Pyramiding disease resistance genes in elite winter wheat germplasm for Western Canada
André Lroche*, Michele FrickRobert J. Grf Jmie Lrsenb, John D. Lurie
aAgriculture and Agri-Food Canada,Lethbridge Research and Development Centre,Lethbridge, Alberta T1J 4B1,Canada
bAgriculture and Agri-Food Canada,Harrow Research and Development Centre,2585 County Road 20,Harrow,Ontario N0R 1G0,Canada
Keywords:Bunt Gene pyramiding Wheat rusts Marker-assisted selection Triticum aestivum
ABSTRACT We report on pyramiding different disease resistance genes against fungal pathogens in Canadian winter wheat germplasm based on available DNA markers and gene sequences.Genetic resistance represents a safe, economical and ecological method for protecting plants, growers and the health of consumers. Major diseases of wheat on the Canadian Prairies are common bunt,rusts (leaf,stem and stripe)and Fusarium head blight.Over the years markers for resistance genes against these diseases have been identified and used by the international wheat community. We describe markers that we have used to pyramid different resistance genes and indicate their presence in Canadian winter wheat cultivars issued from the winter wheat breeding program at the Agriculture and Agri-Food Canada,Lethbridge Research and Development Centre, the only winter wheat breeding program in Western Canada actively delivering new varieties for all regions of the Canadian Prairies.The sources of resistance and identities of PCR primers and amplification conditions are indicated to enable the transfer and pyramiding of different resistance(R)genes to breeding lines. We conclude by reviewing new tools for identifying R genes in wheat and indicate how mutagenesis and gene editing can help future efforts to extend the protection offered by known R genes.
Contents
1. Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 740
2. Marker-assisted selection in Western Canada winter wheat germplasm. . . . . . . . . . . . . . . . . . . . . . . . . 742
2.1. Incorporating and maintaining the presence of common bunt resistance gene Bt10 in wheat germplasm . . 742
2.2. Incorporating and maintaining the presence of rust resistance genes in wheat germplasm . . . . . . . . . . . 742
2.2.1. Stripe rust resistance genes. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 744
2.2.2. Leaf rust resistance genes. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 745
2.2.3. Stem rust resistance genes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 745
3. Future prospects. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 745
4. Novel sources of R genes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 745
Conflict of interest . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 746
Acknowledgments . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 746
References. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 746
1. Introduction
Canada is a large country spanning more than 5000 km between the Atlantic and Pacific Oceans with many different agro-ecological zones in which the challenges posed by wheat pests and pathogens vary but are always present. It has been consistently demonstrated that genetic resistance against pests and pathogens is the most sustainable contribution to modern agriculture. Genetic resistance is economical(no need to use chemicals for plant protection),ecological (limits large scale introduction of chemicals to the environment and minimizes issues of leaching and residues on non-target organisms), safe (limits exposure of chemicals to humans), and reduces development of resistance by pathogens and pests to the active ingredients of different fungicide and pesticide formulations. However,release of resistant varieties with durable resistance is challenging and time consuming. When effective genetic resistance is not available, fungicides are very useful for preserving crop yields during epidemics.
Wheat was introduced to Canada via several waves of settlement. Reports suggest that spring wheat was first cultivated in North America by Louis Hébert in 1606 at the beginning of European settlement [1]. It was reported that yield losses due to diseases (rust, blight) and pests (caterpillars)in 1742 and 1743 resulted in an interruption of wheat exports to other colonies during those years[2].At that time,white spring wheat originating from Scandinavian countries was grown as it was better adapted to the shorter growing season in eastern North America than the wheat varieties originating from France and continental Europe[2].During the period 1900-1960, epidemics of wheat stem and leaf rusts on the Canadian Prairies jeopardized farm economic sustainability that lasted until adequate resistance was incorporated [3]. Effective resistance against wheat stem and leaf rusts (caused by Puccinia graminis Pers.: Pers. and P. triticina Eriks., respectively) has been available since the late 1950s-early 1960s in Western Canada[3].Ninety percent of Canadian wheat production occurs on the Prairies encompassing the provinces of Alberta, Saskatchewan and Manitoba[4].
Today,about 9.8 million ha(24 million acres)of spring and winter wheat is harvested in Canada, making it the largest crop grown in terms of area [5]. Winter wheat represents about 6% of the total wheat production, with the greatest concentration being in Ontario [5]. The research program at the Lethbridge Research and Development Centre(LeRDC)has recently emerged as the only winter wheat breeding program actively registering new varieties in Western Canada and has breeding objectives for all of the various agro-ecological regions of the Canadian Prairies. Lines with high yield,excellent cold tolerance,early maturity,resistance to lodging,and various types of disease resistance are required for these regions. The area around Lethbridge in southern Alberta is known as the traditional zone for winter wheat production on the Canadian Prairies, but production of winter wheat has expanded across Western Canada over the past 40 years.This expansion in winter wheat area resulted from improved agronomic systems that helped to ensure survival, advances in farm machinery for straw management, and new varieties with short, strong straw and enhanced disease resistance traits.
The genetic background of the LeRDC winter wheat breeding program originates from areas in which winter wheat production predominates,most importantly from the Central Great Plains and Pacific Northwest regions of the USA, with contributions from Eastern and Western Europe.When required,specific genes have been moved from spring wheat germplasm. The overarching factor in the selection and development of new germplasm is the required high level of cold tolerance for successful survival to the cold Canadian winter.A second critical factor is excellent milling performance for which the major Western Canadian winter wheat class Canada Western Red Winter (CWRW) is well known. In contrast to spring wheat, maturity is somewhat less of a concern. Due to farm operational limitations, it is preferable that winter wheat is ready to harvest by August 15,generally two weeks ahead of spring wheat.
The strategy for deploying disease resistance (R) genes in winter wheat lines is based on the anticipated region of cultivation within the Canadian Prairies. Currently, for a new winter wheat variety to be endorsed for registration by plant pathologists, it must be rated at least “Intermediate” in resistance to all three rusts and common bunt (Tilletia ssp.)and “Moderately Susceptible” to Fusarium head blight (FHB,Fusarium graminearum Schw.). It is anticipated that the required rating for FHB will be raised to “Intermediate” in the next few years. From a practical standpoint, the critical disease resistances required in regions west of Saskatoon are for stripe rust and common bunt; and for regions east of Saskatoon the required resistances are for stem rust,leaf rust,FHB, and common bunt. Due to the overlap of spring and winter wheat production, protection from wheat streak mosaic in the form of resistance to either the wheat curl mite vector (Aceria tosichella Keifer) or to the virus is desirable in all areas.
From an epidemiological perspective, urediniospores from the three wheat rusts are airborne to Canada from the USA.For leaf rust and stem rust,the pathogens enter Canada via the Puccinia pathway that follows the Mississippi River valley moving from south to north with wheat maturation and eventually make their way into Manitoba and eastern Saskatchewan [3]. Stem rust is sometimes observed on susceptible winter wheat in the western Canadian Prairies but when crops are well managed,infection is usually too late to cause economic loss. Leaf rust is rarely observed at economic threshold levels on the western Canadian Prairies.Leaf rust and stem rust resistance genes deployed in the winter wheat breeding program are similar to those that are effective and have been deployed in the Central Great Plains of the USA,as races identified in Canada originate from that region.It is generally accepted that leaf rust and stem rust do not overwinter on the Central Great Plains and each year the pathogens originate from Mexico and southern States of the USA [3]. There are two transmission pathways for the stripe rust pathogen; the one mentioned above along the Mississippi River valley, and the Pacific Pathway with urediniospores originating from Mexico and USA States of California,Oregon and Washington[6].Although P.striiformis(Pst)urediniospores are detected and observed in most years in the eastern Canadian Prairies, economic losses to date have been rare. The situation is vastly different in the western Canadian Prairies, where epidemics initiated by urediniospores originating from the USA Pacific Northwest(Oregon and Washington),cause significant losses in susceptible wheat cultivars. Though infrequent, the stripe rust pathogen can also overwinter in Alberta [7]. Spore showers originating from the Pacific Northwest inoculate susceptible wheat cultivars during the spring in southern Alberta and pathogen populations often multiply before spreading to central and northern Alberta and western Saskatchewan.Stripe rust causes economic losses in susceptible wheat varieties nearly every year.Although breeders from both the Pacific Northwest and Western Canada are deploying the same effective stripe rust resistance genes in their germplasm, it is important to point out that no US winter wheat cultivars are grown in Canada due to lack of sufficient cold tolerance for winter survival. Other critical factors are maturity (either too late or too early) and end-use applications.
Sixteen race-specific R genes effective against common bunt have been identified and characterized in wheat germplasm [8,9]. Bunt is a difficult and laborious disease to screen and characterize.Furthermore,the level of resistance and susceptibility in the host plant can be dramatically affected by environmental conditions as observed in both North America and continental Europe [9-13]. Given the challenge of effective bunt rating, utilization of molecular markers would facilitate the incorporation and maintenance of multiple bunt R genes in wheat germplasm.
Wheat leaf rust and stem rust are still potentially important disease issues and are priority #1 diseases for the eastern Prairie region (Manitoba and eastern Saskatchewan) and effective resistance must be present in all new wheat cultivars [14]. Before 2000, wheat stripe rust was considered a regional disease confined to the irrigation districts of southern Alberta and would generally disappear during the warm days of summer. The stripe rust pathogen is ever-evolving and a major race shift with a new virulence combination was detected during the summer of 2000. New races were more aggressive and better adapted to higher temperatures [6,15]. In Canada and parts of North America,many wheat classes did not carry specific resistance genes against stripe rust, as it had not been a significant threat.Resistance genes effective against stripe rust were serendipitously present in some cultivars, having been inherited along with resistance to leaf rust and stem rust;for example,Yr18/Lr34/Sr57 or Yr36 which is associated with the high grain protein gene from Triticum dicoccoides[16].
Interaction between plants and pathogens has been described as an on-going arms race between the ability of the pathogens to mutate to defeat the host defense system and the ability of host to maintain a repertoire of effective defense genes capable of restraining the enemy [17].Multiple examples of pathogens overcoming single host R genes have been documented. In parallel, stacking of R genes for effective and durable resistance has long been considered a successful strategy for disease control [18].Pathogens can reproduce both sexually and asexually.Gene recombination usually occurs during meiosis and is believed to be a major contributor to genetic variation and the ability of the pathogen to defeat R genes or develop resistance to pesticides. In the case of P. graminis (Pgt), an alternate host is needed for meiosis to occur and barberry(Berberis vulgaris L.) has been identified as the host for sexual reproduction. The identification of the alternate host led to the barberry eradication program on the North American Great Plains initiated in 1918 [19]. Like stem rust,the alternate host for P. striiformis is also barberry [20].Inability to identify the alternative host for more than a century led to the suggestion that genetic recombination during meiosis might not be the main mechanism of development of new Pst races [20]. Pst has been known to defeat single,all-developmental stage R genes very rapidly,and R gene stacking is required to obtain stable, lasting(durable) resistance [6,18]. Somatic recombination is also considered to be an effective means of generating new races [21]. It is thus predicted that multiple R genes help to extend the durability and effectiveness of each gene pyramided within a cultivar [22,23]. Metabolic costs imposed by the presence of multiple R genes have been considered [24]but this concept is difficult to evaluate precisely. Given the fact that pathogens can reduce yield,the question becomes whether there is a point where the number of stacked R genes has a negative impact on yield in the absence of the pathogen? Given the type of protection offered by different R genes, it was recently suggested that active or inactive states of an R gene depending on the presence/absence of a pathogen may have different impacts on plant fitness costs [23,24].
Traditionally, a challenge with gene pyramiding was the inability of detecting the presence of multiple genes conferring resistance against the same disease, as analyses of segregation between the R line and a susceptible line had to be carried out over two or more generations to confirm the presence of multiple R genes in the resistant plant.Molecular markers linked to R genes [25-39]alleviated this issue. The cloning of R genes [40-56]now provides perfect probes for marker-assisted selection. As R genes become defeated by ever-evolving pathogens, the number of available genes providing resistance decreases as efforts to identify new effective genes is limited. Of the over 80 known R genes conferring resistance to stripe rust, only two all-stage resistance genes, Yr5 and Yr15, are currently effective worldwide [55-57]. However, many more genes remain effective in specific geographical regions [57]. A similar situation exists with R genes effective against stem rust[58-61].
DNA tags are needed to identify genes to enable their detection and to determine whether one or more genes present in individual plants will contribute to resistance.These tags, whether on the genes themselves or closely linked to the target gene, are also needed to identify plants with R genes originating from one or more parents in elite germplasm. At our Research Centre in the winter wheat breeding program,we have used molecular markers to stack various R genes in elite germplasm and new wheat cultivars.
PCR technology has revolutionized the identification of DNA fragments linked to, or part of, an R gene. It greatly simplifies and accelerates identification of DNA fragments compared to earlier technologies. From the original report of random DNA primers [62]to Kompetitive Allele Specific PCR(KASP) markers based on SNPs identified in sequencing information [63]or Golden Gate assays [64], currently available tools have simplified and accelerated the identification and selection of R genes in host plants.
2. Marker-assisted selection in Western Canada winter wheat germplasm
The LeRDC winter wheat breeding program generally designs three-way or more complex crosses to facilitate markerassisted selection at the F1generation to enrich the population for genes of interest. These plants are used for the production of doubled haploids(DHs),but the seed produced will also be used in a conventional bulk pedigree breeding system. In the first year of field screening at Lethbridge, DH lines are screened as individual rows and selected on the basis of cold tolerance, plant vigor, tillering capacity, plant architecture,and resistance to prevalent diseases.Following the harvest of selected rows, small amounts of seed are allocated to several inoculated disease nurseries based on the disease responses of the parents. Although this delays the testing of DH lines in yield trials by one year,it increases the probability that the lines under test will have the desired combination of disease resistance traits.In the conventional breeding program, a shuttle breeding approach is used to select for broad adaptation. Selection concentrates on plant type in all early generations in addition to cold tolerance and disease resistance in numerous biotic and abiotic stress nurseries located in different Prairie provinces each year.This approach has resulted in a high percentage of broadly adapted, multiple disease resistant breeding lines entering yield trials at the F6to F8generations.Following two years of replicated, multi-location yield trials in Alberta, Saskatchewan and Manitoba,lines with superior combinations of yield,agronomic characteristics, disease resistance, and end-use quality attributes are entered into registration trials where they will be evaluated for three years.Following this scrutiny,a line may be proposed for registration and commercial release. The presence of various molecular markers will usually be determined prior to entry into registration trials and may also be verified at the time of Breeder Seed production.
2.1. Incorporating and maintaining the presence of common bunt resistance gene Bt10 in wheat germplasm
Based on initial work by Demeke et al. [27]in which polymorphic DNA fragments linked to Bt10 were identified,SCAR primers(Table 1)based on a single nucleotide polymorphism were developed to provide a more robust assay [28].The Bt10-specific primers were used initially to identify the presence of this resistance gene in winter and spring wheat germplasm [27,28]and have since been used to detect the presence of the gene in Canadian germplasm and cultivars.This assay has been well described [28]. The diagnostic SCAR primers appear to be located in a unique sequence resembling a repetitive element. The intensity of the southern blot hybridization signal confirmed that the 604 bp sequence hybridized to a repetitive element (A. Laroche, unpublished data). Although we have done in silico screening of several wheat genomic databases with this DNA fragment, we have not been able to identify a perfect match for this sequence encompassing the polymorphic DNA fragment. This further suggests that the sequence encoding Bt10, which originates from accession PI 178383, is not widely present in wheat germplasm. A cautionary note is that the DNA for the sequence appears to be prone to rapid degradation after extraction.Over the last 20 years,we have used different DNA extraction protocols and have found that the assay must be run quickly after DNA extraction (less than 1 week) as the amplification product for the resistance allele will disappear.This issue will not be solved until we identify a much larger DNA fragment that can be used to design new primers located closer to or at the Bt10 locus. The reason for this instability remains unknown. Bt10 has been mapped to chromosome 6DS [65]. Promising markers linked to the bunt R genes Bt11[11]and Bt9 [12]were reported more recently. These markers will facilitate pyramiding of bunt resistance genes. The effectiveness of different bunt R genes varies with environmental conditions[66]and therefore,this should be taken into consideration when contemplating single gene deployment or gene pyramiding.
2.2. Incorporating and maintaining the presence of rust resistance genes in wheat germplasm

Rusts are a very important issue for wheat cultivation and have serious impacts on wheat production worldwide as they pose recurring threats to world wheat stocks.In recent years,Pgt race TTKST, also known as Ug99, and its derivatives attracted worldwide attention [58-61]. Given the disastrous impact of these races in defeating long-established, stable R genes, ongoing efforts since 2000 to identify new effective stem rust R genes continue,as very few known genes are still effective [58-61]. The threat of these virulent Pgt races led to national and international surveillance programs for their early detection and mitigation [61]. These and other widely virulent races caused severe crop losses on the African continent and were recently tracked to the Arabic peninsula and Europe. The necessity of effective international surveillance for crop diseases to protect global food supplies was again reiterated very recently[67].
2.2.1. Stripe rust resistance genes
Using PCR primers (Table 1) derived from the Yr10 gene [49],we identified the presence of this gene in the stripe rust resistant winter wheat cultivar Radiant, which was shown to have good resistance to the prevalent races of stripe rust at the time it was released[68].These PCR primers have worked with all germplasm to date. Fig. 1 shows the characteristic dominant 1100 bp DNA fragment from Yr10 observed in winter wheat cultivar Radiant released in 2001 and AAC Wildfire released in 2015 [69]. This assay is very specific because the forward primer is located in the intron of Yr10.However, the same Yr10 primers did not amplify the Yr10 sequence identified in T. vavilovii [70], indicating their specificity for the gene originating from Moro wheat [49].Other PCR primers covering the 5′ and 3′ regions of the gene were also evaluated for identification of Yr10,but were not asspecific or robust as the primer set described in Table 1 (data not shown).Other primer sets for detecting Yr10 based on the GenBank sequence [49]have been reported [71-75]but they were not evaluated in our laboratory.Although Yr10 has been defeated since 2010 in Southern Alberta, this all stage R gene remains effective in Central and Northern Alberta and in Saskatchewan [76]. This poses questions on the ability of different isolates of Pst to adapt to different agro-ecological zones.
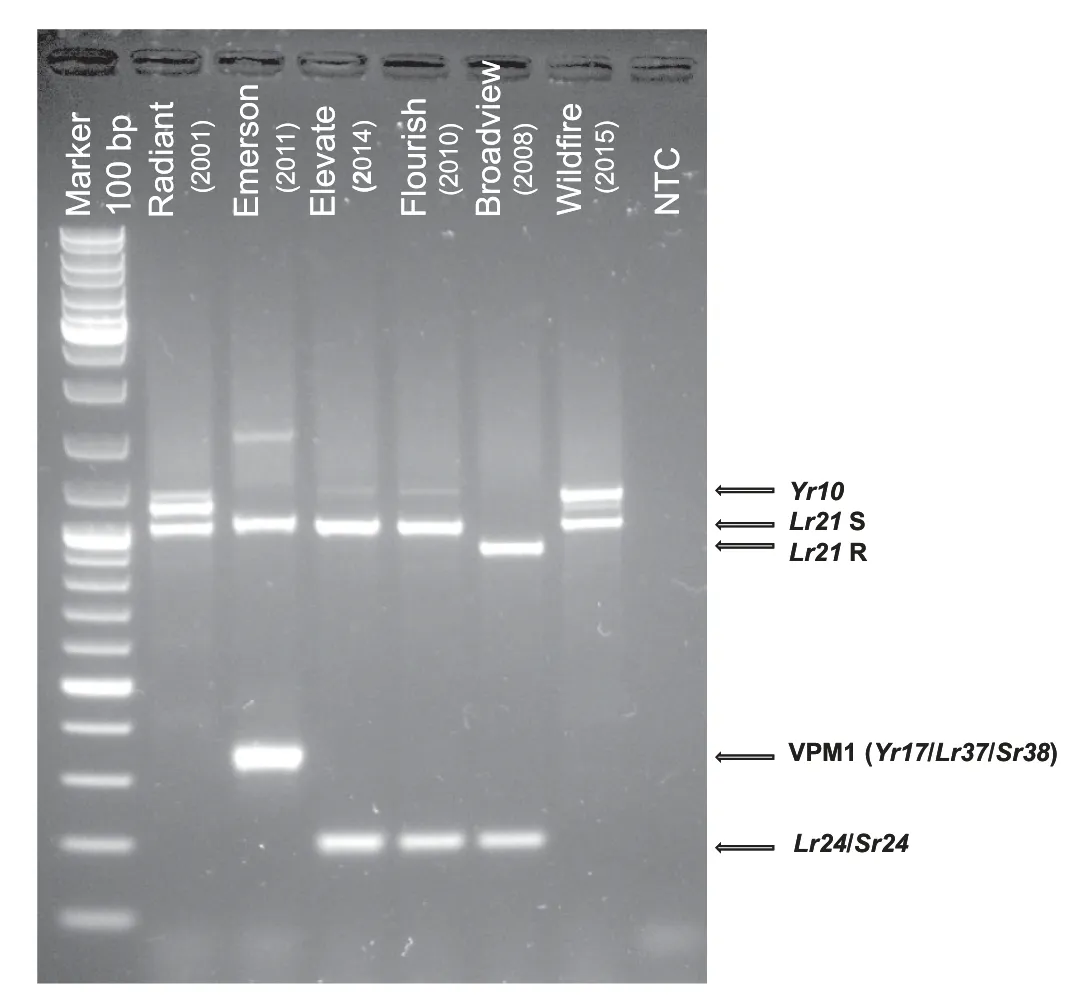
Fig.1- Detection of PCR amplification products for markers for R genes Yr10,Lr21,VPM1,and Lr24/Sr24 in different Canadian winter wheat cultivars.The year indicates the registration date.NTC,negative control.
VPM1 with a wheat-Aegilops ventricosa 2AS/2NS translocation carries Yr17/Lr37/Sr38[29].These linked genes have been deployed in a wide variety of germplasm around the world.Although Yr17 has been defeated in Europe and Australia,it is still widely effective in North America [57]. We previously reported [36]PCR primers to detect the 2NS chromosome fragment of VPM1. These primers were adapted from the Vrga1D sequence information provided by Seah et al.[29]and have worked well with all of the winter wheat germplasm that we have evaluated.The VPM1 locus contributes effective resistance to stripe rust, leaf rust and stem rust in several winter wheat cultivars including Emerson (2011) [36](Fig. 1)and AAC Icefield(2015)[77],the latter being a new hard white winter wheat cultivar. The levels of resistance expressed by these cultivars suggest that the effectiveness of the individual genes depends upon the germplasm in which they are deployed.
Because of the requirement of leaf rust resistance for the eastern Prairies [14], various leaf rust resistance genes,including Lr34, have been incorporated into the winter wheat germplasm. This R gene confers adult plant resistance against wheat rusts and provides up to ‘moderate resistance' (MR) in different wheat genetic backgrounds[16]. Different combinations of primers have been used but under our conditions, the Lr34Δ primers [44](Table 1) have been the better set for evaluating the presence of Yr18/Lr34/Sr57.
The presence of Yr36 linked to the high grain protein gene Gpc-B1 from T.dicoccoides[31]led to a serendipitous addition of the stripe rust resistance gene Yr36 to Canadian wheat germplasm [16]following selection for high grain protein content (HGPC). To actively select for Yr36, the PCR primers(UCW 71F and UWC 71R, Table 1) enabling detection of the HGPC gene worked well to identify the characteristic 100/150/400 bp DNA fragments in different wheat lines. Since the cloning of Yr36 [43], we have used PCR primers based on its sequence[43](Table 1).
Only two stripe rust R genes in wheat are still widely effective worldwide, Yr5 and Yr15 [57]. Primers based on KASP technology [63]have been developed [37,39]and are now widely used. These primers linked to the all stage R genes Yr5 and Yr15 have worked very well in all the winter wheat germplasm evaluated (Table 1). With the cloning of Yr5, new PCR primers were designed that enable the distinction between the related Yr5 and YrSP sequences [55]. These primers also work very well on Canadian winter wheat germplasm (Table 1). Due to intellectual property concerns regarding Yr15 [56], without a specific agreement it might be more prudent to use the linked marker [37]rather than a sequence from the gene [56].
2.2.2. Leaf rust resistance genes
Screening for the presence of adult plant resistance genes Lr34 and Lr37 is covered in the stripe rust section above.Like VPM1 resistance gene Lr24 is linked to Sr24 [32]in an alien segment. Primers Sr24#50 For and Sr24#50 Rev (Table 1) that yield a monomorphic DNA fragment of 200 bp [32]are used for screening all germplasm derived from the two main sources of resistance; the Agent Lr24 source, a larger introgressed alien fragment from Thinopyrum ponticum[26,32]and the Abilene source, a smaller introgressed DNA segment [32]. Lr24 and Sr24 are present in Broadview (2008)[78],Flourish(2010)[79],AAC Gateway(2012)[80],AAC Elevate(2014) [81], and AAC Goldrush (2016) [82](Fig. 1) and contribute to the leaf rust and stem rust resistance in these cultivars.
Detection of the presence of Lr21 is carried out using primers KSU14F and Lr21AR which yield co-dominant DNA fragments of 900 (R) and 1000 (S) bp (Table 1) [41]. Lr21 is present and contributes to the leaf rust resistance in winter wheat cultivar Broadview (2003) [78](Fig. 1) having been contributed by germplasm line KS92WGRC15. As mentioned above, the excellent leaf rust resistance expressed by this cultivar is conditioned via an Lr21 + Lr24 gene pyramid.
The presence of resistance gene Lr22a is detected with primers gwm296 For and gwm296 Rev (Table 1) that amplify DNA fragments of 121 and 131 bp [33].This R gene originated from cultivar AC Minto[33].The LeRDC winter wheat breeding program has advanced breeding lines in which this gene is combined with other leaf rust resistance genes including Lr21,Lr24, Lr34, and Lr37. Spring wheats with known leaf rust R gene pyramids and other genes of interest have also been used as parents in building durable resistance pyramids.
2.2.3. Stem rust resistance genes
Screening for the presence of stem rust R gene Sr24 was described above. Detection of Sr38 and Sr57 was covered by discussion of Yr17 and Yr18, respectively. Sr2 is one of few stem rust R genes effective against Ug99 and derivatives[35,58-60].Consequently,this gene is widely incorporated into wheat germplasm around the world. Primers Sr2F08 F and Sr2F08 R that amplify a dominant 800 bp DNA fragment [35]are very efficient in detecting Sr2 originating from US hard red winter cultivar Scout 66 in crosses with our winter wheat germplasm (Table 1).
Different pairs of PCR primers have been evaluated for detection of Sr36, a gene that has been effective for a long period of time in North America.The most reliable primer set is STM773-2F and STM773-2R that yields a characteristic 155 bp DNA fragment when the gene is present(Table 1) [34].This gene is present in elite winter wheat lines being advanced in selection trials.
The recent cloning of the stem rust R genes Sr22, Sr33,Sr35, Sr45, and Sr50 providing resistance against Ug99 and derivatives [46,47,50,51]will accelerate incorporation and pyramiding of these R genes into wheat germplasm.We have several hundred DH lines in the field at Lethbridge for identification of Sr22 and Sr39. We are planning to utilize the S22GMF and S22GMR primers that amplify a 176 bp diagnostic DNA fragment as described by Steuernagel et al.[51].
3. Future prospects
As winter wheat germplasm increases in complexity due in part to the presence of different R genes, the ability to maintain the presence of 3-4 R genes for the different rusts and against other pathogens will remain a priority.As use of a single effective R gene accelerates selection of mutated pathogenic isolates [23], it has become imperative to ensure that pyramiding of effective R genes is maintained following crosses to minimize their individual vulnerability leading to loss of effectiveness. Marker-assisted selection to pyramid different genes as described above is necessary to protect effective R genes.Although presence of a single R gene in elite germplasm is available in all breeding programs,discipline is and will be required to prevent the release of new varieties containing only one effective R gene.
4. Novel sources of R genes
Screening germplasm and landraces against specific isolates of pathogens is very useful in identifying and then characterizing novel R genes. In the case of stripe rust, both adult and all-stage resistance genes have been identified. Adult plant resistance genes that are not race specific,are less susceptible to erosion. However, they nevertheless contribute to the inoculum load in the environment and indirectly contribute to development of mutant races with new virulence patterns.This impact can be minimized when multiple adult R genes are combined [6]. On the other hand, wild relatives of wheat are a reservoir of numerous wheat rust and other disease R genes [83]. Many of these genes have provided durable resistance [59,60,83]. Intermediate wheatgrass and other Thinopyrum species, have been important contributors of R genes for wheat [57,83-86]. However, transferring genes from Thinopyrum and other genera is difficult, tedious and timeconsuming. This approach has not been widely used during the last 40 years due to these challenges[84-86]. The efficacy of many R genes against rust and other diseases of wheat have been overcome and relatively few effective genes remain. Given the high level of resistance of wheatgrasses to most cereal pathogens, we have initiated a new research project to identify R genes based on the RenSeq protocol[51,87].A variant of this protocol,MutRenSeq[88]was used to identify sequences of Yr5, Yr7, and YrSP in wheat [55]. The approach was further adapted to identify four novel stem rust R genes effective against Ug99 and derivatives from a wheat wild relative [89]. Their cloning provides unique tags to efficiently move them into wheat germplasm.These complementary approaches are much needed to accelerate identification of new R genes needed to protect wheat against different diseases and pests.
In recent years, the development and adoption of KASP technology has accelerated the selection and incorporation of useful alleles to protect plants [37,39,63]. This method accelerates the availability of results and circumvents the need for gels. We have used KASP primers when available.Alternatively to reduce screening time for standard PCR, we have used the QIAxcel instrument (www.qiagen.com) following completion of the PCR step(Table 1).
A challenge with the identification of loci encoding novel R genes in cereals and wild relatives remains the identification of the R gene sequences due to the very large genomes of wheat (17 Gb) and its related species, although availability of linked markers facilitates transfer of novel genes. R gene enrichment methods such as RenSeq [87]and derived MutRenSeq [88]and AgRenSeq [89]accelerate the identification and characterization of new genes. This novel approach is going to be extremely helpful as few sequences are currently known for effective R genes.In the case of resistance to stripe rust, limited sequences related to all stage [49,55,56]and adult stage [43,44]R genes have been identified. SNP variants (haplotypes) of susceptible alleles at the Yr15, Yr18,and Lr21 loci have been identified[44,56,90,91].The impacts of introduced SNPs and domain swapping were investigated to determine the role of the coiled-coil (CC) and leucine-rich repeats (LRR) in different flax rust R genes [92]. Investigation on swapping domains and introduction of SNPs in these domains to evaluate their impact of gene functionality against different pathogen isolates remain to be performed. We anticipate that this should occur within the next few years as the number of identified R genes related to rust resistance in wheat increases.Identification of sequenced novel R genes in wheat and its wild relatives will provide information about genetic variation in CC and LRR domains and will help toward designing SNPs and domain swapping in current known R genes to increase the availability of effective R genes.It is not currently known if the non-expressed pseudogene linked to Yr10 [49]could provide resistance against different Pst isolates, including isolates virulent to Yr10. The availability of newly sequenced R genes effective against Pst would enable genome editing of different R genes. Since CRISPR/Cas9 technology is functional in wheat [93,94], this approach would enable introduction of resistance haplotypes into elite germplasm greatly accelerating the delivery of resistant cultivars by alleviating the 10-20 years delay in moving a resistance allele from a wild relative into a new resistant wheat line. Susceptible alleles of Yr18 and Lr21 could be rapidly mutated to produce effective resistance alleles in elite germplasm. The same principle would apply toward giving a new life to defeated R genes where modification of either or both the CC and LRR domains could be evaluated using Pst isolates virulent against Yr10 or other R gene sequences when available.
Finally, a word of caution must be added against pyramiding genes. It was very recently reported that incompatibility in hybrids can be caused by NBS-LRR domains R genes[95]demonstrating that pyramided R genes could cause necrosis and other unfavorable syndromes. Such effects are thought to be caused by R gene-R gene interactions. The pleiotropic R gene Lr34/Yr18/Sr57/Pm38 has been reported to cause leaf tip necrosis in some genetic backgrounds[23,44]but this impact was absent when the gene was transformed into barley (Hordeum vulgare L.) [96]. Stacking R genes, therefore,needs to be monitored closely in different genetic backgrounds for adverse effects. Stacking of compatible R genes will ensure durable resistance and crop sustainability in the future.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgments
We acknowledge the enthusiastic contributions of numerous students and casual technicians who contributed to selection of various R genes in winter wheat germplasm.Our thanks go to Jim Prus and David Quinn for growing all the wheat lines for screening.Funding from the Agriculture and Agri-Food Canada Peer Review and Growing Forward programs, and Ducks Unlimited Canada is greatly appreciated.
- The Crop Journal的其它文章
- Meta-analysis of QTL for Fusarium head blight resistance in Chinese wheat landraces
- The Crop Journal 作物学报(英文版) (Started in 2013, Bimonthly)
- Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat
- Wheat breeding in northern China: Achievements and technical advances
- Breed ing w heat for resistance to Fusarium head blight in the Global North: China,USA,and Canad a
- Herbicide resistance: Development of wheat production systems and current status of resistant weeds in wheat cropping systems

