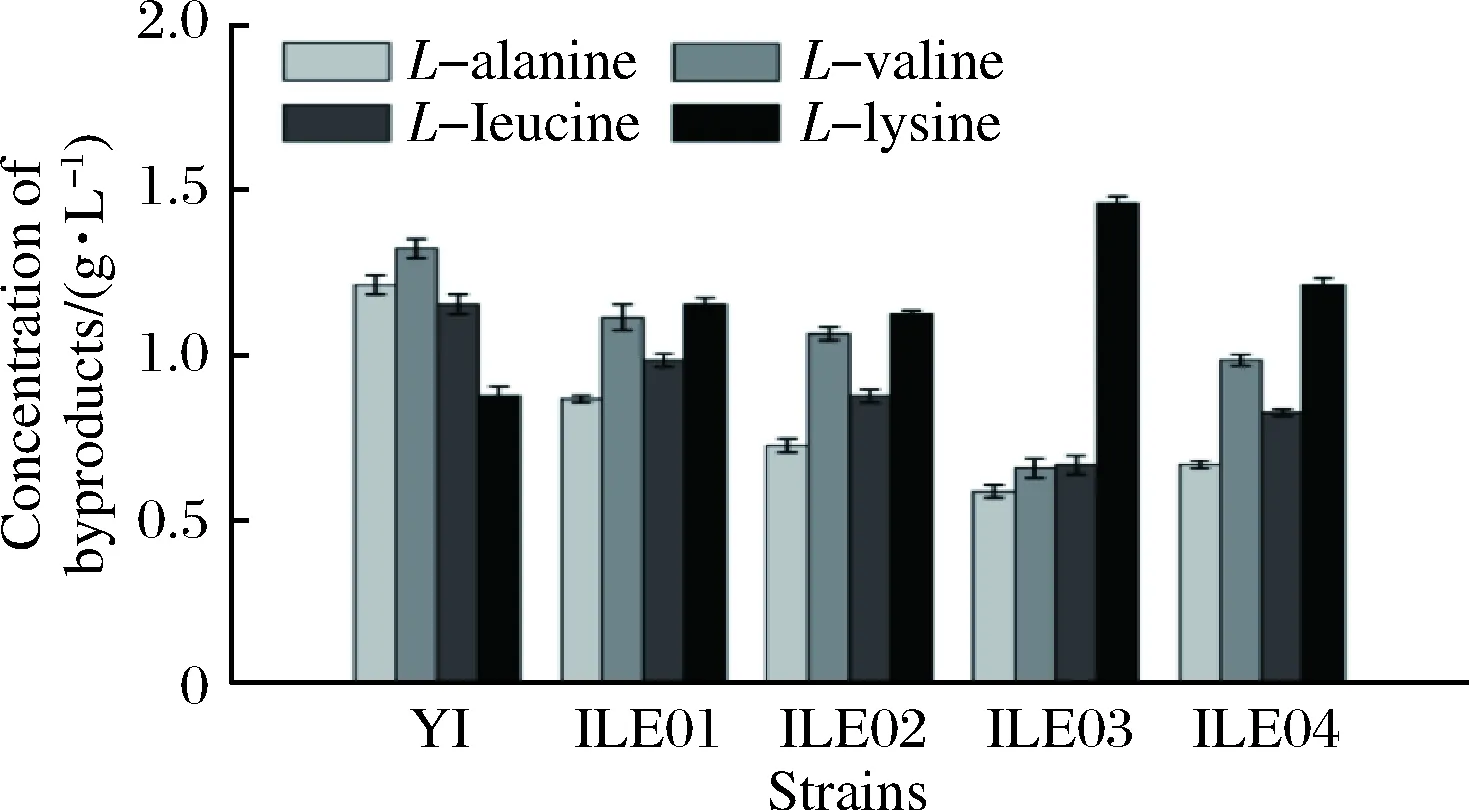
Fig.5 The final concentrations of byproducts in the fed-batch fermentations
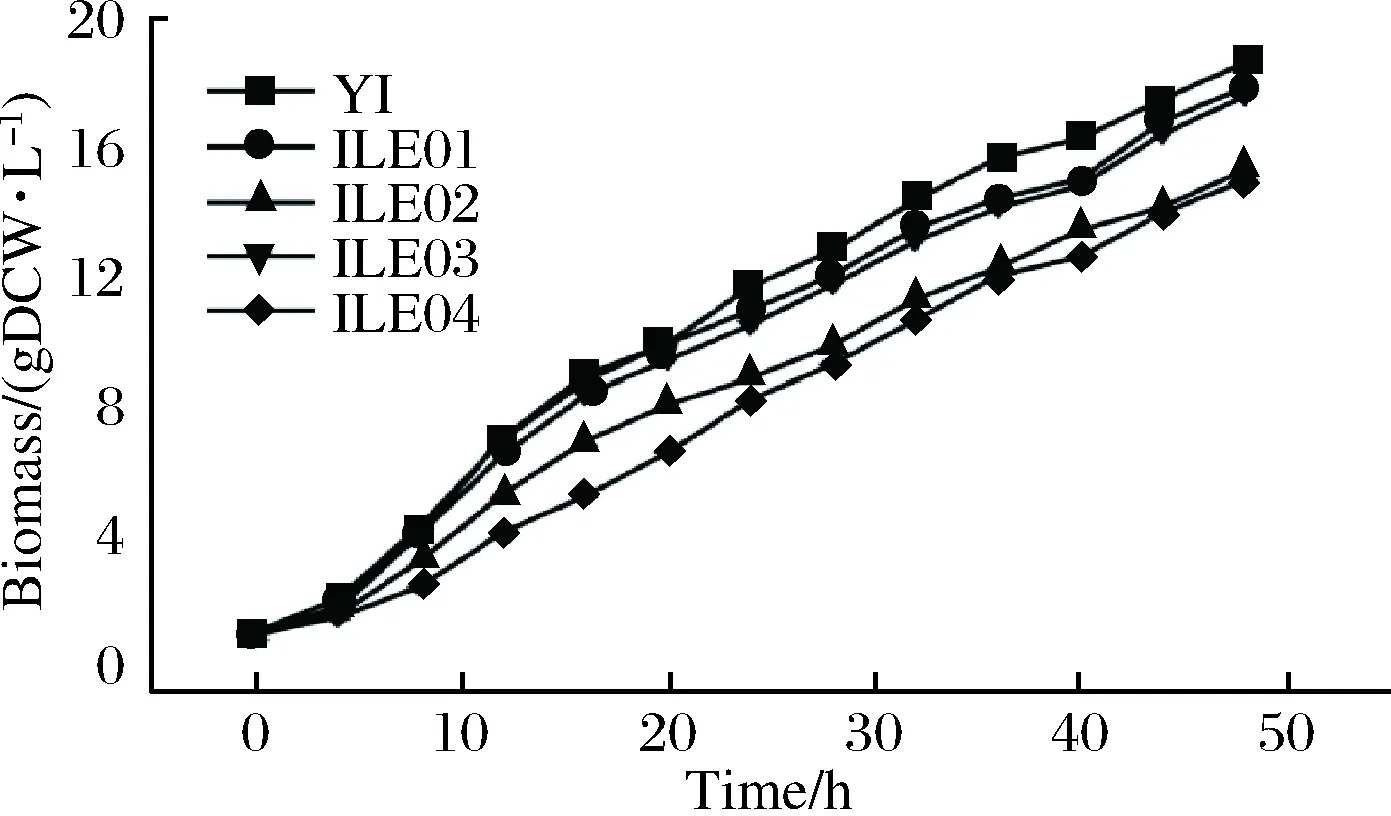
Fig.6 The biomass of the constructed strains in the fed-batch fermentations
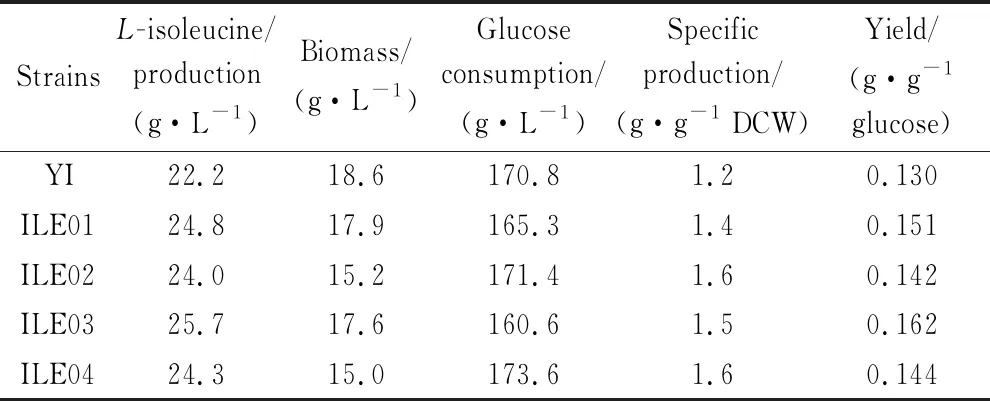
Table 4 Fermentation parameters of strains in the fed-batch fermentations
3 Discussion
Our results indicate that overexpression ofpycandppcboth resulted in remarkable promotion ofL-isoleucine production and yield. Together with the results that pyruvate-derived byproducts (L-alanine,L-valine andL-leucine) accumulations were significantly decreased and those ofL-lysine were increased, it is definitely thatpycandppcoverexpression could increase the carbon flux to oxaloacetate for biosynthesis ofL-isoleucine. But it is surprising to find that effect ofppcoverexpression was more significant, achieving a 15.8% increase inL-isoleucine production. Nevertheless, PEPC contributes only 10% of the total flux for oxaloacetate synthesis inC.glutamicumaccording to the previous report[23]. InC.glutamicum, PEPC is feedback inhibited byL-aspartate and α-ketoglutarate[24]. So far, certain mutations in PEPC have been revealed to release the feedback inhibition and contribute to increased production ofL-glutamate andL-lysine[25]. However, these reported mutations (D299N, R620G, K653G, K813G, S869G, R873G and N917G) did not happen in the PEPC ofC.glutamicumYI. Obviously, the enhancedL-isoleucine biosynthesis in ILE03 and ILE04 indicates that the PEPC is most likely to have already been deregulated of feedback inhibition and functions well to supply oxaloacetate. This may be due to the point mutations of G241D, K383N, T829R, S837A existing in PEPC ofC.glutamicumYI, which may individually or in combination lead to the desensitized feedback inhibition. The inhibited effect ofL-aspartate on PEPC from crude cell extracts ofC.glutamicumYI, ILE03 and ILE04 was tested and it was found that the inhibition was deregulated (data not shown). Furthermore,ppcfromC.glutamicumATCC 13032 was overexpressed inC.glutamicumYI, which resulted in limited production ofL-isoleucine (5.5 g/L in batch fermentation, lower than that in ILE01, ILE02, ILE03 and ILE04). These results indicate that overexpression ofppcencoding PEPC released from feedback inhibition is more beneficial for upgrading production ofL-isoleucine.
Notably, higher specific activities of PC and PEPC in ILE02 and ILE04 did not lead to promoteL-isoleucine production compared with ILE01 and ILE03 in both batch and fed-batch fermentations, but resulted in declined biomass, which might due to the burden brought by plasmids. The results indicate that the genome-integration overexpressing strategy is more suitable for metabolic engineering of strains than the plasmid-based strategy.
In conclusion, overexpression ofpycandppcresults in enhanced production and yield ofL-isoleucine, and the genome-integration overexpressing strategy is more profitable. This study first reports that expression of PEPC released from feedback inhibition, rather than PC is more advantageous for metabolic engineering modification ofL-isoleucine producing strains.

