Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat
Fang Chn, Haiyan Jia, Xiaojun Zhang, Linyi Qiao, Xin Li, Jun Zhng,Huijuan Guo,Carol Powrs, Liuling Yan, Zhijian Chang,*
aCollege of Life Science,Shanxi University, Taiyuan 030006,Shanxi,China
bThe Shanxi Province Key Laboratory of Crop Genetics and Gene Improvement, Institute of Crop Science, Shanxi Academy of Agricultural Sciences,Taiyuan 030031, Shanxi,China
cDepartment of Plant and Soil Sciences,Oklahoma State University,Stillwater, OK 74078,USA
dThe Applied Plant Genomics Laboratory of Crop Genomics and Bioinformatics Centre, Nanjing Agricultural University, Nanjing 210095,Jiangsu,China
eInstitute of Wheat Research,Shanxi Academy of Agricultural Sciences,Linfen 041000,Shanxi,China
Keywords:Map-based cloning Natural variation Bgt isolate E09 Triticum aestivum Molecular breeding
ABSTRACT Powdery mildew, caused by the biotrophic fungus Blumeria graminis f. sp. tritici (Bgt), is a prevalent disease in common wheat (Triticum aestivum L.) and causes serious yield losses worldwide. We used a map-based approach to clone the major broad-spectrum powdery mildew resistance gene PmCH1357 from wheat breeding line CH1357. PmCH1357 was mapped to a 526 kb region containing only TraesCS5D01G044600. The TraesCS5D01G044600 sequence of the susceptibility allele in Taichung 29(TC29) was identical to that in Chinese Spring, whereas the sequence of the resistance allele in CH1357 was identical to Pm2a previously cloned from the germplasm Ulka/*8Cc. The susceptibility allele in TC29 contained a 7 bp deletion in exon 1, resulting in loss of 856 of the 1277 amino acids in the predicted nucleotide-binding domain leucine-rich repeat containing Pm2a protein.PmCH1357/Pm2a sequence was also isolated from the Chinese wheat landraces and cultivars that were previously reported to possess the resistance gene Pm2b, Pm2c,PmLX66, or PmND399. The PmCH1357/Pm2a resistance allele was present in 10 of 495 accessions in core germplasm and contemporary cultivars from China and the USA. A newly developed diagnostic marker for the 7 bp InDel in the resistance gene can be used to eliminate the susceptibility allele in wheat breeding programs.
1. Introduction
Common wheat (Tritium aestivum L., 2n = 6x = 42, AABBDD) is ubiquitously threatened by powdery mildew, a foliar disease caused by the biotrophic fungus Blumeria graminis f. sp. tritici(Bgt),which causes significant losses in grain yield worldwide.This disease is more prevalent in crops grown under high fertility conditions[1,2].Average yield losses from this disease range from 5% to 8% but can exceed 35% under conditions of severe infection[3].Susceptible varieties have occupied more than 80%of the wheat area in China over the past decade[4].The area affected by the disease in China in 2017 was estimated at 8 million ha [5]. Grain production losses during 2006-2015 were 200,000-600,000 metric tons annually [6].Resistant varieties are the most effective and preferred means of control of the disease, and a wide diversity of powdery mildew resistant genetic resources have been exploited by breeders.
More than 140 genes and quantitative trait loci (QTL)have been reported to confer resistance to powdery mildew.These include 83 formally and 59 temporarily designated genes/QTL[7,8]. Many of these genes were characterized in diploid and tetraploid wheat species or putatively transferred from relative species. Most of the described resistance genes are effective against only some Bgt isolates [9], and only a few have been cloned,including Pm3b[10],Pm8[11],Pm21[12-14],Pm60[15], and Pm2a[16].
The Pm3b gene on chromosome 1AS was the first cloned powdery mildew resistance gene. It was cloned by chromosome walking using genetic resources available for diploid and tetraploid wheat species [10]. Pm3b encodes an NLR protein comprising a nucleotide-binding (NB) domain and a leucine-rich repeat(LRR)domain. The susceptible Pm3b allele differed from the resistance allele by a single base-pair deletion in the coding region [10], and availability of its sequence facilitated identification of 17 functional Pm3 alleles in wheat, some of which have been used in wheat improvement[17-20].Pm60,conferring the resistance to Bgt isolate E09[15], a previously prevalent race in North China [21], was cloned from diploid T. urartu Thumanjan Ex Gandilyan(accession PI 428309), the donor of the A subgenome of common wheat; its susceptible allelic variant lacked an 80-amino acid motif[15].
Unlike Pm3b and Pm60, which were cloned from Triticum species accessions, two other Pm resistance genes were cloned from chromosomes of wheat relative species. Pm8 had been introgressed from cereal rye (Secale cereale)chromosome 1RS into wheat and was cloned by homology-based cloning and subsequent physical and genetic mapping [11].Pm21, located in a 6VS segment from Dasypyrum villosum,currently confers a high level of resistance to all known Bgt races [12]. Initially, several different genes, including Stpk-V and DvUPK, were suggested as candidates for Pm21; but recently it was shown to encode a protein with coiled-coil and NB-LRR domains by an integrative resistance gene analog(RGA)strategy combined with comparative genomics[13].The identity of Pm21 was validated in an independent study involving map-based cloning[14].
Pm2a, located on chromosome 5DS, was cloned using a new method called mutant chromosome sequencing(MutChromSeq), in which mutated chromosomes are sequenced and compared to the wild-type chromosomes to identify the target gene [16]. AvrPm2 from the specialized forms of wheat and rye powdery mildew fungus were identified to encode an RNase-like avirulence effector [22].Many other powdery mildew resistance genes have been mapped to chromosome arm 5DS of resistant Chinese wheat cultivars, landraces, and breeding lines, including Pm2b in KM2939 [23], Pm2c in Niaomai [24], PmLX66 in Liangxing 66[25],PmND399 in Nongda 399[26],PmW14 in Wennong 14[27],PmYB in Yingbo 700 [28], and PmZ155 in Zhongmai 155 [2]. In addition, genes MlBrock [29], PmFG-1 [5], Pm46/Pm48 [30,31],and PmSub [32]were reported to be located on chromosome arm 5DS of the British cultivar Brock, French cultivar FG-1,German cultivar Tabasco, and European cultivar Substil,respectively. These various studies reported a few molecular markers for mapping, but no marker was developed for specific identification of Pm2a. It was not known whether Pm2a was allelic with any of these genes.We decided the best way to solve these issues was to clone the genes that had been mapped to chromosome 5DS by positional cloning.
Although positional cloning had been successful for some genes[33],it is a challenging procedure due to the large genome size and polyploid nature of the common wheat. Using the recently released wheat genome sequence [34]and newly developed high-throughput single nucleotide polymorphism(SNP)genotyping platform[35]we performed positional cloning of PmCH1357 on chromosome arm 5DS in the wheat line CH1357. We found that PmCH1357 was identical to Pm2a; we expanded knowledge of this gene by exploring variation in its sequence among 495 wheat genotypes. We found that Pm2a was present in several Chinese cultivars/lines and developed a diagnostic marker for breeding programs.
2. Materials and methods
2.1. Plant materials
CH1357 was resistant to powdery mildew, whereas the common wheat cultivar ‘Taichung 29' (TC29) was susceptible[36]. TC29 was crossed with CH1357 and the F1hybrid was backcrossed with TC29 to yield a BC1F1population.The F2,F2:3,and BC1F1populations were phenotyped for powdery mildew response. The F2population was also used for preliminary mapping of the powdery mildew resistance gene(s),and the F2plants and F2:3families that were heterozygous at the resistance locus were further used to generate a cloning population of 2252 F3:4plants.
A diagnostic marker for identifying the presence or absence of the resistance allele was used to genotype three sets of genetic and breeding materials. The first set included 261 accessions from the Chinese mini-core collection; it comprised 157 landraces, 88 modern cultivars, and 16 introduced lines[37].The second set included 164 contemporary Chinese cultivars/lines, and the third set included 70 varieties from the southern Great Plains of the USA used in a Wheat Coordinated Agricultural Project[38,39].
2.2. Evaluation of powdery mildew response
The powdery mildew responses of 14 F1, 98 BC1F1, and 483 F2plants, and 483 F2:3families were evaluated. These assays were determined in a glasshouse,with a 12 h photoperiod set at 22 °C during the day and 16 °C at night, and 65%-70%relative humidity. Seeds were germinated on filter paper in darkness at 4 °C, and the germinated seedlings were transferred into soil.When the first leaves were fully unfolded,the seedlings of each plant were inoculated by brushing them with fresh Bgt conidiospores.
Thirty-two Chinese Bgt isolates were used for inoculation of seedlings(Table 1).These isolates were kindly provided by Dr. Diaoguo An, Center for Agricultural Resources Research,Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, China. Bgt isolate E09 was used to inoculate the F1and segregating materials. This isolate was known to be virulent on control lines with Pm1a,Pm3a, Pm3b, Pm3c, Pm3d, Pm3e, Pm3f, Pm5a, Pm6, Pm7, Pm8,Pm17,Pm19,and Pm40,but avirulent on lines with Pm1c,Pm2a,Pm4a,Pm4b,Pm4c,Pm5e,Pm12,Pm16,Pm20,Pm21,Pm24,Pm33,and Pm43[40-43].
Host reactions visually evaluated at 12-14 days postinoculation when conidia were fully developed on susceptible controls. Infection types were recorded on a 1 to 6 scale [44],where 1 = immune(no visible symptoms);2 = nearly immune(showing hypersensitive necrotic flecks); 3 = highly resistant(minute colonies); 4 = moderately resistant (moderate-size pustules with chlorosis); 5 = susceptible (large size conidia without necrosis or chlorosis), and 6 = highly susceptible(large conidia lacking necrosis or chlorosis) (Fig. S1). Tests on F2:3families were repeated three times with 10 seedlings per family in each test.
A total of 495 germplasm accessions were tested for reaction types to Bgt isolate E09 and were genotyped at TraesCS5D01G044600;these included a mini-core collection of 261 Chinese accessions(Table S1),164 contemporary Chinese cultivars/lines (Table S2), and 70 wheat cultivars/lines from the USA(Table S3).
2.3. Molecular mapping of SSR and SNP markers
Genomic DNA was extracted from the parental lines and individual progeny plants using a previously described method, and a PCR for each marker was performed using a reported protocol[36].Previously reported CFD and GWM SSR markers [45]were used to genotype the wheat populations.The BWM series SSR markers on chromosome 5D used to test the parental lines and the contrasting DNA bulks were also described previously[46].
A 90 K SNP array of 81,587 single nucleotide polymorphism(SNP) markers was further used to test F2:3family bulks and parents in order to increase marker density in the targeted PmCH1357 region [47]. Based on the SNPs and putative genes identified, six markers specific to chromosome 5DS weredeveloped, including SXAC-UP10, SXAC-UP12, SXAC-UP21,SXAC-DN1,SXAC-DN3,and SXAC-DN7(Table 2).The sequences were used as queries to search the IWGSC CS wheat scaffold sequences of chromosomes 5A, 5B, and 5D (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php). The BLASTN search was limited to the top hit with an E-value cut-off of at least 1E-10. The resulting scaffolds were used to design chromosome 5D-specific primers to that were used to amplify DNA fragments covering the SNP marker sequence regions.The amplified PCR products were sequenced to confirm their location on chromosome 5D.
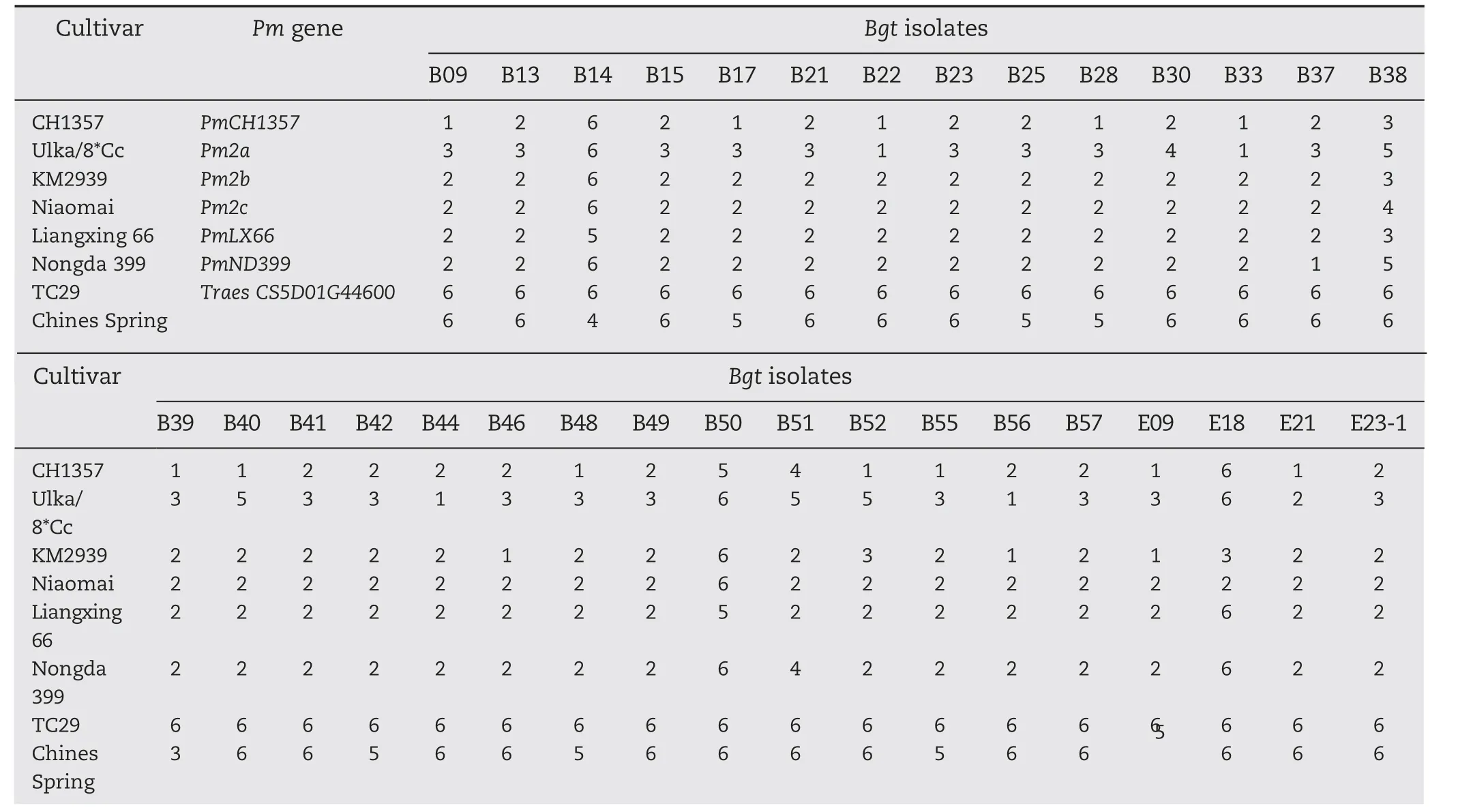
Table 1-Infection types produced by wheat cultivars/lines when inoculated with 32 Chinese Bgt isolates including E09.
Variation in powdery mildew response between the parental cultivars facilitated the mapping of the resistance trait based on segregation in the mapping population.Heterozygous alleles outside the targeted region were used to determine the chromosomal location of the resistance gene.A total of 16 SSR and SNP markers were assembled in a linkage group using JoinMap 4.0 [48].
2.4. Identification of TraesCS5D01G044600 alleles
The sequences associated with the two closest markers flanking PmCH1357 were used as queries in search of the CS chromosome 5D sequence available from IWGSC to determine the boundaries of the interval. The gene annotation information for the interval was then retrieved from IWGSC RefSeq v1.0 annotation (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/).
The TraesCS5D01G044600 gene on chromosome 5DS was associated with variation in powdery mildew response in the mapping populations.Specific primers for different alleles of the gene were used to amplify genomic DNA fragments of TraesCS5D01G044600 from CH1357 and TC29. Primers PmCH1357-F1 (5′-CTCCACACAACTGCAGCTCCAGT-3′) and PmCH1357-R1 (5′-CGTTTCTTCCACTAGTGCAACGACGACT-3′) were specific to PmCH1357/Pm2a, whereas primers PmCH1357-F2 (5′-CCATTTCTCCCTCTCCACAGCTT-3′) and PmCH1357-R1 were specific to the TraesCS5D01G044600 allele in TC29.PCR was performed using Taq DNA polymerase(New England BioLabs, Ipswich, MA, USA) and the following thermal cycling conditions: initial denaturing at 95 °C for 5 min;40 cycles of 95 °C for 30 s,55 °C for 30 s,and 72 °C for 4 min; with a final extension at 72 °C for 10 min. PCR products with the expected size were either directly sequenced or purified,cloned,and then sequenced.
PmCH1357 sequences were used as BLASTN queries against assembled chromosome sequences of T. tauschii, with an E-value cut-off of at least 1E-10, to determine the physical position of the markers on chromosome 5DS[49].
2.5. Expression of PmCH1357
Total RNA was extracted from the shoots and roots of wheat plants using TRIzol reagent (Thermo Fisher Scientific, Waltham,MA,USA)according to the manufacturer's instructions.One μL of RNA was treated with DNase I (Thermo FisherScientific), and cDNA was synthesized using a RevertAid Premium Reverse Transcriptase kit(Thermo Fisher Scientific)with random primers p(dN)6. qRT-PCR was carried out using SG Fast qPCR Master Mix(High Rox;Thermo Fisher Scientific)in an ABI Stepone Plus Thermocyler (Thermo Fisher Scientific). A two-step cycling program was performed to test the expression of TraesCS5D01G044600, with an initial denaturation step at 95 °C for 3 min,followed by 45 cycles at 95 °C for 30 s,and 60 °C for 30 s.

Table 2-Newly developed SSR markers detected polymorphism between the mapping parents.
Primers SXAC-Fn (5′-TTGGAACCACTACAACGGGAC-3′)and SXAC-Rn (5′-CCTTTCGACTCCCATCACCT-3′) were designed to amplify a 213 bp fragment of TraesCS5D01G044600 cDNA.The CDCP gene was used as endogenous control(CDCPF: 5′-CAAATACGCCATCAGGGAGAA-3′ and CDCP-R: 5′-GCTTCAGGGTTGTCCTTCCTC-3′) [50]. Gene transcript levels were described using values calculated by the 2-ΔΔCTmethod[39], where CT is the threshold number of cycles.All samples were subjected to six technical/biological replicates. The mean transcript level was statistically assessed using an analysis of variance(ANOVA),and a t-test was used to assess the difference between the two alleles.
3. Results
3.1. Powdery mildew reactions in CH1357 and TC29 and mapping of the gene conferring resistance to Bgt isolate E09
CH1357 was(nearly)immune(IT 1-2)to 27 isolates(including E09),highly resistant(IT 3)to isolate B38,moderately resistant(IT 4)to B51,and susceptible(IT 5-6)to three isolates(B14,B50,and E18). In contrast, TC29 was susceptible to all the Bgt isolates(Table 1).
Isolate E09 was used in all subsequent powdery mildew tests.CH1357 × TC29 F1plants were resistant(IT 1-2)indicating dominance,or near dominance,of resistance.Segregation of the BC1F1was 51 resistant: 47 susceptible consistent with an expected 1:1 ratio (χ2= 0.163, P = 0.686, Table 3). The F2population segregated 351 resistant (IT 1-4): 132 susceptible(IT 5-6) plants and the derived F2:3families were distributed 120 homozygous resistant: 231 segregating: 132 homozygous susceptible, clearly supporting segregation at a single locus(χ2= 1.509, P = 0.470, Table 3). The resistance allele was designated PmCH1357.
3.2. Mapping of PmCH1357
In an initial mapping effort we screened 767 simple sequence repeat (SSR) markers for polymorphism between CH1357 and TC29;406 markers(52.9%)were polymorphic between the two lines,but only three markers,Xcfd18,Xgwm190,and Xcfd81 on chromosome arm 5DS, showed an association with the differences in powdery mildew response between both the parents and contrasting DNA bulks(Fig.S2).Next,we used the 91 BWM series SSR markers on chromosome arm 5DS to test the parents and contrasting DNA bulks.Five markers,Xbwm8,Xbwm9,Xbwm20,Xbwm21,and Xbwm25,mapped in the same linkage group as Xcfd18, Xgwm190, and Xcfd81 (Fig. S2). The data showed that PmCH1357 was in chromosome arm 5DS.
To increase the density of molecular markers in the targeted PmCH1357 region, we tested the parents and the contrasting DNA bulks using the 90 K SNP array. A total of 8407 SNP markers were polymorphic and among them SNP43494377,SNP43556851 and SNP44567004 were associated with PmCH1357 (Table 2). We also developed five new SSR markers (Table 2, Fig. S2): SXAC-5DS-1, SXAC-5DS9, SXAC-5DS17, SXAC-5DS18, and SXAC-5DS-27. The 16 SSR and SNP markers and PmCH1357 formed a linkage group covering a genetic distance of 35.4 cM. The PmCH1357 locus was delimited to a 7.0 cM interval flanked by markers Xbwm8 and Xcfd81(Fig. 1).
3.3. Positional cloning of PmCH1357
In the initial mapping population of 483 F2:3lines PmCH1357 was mapped and placed between markers Xbwm8 and SXAC-5DS1(Figs.1,2-a).Line#156 was heterozygous at SXAC-5DS1,homozygous at Xbwm8, and not segregated in resistance.Therefore, PmCH1357 was located on the proximal side of Xbwm8 (Fig. 2-a). Forty-five crossovers were identified between Xbwm8 and PmCH1357, but only one (line #215) was identified between PmCH1357 and SXAC-UP21(Fig.S3), a newSNP marker at physical position 43,300,735(Fig.2-a).Line#215 was homozygous at Xbwm8,heterozygous at SXAC-5DS1,and segregated in phenotype, placing PmCH1357 distal to Xbwm8(Fig. 2-a). In the physical map (Fig. 2-b) PmCH1357 was between SXAC-UP21 at position 43,301,997 and SXAC-5DS1 at position 43,971,840, hence within a physical interval of 669,843 bp according to the IWGSC CS genomic sequence(GenBank accession number LS992094).

Table 3-Numbers of plants/lines in response to Bgt isolate E09 in six generations(P1,P2,F1,F2,BC1F1,and F2:3)of the cross between TC29 and CH1357.

Fig.1-Genetic linkage map of the PmCH1357 locus on 5DS.A population of TC29 × CH1357 F2:3 lines was inoculated with the Bgt isolate E09,and the PmCH1357 locus is indicated by the red rectangle.Molecular markers flanking the PmCH1357 locus are indicated on the right side of the chromosome,and genetic distance(cM)between Xcfd18(the first marker on the top)and each of the mapped markers are indicated on the left side of the chromosome.
To narrow down the 669,843 bp region containing PmCH1357 we generated 2252 F3:4individual plants to screen for new crossovers between SXAC-UP21 and SXAC-5DS1. Fifteen crossovers were identified between the two flanking markers. We developed four internal markers,SXAC-UP10,SXAC-UP12,SXACDN1,and SXAC-DN3(Table 2)for fine mapping of all crossovers in the F2:3and F3:4populations. Fourteen of the crossovers were mapped to a region of 81 kb between SXAC-UP21 and SXAC-UP12;however, no crossovers were observed within a region of up to 565 kb between SXAC-5DS1 and TraesCS5D01G044600(located at 43,406,783 bp) (Fig. 2-b). This indicated a very uneven crossover distribution in the genomic region containing PmCH1357.
Two crossovers occurred between SXAC-UP10 and TraesCS5D01G044600 were represented by F3:4lines #10 and#77 (Fig. 2-b). The progeny derived from these lines were homozygous at SXAC-UP10 and heterozygous at TraesCS5D01G044600 but were segregating for powdery mildew response; therefore, PmCH1357 was co-segregating with TraesCS5D01G044600. The comparative phenotypes of typical plants carrying the homozygous resistance or susceptibility alleles are shown in Fig. 2-a. We also determined the precise location of the crossover in line#156 using additional markers.It was located between SXAC-DN3 and SXAC-DN7 representing TraesCS5D01G044700 (Fig. 2-b). TraesCS5D01G044700 was excluded as the PmCH1357 candidate because it was homozygous at SXAC-DN3 and heterozygous at SXAC-DN7(Fig.2-b),but the phenotype of its offspring was non-segregating for powdery mildew response.
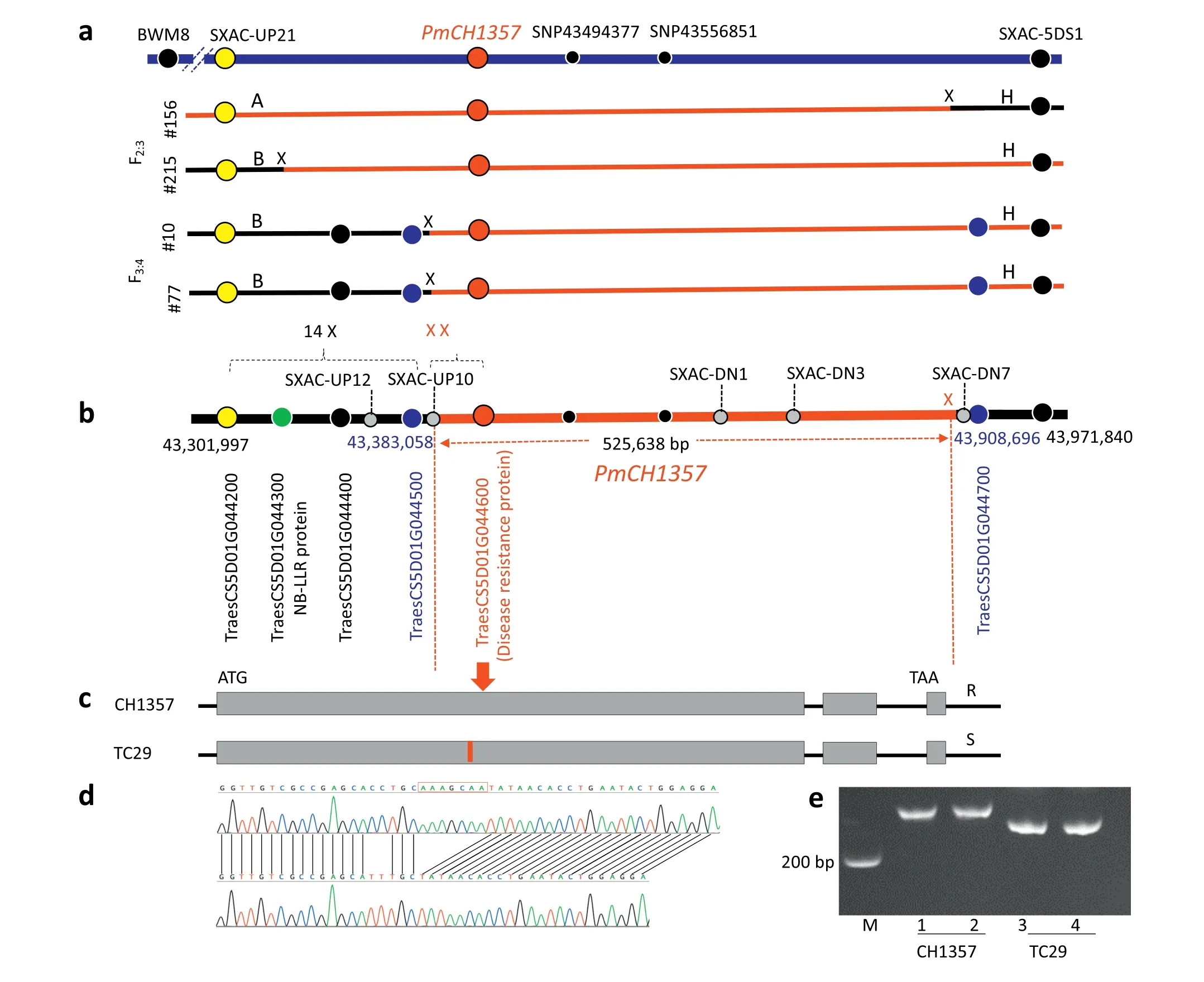
Fig.2- Cloning and allelic variation of PmCH1357.a) Diagrammatic representations of the phenotypes(reactions to the Bgt isolate E09)and genotypes of four critical recombinant lines with crossovers. ‘A',CH1357 allele; ‘B',TC29 allele; ‘H',heterozygous.Three candidate genes are highlighted with circles.‘X' indicates a crossover detected between neighboring markers,‘14 X'represents 14 crossovers between SXAC-UP21 and SXAC-UP12.b) Physical map of the PmCH1357 region.PmCH1357 was delimited by markers for genes TraesCS5D01G044500 and TraesCS5D01G044700.The markers used for fine mapping included SXAC-UP12,SXAC-UP10,SXAC-DN3,and SXAC-DN7.c)Aallelic variation in candidate gene TraesCS5D01G044600.The 7 bp deletion in the first exon was used to develop a diagnostic marker.Exons are depicted by boxes;introns and untranslated regions are indicated by lines.R,resistant phenotype; S,susceptible phenotype.d)Chromatogram images of the 7 bp InDel region in the TraesCS5D01G044600 alleles in CH1357(upper)and TC29(lower).e)The 7 bp InDel produced a diagnostic marker.Primers TaPM2-F1(5′-GGTTTGAATCCAAGAGATGATGCATATT-3′)and TaPM2-R1(5′-TAAGTGGTAATAGCTCAATCTGAGAA-3′)were used to amplify the 219 bp(CH1357)and 212 bp (TC29 fragments).The PCR products were separated in a 6%acrylamide/bisacrylamide gel(19:1)at 300 V for 2.5-3.0 h.
PmCH1357 was finally mapped to the region between TraesCS5D01G044500 and TraesCS5D01G044700, in a 525,638 bp genomic region based on the CS genome sequence(Fig. 2-b). Only one candidate gene, TraesCS5D01G044600, was annotated within this region (Fig. 2-b) and its annotation is that it encodes an NBS-LRR resistance protein of 421 amino acids. We concluded that TraesCS5D01G044600 was the only reasonable candidate for PmCH1357.
3.4. Allelic variation in TraesCS5D01G044600 between CH1357 and TC29
We amplified the complete sequence of TraesCS5D01G044600 from both CH1357 and TC29 using a single PCR. The genespecific primers PmCH1357-F2 and PmCH1357-R1 were designed based on the CS genomic sequence. The PCR product amplified from TC29 was the expected size, and sequencing revealed that it contained the same TraesCS5D01G044600 sequence as CS. However, despite excellent quality of the TraesCS5D01G044600 sequence amplified from TC29,the same primers failed to amplify a product in CH1357.
TraesCS5D01G044600 was used to search for allelic sequences GenBank. It was found to share 98% identity with gene accession LN999386, the Pm2a sequence cloned using a MutChromSeq approach[16].The Pm2a gene present in wheat line Ulka/8*Cc (CI 14118) consisted of three exons and two introns and was predicted to encode a protein of 1277 amino acids(GenBank accession number CZT14023).We next tested whether CH1357 contained the Pm2a sequence.
Primers PmCH1357-F1 and PmCH1357-R1 were designed to amplify Pm2a, and a PCR product of the expected size was amplified from CH1357. Sequencing showed that PmCH1357 contained the same coding sequence as Pm2a in Ulka/8*Cc.The directly sequenced PCR product was of excellent quality,indicating that CH1357 contains a single copy of Pm2a.
Next, we tested whether Pm2a in CH1357 was allelic to TraesCS5D01G044600 in TC29. Their sequences were identical, except that the TC29 susceptibility allele contained a 7 bp deletion that caused a premature stop codon in exon 1(Fig. 2-c, d). The TC29/CS allele was predicted to encode a truncated protein comprising 421 amino acids [22]. In comparison with the predicted 1277-amino-acid protein in CH1357, the truncated TC29 protein lacked most of the NBS domain and all of the LRR domain (Fig. 2-c, d). We developed a diagnostic marker for the 7 bp insertion/deletion (InDel) in TraesCS5D01G044600 to distinguish between the CH1357 and TC29 alleles (Fig. 2-e). The 7 bp insertion in TraesCS5D01G044600 co-segregated with the resistance gene in CH1357. We concluded that PmCH1357 was identical to Pm2a, and was allelic to TraesCS5D01G044600 in TC29.
The expression of the CH1357 allele but not the TC29 allele of TraesCS5D01G044600 was induced by Bgt isolate E09(Fig. 3-a). Before inoculation, the gene showed similar low levels of expression in both CH1357 and TC29;however,the expression of the CH1357 allele showed a 3.5-fold increase at 24 h postinoculation and a 13.6-fold increase at 30 post-inoculation,whereas the TC29 allele showed similar expression levels before and after inoculation (Fig. 3-b). The result supported the finding that the CH1357 allele of TraesCS5D01G044600 conferred resistance to isolate E09.
3.5.Identities of previously designated alleles at the Pm2 locus
We sequenced TraesCS5D01G044600 in each of four accessions that were previously reported to carry Pm2 alleles distinguished by differential responses to arrays of Bgt isolates.Primers PmCH1357-F1 and PmCH1357-R1 specific to PmCH1357/Pm2a successfully products from KM2939 (Pm2b),Niaomai (Pm2c), Liangxing 66 (PmLX66), and Nongda 399(PmND399). Conversely, primers PmCH1357-F2 and PmCH1357-R2, specific to the TC29 allele of TraesCS5D01G044600, failed to amplify any sequence from these cultivars/lines. These results showed that all five lines carried PmCH1357/Pm2a(Fig.4).
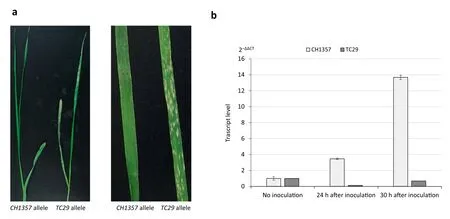
Fig.3-Phenotypes of plants containing different alleles of TraesCS5D01G044600 and TraesCS5D01G044600 expression patterns following inoculation with Bgt isolate E09.a)Phenotypes of inoculated plants containing the CH1357 and TC29 alleles.b)Induced expression of TraesCS5D01G044600 following infection.Transcript levels were determined by the 2-ΔΔCT method,where CT is the threshold number of cycles.The bars represent the mean expression levels relative to that of the endogenous control gene CDCP(n = 6). Error bars indicate standard errors.

Fig.4-Comparison of TraesCS5D01G044600 proteins in various wheat cultivars and Ae.tauschii.The Pm2a protein previously reported in Ulka/*8Cc consisted of NB-ARC and LRR conserved domains.This protein was identical those in CH1357,KM2939,Niaomai,Liangxing 66,and Nongda 399.TC29 has the same truncated TraesCS5D01G044600 protein sequence as CS,whereas the Ae.tauschii accession AL8/78 has a different truncated protein.X indicates the same point mutations in Chinese Spring and Ae.tauschii.* indicates different point mutations in Ae.tauschii relative to wheat cultivars. Red dots indicate missing amino acids when compared with other cultivars.
The CH1357 TraesCS5D01G044600 sequence was used to search the Aegilops tauschii genomic sequence database. The orthologous gene was located on chromosome 5D, with the start codon at position 46,999,777 and the stop codon at position 47,004,133. The orthologous gene in this diploid wheat accession encoded a truncated protein of 513 amino acids,due to the presence of a premature stop codon in exon 1(Fig. 4).
When inoculated with the 32 Bgt isolates, CH1357 and wheat cultivars/lines carrying the putative Pm2 alleles showed similar overall responses but with slightly different infection types reactions whereas TC29 and CS were susceptible to all isolates(Table 1).
3.6. Distribution of the CH1357 allele of TraesCS5D01G044600 in wheat cultivars
We used the diagnostic marker TaPM2 for the 7 bp InDel(Table 4) in TraesCS5D01G044600 to facilitate identification of powdery mildew resistance in the genotypes used in wheat breeding. We genotyped the 261 accessions that wereextensively used as a core collection for Chinese breeding programs, and phenotyped them using the isolate E09 (Table S1). Most (253) of these accessions were susceptible and had the 7 bp deletion.Among the eight resistant accessions Xingyi 4 (ZM023315), Hongchunmai (ZM005294) and Hongdongmai-1(ZM005188) were predicted to contain the CH1357 resistance allele on the basis of amplification product,whereas the other five (Fuzhuang 30 (ZM017213), Huomai (ZM020632), Guinong 10 (ZM023371), Yanzhan 1 and Zijiehong (ZM002272)) were predicted to carry other resistance genes since they generated no amplification product from the CH1357 resistance allele.
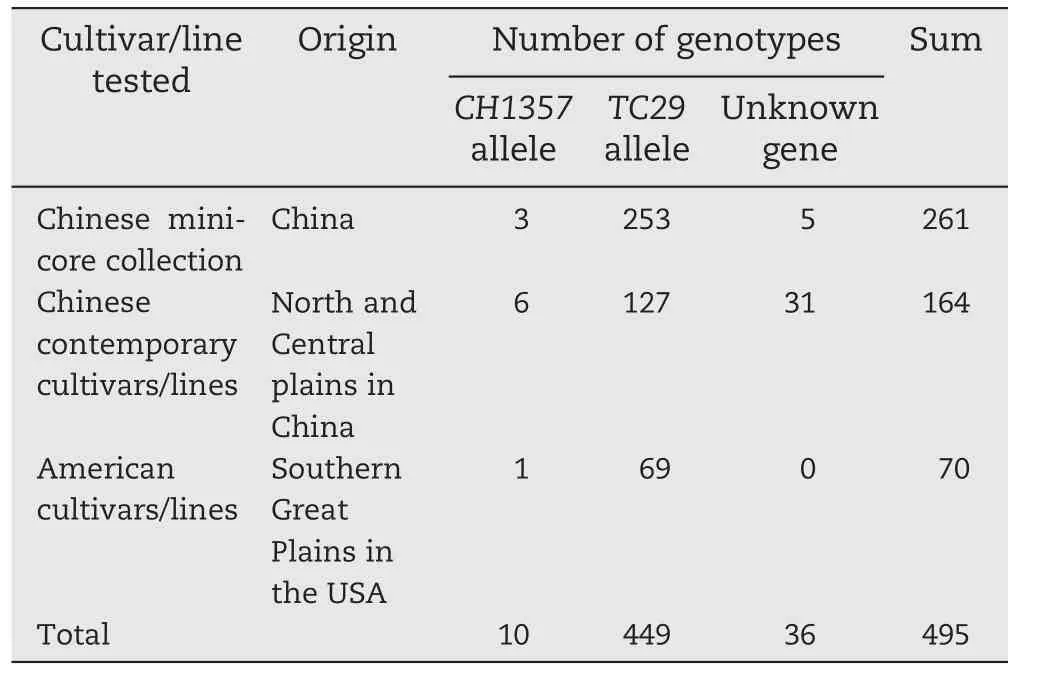
Table 4-Distribution of the CH1357 allele of TraesCS5D01G044600 in a total of 495 of wheat cultivars/lines.
One hundred and sixty-four cultivars/lines currently grown in China were also genotyped and phenotyped (Table S2).Only six cultivars/lines(Liangxing 66,Heze 2,Mianmai 41,Nongda 399, Zhongmai 175, and Tai 1113) lacked the 7 bp deletion and showed resistance to isolate E09; 127 cultivars/lines had the 7 bp deletion and were susceptible. The remaining 31 cultivars/lines possessed the 7 bp deletion displayed variable resistant responses presumably due to genes other than Pm2. Molecular genotyping and powdery mildew tests on 70 cultivars from the USA southern Great Plains detected only one line,GRN*5/ND614-A,possessing the CH1357 resistance allele(Table S3).
4. Discussion
Wheat lines CH1357 and Ulka/8*Cc showed some differences in reaction to 32 Chinese Bgt isolates (Table 1). We therefore initiated cloning of the resistance allele in the locally adapted line CH1357.PmCH1357 was isolated by positional cloning and shown to be Pm2a, which had been cloned previously by MutChromSeq [16]. We concluded that the differences in the powdery mildew response between CH1357 and Ulka/8*Cc were caused by minor variations in genetic background rather than different genes/alleles on chromosome 5DS.
Although Pm2a had been cloned previously, two issues are needed to be addressed in regard to powdery mildew resistance in China. First, the allelic variation between resistance and susceptibility alleles was not revealed in the previous study.Second, the genetic relationship between the reference Pm2a allele and other genes/alleles located on chromosome 5DS was not known.Isolation of PmCH1357 not only confirmed the results of isolation of Pm2a in the previous study[16],but also revealed difference between the resistance and susceptibility alleles of gene TraesCS5D01G044600.Our study did not duplicate previous experiments that examined the function of the PmCH1357/Pm2a gene using mutant and transgenic wheat lines.Instead,with the now-available IWGSC CS sequence we identified a 7 bp InDel causing the different response between the two alleles, and designed a diagnostic marker enabling the detection of the functional resistance allele.
Detailed pedigree analyses of cv.CH1357(Zhong 8701//Jimai 26/Xiaoyan 7430)and other lines with Pm2a/PmCH1357 failed to provide any clues as to the origin of the gene in Chinese wheat germplasm. The origin of the gene in materials used in European countries seems to be Ulka that originated from the former USSR. During the 1970s a range of germplasm was introduced to China from the USSR and some accessions might have carried Pm2a.About that time the gene was also used in a series of wheat cultivars (including Brock) in the UK. The extended pedigree of CH1357 includes Xiaoyan 7430, a Thinopyrum ponticum-derived partial amphiploid (2n = 56), and it is possible that Pm2a/PmCH1357 was originated from Xiaoyan 7430.Since at least one of the lines(Niaomai)is documented as a landrace [24], it is possible that the gene might have been present in older Chinese germplasm.Therefore,it is very likely that Pm2a has multiple origins in wheat.
We also investigated the TraesCS5D01G044600 sequence in AL8/78 genome sequence of Ae. tauschii [49]. The sequence in this accession had a premature stop codon causing a truncated protein of 513 amino acids. However, Pm2a was identified Ae.tauschii accession‘Braunschweig BGRC 1458'[51]and is known to be present in a historic AABBDD synthetic wheat produced by E.R. Sears (R.A. McIntosh, personal communication).
Twelve powdery mildew resistance genes/alleles have been reported on chromosome 5DS in common wheat. Some were identified in Chinese cultivars/lines[2,23-28]and others in European ones [5,29-32], but were located at different positions in chromosome 5DS [32]. In the present study cloning and sequencing of the same allele in four additional Chinese cultivar/lines (KM2939, Niaomai, Liangxing 66, and Nongda 399) indicated that previously designated alleles Pm2b, Pm2c, PmLX66, and PmND399 in other Chinese and European lines are the same.However,it is also possible that there are additional genes on chromosome 5DS.For example,the germplasm Subtil was reported to contain a sequence identical to the Pm2a sequence,but was also reported to differ from the cloned Pm2a sequence [32]. Pm46/Pm48 in Tabasco was reported as not allelic to Pm2a, and produced a different array of powdery mildew responses with 15 Bgt isolates [30].This work raises issues in interpretation of disease response data based on multi-pathotype tests and shows that not only must the individual pathogen isolated be pure and correctly identified,but also that a degree of flexibility must be allowed for genetic background differences in such studies. In other situations minor difference in environment will also be important [52,53]. Obviously, gene cloning should provide the final evidence of gene identity.
PmCH1357/Pm2a is a typical NLR gene. Similar resistance(R) genes in plants often have multiple sequence-related paralogs arranged in tandem as a gene cluster, as is the case for Pm3 [10]. The Pm3 locus contains a series of alleles that protect against different pathogen isolates. This feature of R genes causes difficulties in distinguishing paralogous R genes and their functions. However, the PmCH1357 locus in the CS genome consists of only one copy, namely TraesCS5D01G044600, making it easier to characterize the candidate gene. TraesCS5D01G044500 in the adjacent region also encodes a NBS-LLR protein but it was excluded as a candidate gene by the mapping of the two critical crossovers.
The PmCH1357/Pm2a allele has been introduced into many commercial wheat cultivars in China [54-56], but it is surprisingly uncommon in the core germplasm or contemporary cultivars/lines.The PmCH1357/Pm2a was observed in only three of the 261 mini-core germplasm accessions evaluated.Five other resistant accessions possessed resistance genes other than PmCH1357 (Table S1); Fuzhuang 30 has Pm5e,located on chromosome 7BL [57]; Guinong 10 possesses a resistance gene on chromosome 2A [58]; and Yanzhan 1 contains an unknown powdery mildew resistance gene that is different from PmCH1357/Pm2a. Only six of the 164 cultivar/lines currently grown in the central plains of China contain the CH1357 allele (Table S2), and just one of the 70 cultivars from the southern Great Plains of the U.S.A. had this gene(Table S3).
Additionally, we also found that among the 164 tested contemporary cultivars/lines in China, 31 contained the 7 bp deletion, but had high or moderate resistance to Bgt isolate E09 (Table S2). These lines obviously carried other genes for resistance; for example, Jimai 22 and Liangxing 99 contain Pm52 that has been extensively used in breeding programs of the northern and central plains [59]. It would be a good strategy to pyramid Pm2 and Pm52 in new wheat cultivars.
5. Conclusions
PmCH1357 is allelic to TraesCS5D01G044600 on chromosome 5DS in CS wheat. PmCH1357 conferred resistance to the Bgt isolate E09, which was collected in the central and northern areas of China. PmCH1357 is identical in sequence to the previously named Pm2a. The CS TraesCS5D01G044600 allele has 7 bp deletion in the first exon relative to the Pm2a allele. This deletion leads to a much shorter protein that confers no powdery mildew resistance. A marker based on the deletion allows identification of lines that do not carry Pm2a/PmCH1357.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2019.08.004.
Declaration of competing interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Acknowledgments
We thank Dr. Diaoguo An, Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology,Chinese Academy of Sciences (CAS), Shijiazhuang for assistance with powdery mildew tests and for providing wheat genotypes carrying Pm2a and other mapped resistance genes on chromosome 5DS. The mini-core collection and other germplasm were kindly gifted by Dr.Xueyong Zhang,Institute of Crop Sciences, Chinese Academy of Agricultural Sciences,Beijing. The Bgt isolates were provided by Prof. Yilin Zhou,Institute of Plant Protection,Chinese Academy of Agricultural Sciences. The work was supported by funding from the National Key Research and Development Program of China(2016YFD0102000),Shanxi Provincial Key Platform for Science and Technology Innovation Program(201605D151002),Shanxi Provincial International Cooperation in Science and Technology Program (201803D221018-5), Shanxi Provincial Key Research and Development Project (201803D421020 and 201703D211007),and Shanxi Academy of Agricultural Sciences(Project YGG17123).
- The Crop Journal的其它文章
- Meta-analysis of QTL for Fusarium head blight resistance in Chinese wheat landraces
- The Crop Journal 作物学报(英文版) (Started in 2013, Bimonthly)
- Wheat powdery mildew resistance gene Pm64 derived from wild emmer(Triticum turgidum var.dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5
- Wheat breeding in northern China: Achievements and technical advances
- Breed ing w heat for resistance to Fusarium head blight in the Global North: China,USA,and Canad a
- Pyramiding disease resistance genes in elite winter wheat germplasm for Western Canada

