3种白鲫杂交子代的转录组学分析
周大颜 张志新 黄彩林 招志杰 莫飞龙




摘要:【目的】研究3种白鲫杂交子代转录组学特征,为揭示鲫鲤杂交优势的分子机理提供理论依据,同时为在生产上培育出生长速度快、肉质好、适应能力强的杂交品种提供技术参考。【方法】以白鲫(♀)×黑龙江野鲤(♂)杂交子代(简称HB)、白鲫(♀)×散鳞镜鲤(♂)杂交子代(简称SB)和白鲫(♀)×兴国红鲤(♂)杂交子代(简称XB)为研究对象,利用RNA-seq高通量测序技术构建3种白鲫杂交子代转录组文库,以HiSeq PE150进行测序分析,原始序列经Trinity组装后进行功能注释(E-value<1e-5);以DESeq2 R鉴定差异表达基因,利用GOseq R和KOBAS分别对差异表达的基因进行GO和KEGG富集分析;并采用MicroSAtellite对转录本中的SSR位点进行挖掘。【结果】共组装得225858条unigenes,平均长度为668 bp,N50为938 bp,有171461条unigenes可注释到蛋白质数据库(Nr)、非冗余核苷酸数据库(Nt)、蛋白质序列数据库(SwissPort)、基因本体论(GO)、直系同源基因簇(COG/KOG)和京都基因与基因组百科全书(KEGG)数据库中,注释比例为75.92%。其中,52630条unigenes注释到NR数据库,43659条unigenes注释到SwissPort数据库,35756条unigenes注释到COG/KOG数据库,包括生化代谢、信号转导机制、防御系统和细胞结构等。差异表达基因KEGG分析结果显示,较多的差异表达基因注释到内吞作用、Jak-STAT信号通路、溶酶体、吞噬体和Wnt信号通路等免疫相关及与生长发育相关的MAPK信号通路、Hippo信号通路和背腹轴形成等通路中。此外,从获得的转录组序列中共鉴定出20272个SSR位点,大多数为二核苷酸重复基元(占62.15%)。【结论】不同白鲫杂交子代间存在较多的差异表达基因,从中获得参与抗氧化、免疫和生长发育相关的通路和基因序列,且挖掘出20272个SSR位点,有助于选择性育种、分子标记开发及开展遗传多样性、遗传图谱构建和QTL定位等研究。
关键词: 白鲫;杂交子代;转录组;信号通路;SSR位点
中图分类号: S965.117 文獻标志码: A 文章编号:2095-1191(2019)06-1328-11
Abstract:【Objective】Transcriptomics characteristics of three Carassius auratus cuvieri hybrids were studied to provide reference for molecular mechanism of hybrid heterosis of the hybrids, and also to offer technical support for breeding hybrids with rapid growth, quality meat and strong adaptability. 【Method】Three white crucian carp hybrids i.e. C. auratus cuvieri(♀)×Cyprinus carpio haermatopterus(♂)(abbreviated as HB), C. auratus cuvieri(♀)×C. carpio L.(♂)(abbreviated as SB) and C. auratus cuvieri(♀)×C. carpio var. singuonensis(♂) (abbreviated as XB) were used as research objectives. RNA-seq high-throughput sequencing was used to construct C. auratus cuvieri hybrids transcriptome library. HiSeq PE150 was used to conduct sequencing analysis, and the function annotation on original sequence after Trinity assembling was carried out(E-value<1e-5). Differentially expressed genes was identified by DESeq2 R, then GO and KEGG enrichment analysis of differentially expressed genes were conducted by GOseq R and KOBAS respectively. MicroSAtellite was used to dig SSR loci in transcript. 【Result】A total of 225858 unigenes, with average length of 668 bp and N50 of 938 bp were generated. Of these, a total of 171461 unigenes could be annotated in protein sequence database(Nr), non-redundant nucleotide database(Nt), protein sequence database(SwissProt), Gene Ontology database(GO), Clusters of Ortho-logous Groups(COG/KOG) and Kyoto Encyclopedia of Genes and Genomes(KEGG) database. The annotation ratio was 75.92%. There were 52630 unigenes annotated in NR database, 43659 unigenes annotated in SwissPort database, 35756 unigenes annotated in COG/KOG database, including biochemical metabolism, signal transduction mechanism, defensive system and cell structure. KEGG analysis of differentially expressed genes showed that more differentially expressed genes were involved in immune-related pathways such as endocytosis, Jak-STAT signaling pathway, lysosome, phagosome and Wnt signaling pathways, and growth and development related pathways such as MAPK signaling pathway, Hippo signaling pathway and dorso-ventral axis formation. In addition, 20272 SSR loci were identified from transcriptome sequence and majority of which were dinucleotide repeats units(accounted for 62.15%). 【Conclusion】There are many differentially expressed genes in different hybrids. Some pathways and gene sequences involved in antioxidation, immunity, growth and development are preliminarily obtained. A total of 20272 SSR loci are dug. This result is helpful for selective breeding, molecular markers development, researches in genetic diversity, genetic map construction and QTL mapping.
Key words: Carassius auratus cuvieri; hybrid; transcriptome; signal pathway; SSR locus
收稿日期:2018-10-13
基金项目:农业部物种品种资源(渔业)保护费项目(171721301354052099);广西海洋和渔业厅预算项目(桂海渔财〔2018〕97号)
作者简介:周大颜(1981-),主要从事水产动物遗传育种与养殖推广研究工作,E-mail:4948738@qq.com
0 引言
【研究意义】我国水生生物多样性丰富,许多种类经人工驯化及繁育后已发展成为重要的经济水产品种。其中,鲤科(Cyprinidae)的多样性尤为突出,在现存的800余种淡水鱼中,鲤科鱼类约占50%。鲤鱼养殖迄今已有2400余年历史,是池塘、稻田和网箱等养殖的主要对象,在天然水域产量中也占有很高比例(朱健等,2000)。遗传组成不同的个体或群体结合后,其遗传物质重新组合而形成新品种的现象称为杂交(Zhang et al.,2014),在自然界中普遍存在。杂交育种作为一种常规的育种手段,在水生生物品种改良和生产中发挥重要作用,是获得新品种的重要途径之一(Arcella et al.,2014),且杂交获得的F1代通常在生产、生活、繁殖和适应性等方面具有优于双亲均值或超过亲本的杂交优势(Jin et al.,2017)。因此,基于转录组学分析技术开展杂交优势分子机理研究,对加速水产动物选择性育种、分子标记开发及遗传多样性分析等具有重要意义。【前人研究進展】目前,国内外关于水产动物杂交的研究已有大量报道,如梭子蟹(Portunus trituberculatus)(Gao et al.,2014)、牙鲆(Paralichthys olivaceus)(Liu et al.,2014)、鲶鱼杂交种(Pseudoplatystoma sp.)(Sinhorin et al.,2014)、石斑鱼(Epinephelus fuscoguttatus ♀×E. lanceolatus ♂)(Firdaus et al.,2016)、鲍鱼(Haliotis discus hannai)(Li et al.,2017)、牡蛎(Crass-ostrea sikamea×C. angulata)(Yan et al.,2017)和中华鳖(Pelodiscus sinensis)(Zhang et al.,2017b)等,其中又以不同鲤鱼品种间杂交的效果最好(楼允东,1999)。我国已培育出具有显著杂种优势的丰鲤(Cyprinus carpio var. singuonensis ♀×C. carpio L.♂)、荷元鲤(C. carpio var. wuyuanensis ♀×C. carpio var. yuankiang ♂)、荷花鲤(C. carpio L. ♀×C. carpio var. singuonensis ♂)和三杂交鲤(Heyuan carp ♀×C. carpio L. ♂)等杂交种(Liu et al.,2017),且均已得到推广养殖,但目前针对杂交鲤的研究仍停留在形态学特征、主要经济性状、遗传变异及亲子鉴定等方面(刘义新等,2007;武耀等,2012;Liu et al.,2018)。随着基因组测序技术的快速发展,转录组测序技术已成为挖掘缺乏基因组信息物种功能基因的重要手段(唐玉娟等,2018),在经济鱼类杂交后代的杂种优势研究中得到广泛应用。Bougas等(2010)对3个美洲红点鲑(Salvelinus fontinalis Mitchill)群体及其杂交后代进行转录组学分析,结果显示不同群体杂交后代表现出不同的基因表达模式和生长优势。Gao等(2013)对杂交河豚(Jiyan-1 Puffer)及其亲本(Takifugu rubripes ♀×T. flavidus ♂)进行转录组测序分析,结果发现杂种优势的形成可能与能量代谢、离子结合和激酶激活等过程有关。Liu等(2018)对白鲫(Carassius auratus ♀)和红鲫(C. auratus red var. ♂)及其杂交F1代的转录组进行测序,GO分析结果显示,F1代的杂合基因与代谢过程、免疫系统和生长发育有关。【本研究切入点】白鲫(C. auratus cuvieri)又称日本白鲫,隶属于鲤形目(Cypriniformes)鲤科(Cyprinidae)鲤亚科(Cyprinae)鲫属(Carassius),原产于日本琵琶湖,于1976年引进我国,体色为白色,具有繁殖力强、食性广、生长速度快、适应性强等优点,但其肉质欠佳(王静等,2015)。黑龙江野鲤(C. carpio haermatopterus)与黄河、长江、辽河等野鲤同属一个亚种,起源于欧洲野鲤,在长期的进化过程中黑龙江野鲤对于黑龙江流域多变的水温环境已完全适应,具有极强的抗寒能力和抗病能力(朱健等,2014)。散鳞镜鲤(C. carpio L.)原产于前苏联,于1959年引入我国,抗逆性强、生长速度快,是杂交选育的重要亲本来源(李盛文等,2014)。兴国红鲤(C. carpio var. singuonensis)主要分布在江西兴国县,已有超过1300年的养殖历史,其温度适应范围广、易于饲养、产卵量大,是重要的杂交亲本,在我国鱼类杂交育种中占据重要地位(岳华梅等,2016)。以白鲫为母本,分别与黑龙江野鲤、散鳞镜鲤和兴国红鲤杂交,均可获得具有明显杂交优势的杂交F1代,但其杂交优势的分子机理尚未明确,因此有必要利用转录组测序技术从基因层面揭示鲤鱼杂交优势的分子机理。【拟解决的关键问题】通过RNA-seq高通量转录组测序、组装和分析,对3种白鲫杂交子代进行转录组学特征研究,以期为揭示鲫鲤杂交优势分子机理提供理论依据,同时为在生产上培育出生长速度快、肉质好、适应能力强的杂交品种提供技术参考。
1 材料与方法
1. 1 试验材料
供试鱼为白鲫(♀)×黑龙江野鲤(♂)杂交子代(简称HB)、白鲫(♀)×散鳞镜鲤(♂)杂交子代(简称SB)和白鲫(♀)×兴国红鲤(♂)杂交子代(简称XB),由广西水产引育种中心武鸣基地提供。试验前,先在室内养殖池中隔离暂养10 d,饥饿48 h后从不同杂交子代中分别挑选200尾体质健康、规格一致的供试鱼。经测量发现,HB、SB和XB供试鱼的平均体重分别为31.9、32.0和32.3 g/尾,并分别在腹腔注射PIT电子标记,记录标记号后放入池塘殖。
1. 2 养殖管理
每天上、下午各投喂1次;每晚开增氧机增氧;每隔25 d泼洒聚维酮碘(水产用)1次,进行水体消毒和疾病预防。养殖周期共90 d(2017年7月25日—2017年10月24日)。
1. 3 样品采集
养殖结束后,从HB、SB和XB供试鱼中随机抽取样品鱼各30尾,用无菌手术刀剪取肌肉、鱼肝和鱼皮,分别装入标识好的无RNA酶试管中,经液氮速冻后置于-80 ℃冰箱中保存备用。
1. 4 RNA提取
取样品鱼的肌肉、肝脏和鱼皮,置于经高压灭菌的研钵中加液氮研磨成粉状,然后参照Invitrogen公司的TRIzol Reagent说明提取组织总RNA。以TURBO DNA-FreeTM Kit(Ambion,Thermo Fisher Scientific,USA)消除总RNA中的DNA后,分别用1%非变性琼脂糖凝胶电泳(120 V,10 min)检测RNA的完整性、NanoDrop 2000超微量分光光度计(Thermo Fisher Scientific,USA)检测RNA纯度和浓度。
1. 5 转录组文库构建及测序
取检测合格的各组织RNA等量混合(总量1 μg)后进行文库构建和测序。用携带Oligo(dT)的磁珠富集含poly(A)尾巴的mRNA,随后加入Fragmentation Buffer打断mRNA,以片段化的mRNA为模板,采用六碱基随机引物合成单链cDNA;再加入缓冲液、dNTPs、RNase H和DNA聚合酶I合成cDNA第二链,经QiaQuick PCR试剂盒纯化并加入EB缓冲液洗脱后进行末端修复及加poly(A)尾巴,并连接测序接头;以琼脂糖凝胶电泳进行片段大小筛选,最后进行PCR扩增,构建好的文库采用HiSeq PE150进行测序分析。
1. 6 序列组装及功能注释
由测序获得的数据称为raw reads,先对raw reads进行质控(QC),去除测序过程中低质量序列及不确定序列;采用转录组组装软件Trinity对过滤后的clean reads进行组装,选取每个transcript cluster中最长的转录本作为unigene。以NCBI蛋白质数据库(Nr)、非冗余核苷酸数据库(Nt)、蛋白质序列数据库(SwissPort)、基因本體论(GO)、直系同源基因簇(COG/KOG)和京都基因与基因组百科全书(KEGG)数据库作为参考,对所获得的unigene进行功能注释(E-value<1e-5)(Zhang et al.,2017a)。
1. 7 差异表达基因分析
采用RPKM(Reads per kilo bases per million reads)衡量基因表达量(Li and Dewey,2011),并以DESeq2 R进行差异表达基因筛选,筛选标准为错误发现率P≤0.05,|log2ratio|≥1(Yan et al.,2017)。利用GOseq R(Young et al.,2010)和KOBAS(Mao et al.,2005)分别对差异表达的基因进行GO和KEGG富集分析,其中,GO富集分析以corrected-pvalue≤0.05为阈值,满足此条件即定义为在差异表达基因中显著富集的GO term;Pathway显著性富集分析以KEGG Pathway为单位,应用超几何检验,当Q value≤0.05即定义为在差异表达基因中显著富集的Pathway。
1. 8 SSR分子标记鉴定
利用MicroSAtellite(MISA,http://pgrc.ipk-gater-sleben.de/misa/)对转录本中的SSR位点进行挖掘,为鉴定二、三、四、五和六核苷酸重复基元,对应的阈值分别被设为 6、5、4、4和4(Zhou et al.,2013)。根据碱基互补配对原则,将所有互补的简单重复序列视为一类,其中,二核苷酸重复基元4种(AT、AG、AC和CG),三核苷酸重复基元10种,四核苷酸重复基元33种,五核苷酸重复基元102种,六核苷酸重复基元350种。
2 结果与分析
2. 1 测序数据的产出和组装结果
为保证数据质量,对各测序样品的原始数据进行质控和过滤。去除低质量、含接头序列的reads后,得到的clean reads如表1所示。各样品测序质量不低于20(Q20)的碱基占总碱基比例均在96.00%以上,质量不低于30(Q30)的碱基占比在90.00%以上,GC含量则低于50.00%。采用Trinity对所有clean reads进行从头组装后,共得到225858条unigenes,总长150956990 bp,平均长度668 bp,N50为938 bp。其中,有102591条(45.42%)的unigenes长度在200~400 bp,42555条(18.84%)的unigenes长度在400~600 bp,长度在1000 bp以上的unigenes有42723条(18.91%)(图1)。
2. 2 功能注释及分类结果
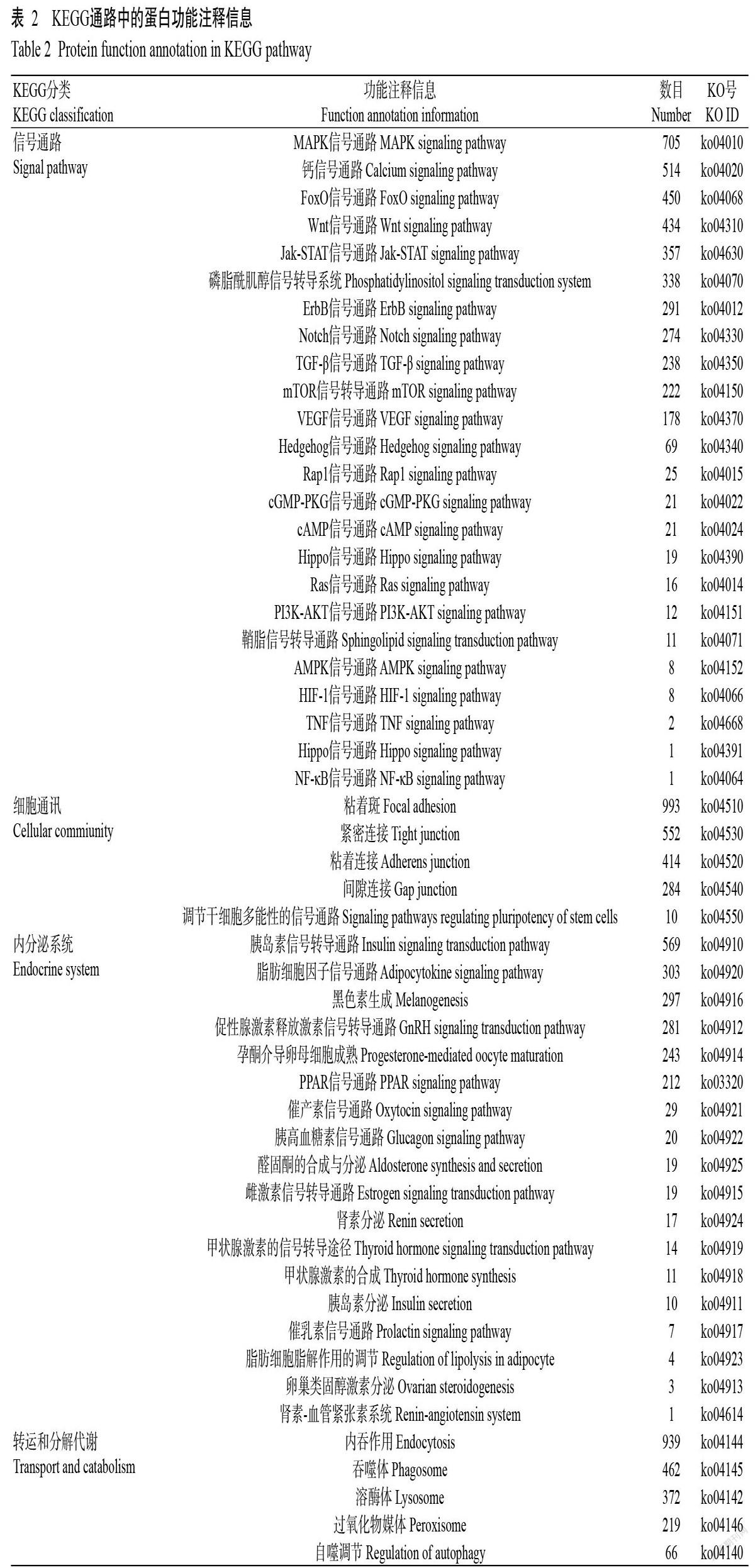
将所有unigenes分别与NR、SwissProt、KEGG和COG/KOG(E-value<1e-5)等数据库中的相关序列进行BLASTx比对分析,找出与指定unigene具有最高序列相似性的蛋白,从而获知unigene的蛋白功能注释信息。结果显示,在225858条unigenes中有171461条获得蛋白功能注释信息,注释比例为75.92%。其中,52630条unigenes注释到NR数据库,43659条unigenes注释到SwissPort数据库,35756条unigenes注释到COG/KOG数据库,包括生化代谢、信号转导机制、防御系统和细胞结构等(图2)。此外,有28164条unigenes注释到不同的KEGG通路中(图3),最有代表性的是信号转导通路(4215条)、细胞通讯(2253条)、内分泌系统(2059条)及转运和分解代谢(2058条)(表2)。
2. 3 差异表达基因的筛选及其功能注释
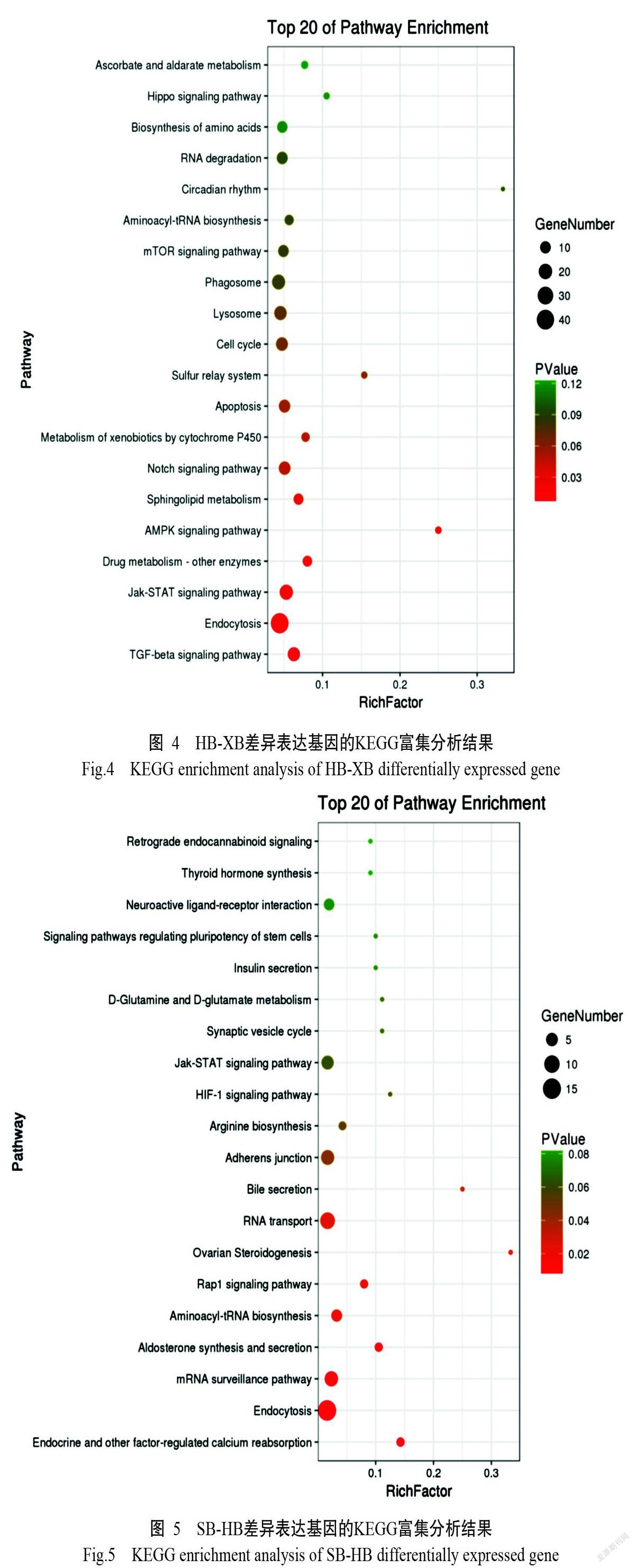
HB、SB和XB 3种杂交子代两两进行差异表达基因筛选分析,结果显示,HB和XB间(HB-XB)的差异表达基因有6448个,其中上调基因2064个、下调基因4384个;SB和HB间(SB-HB)的差异表达基因有1615个,其中上调基因1053个、下调基因562个;SB和XB间(SB-XB)的差异表达基因有4249个,其中上调基因2290个、下调基因1959个。对这些差异表达基因进行KEGG通路分析,结果发现,在HB-XB的差异表达基因中,有较多基因注释到内吞作用、Jak-STAT信号通路、溶酶体和吞噬体(图4);在SB-HB的差异表达基因中,以注释到内吞作用、mRNA监测通路、RNA转运和粘着连接中注释的差异表达基因较多(图5);在SB-XB的差异表达基因中,较多基因注释到内吞作用、吞噬作用、Wnt信号通路和RNA转运通路中(图6)。
2. 4 SSR位点鉴定结果
将拼接所得序列提交至MISA进行分析,共得到20272个SSR位点,其中有2359条序列包含1个以上的SSR位点。根据重复单元的类型和重复次数对SSR位点进行分类,结果发现二核苷酸重复基元的SSR位点数最多,为12594个,占全部重复序列的62.15%;其次是三核苷酸重复基元的SSR位点(5318个),占26.25%;四、五和六核苷酸重复基元的SSR位点较少(1826、371和163个),占比分别为9.00%、1.80%和0.80%。
2. 5 重复性检验结果
在转录组学研究中通常以相关性热图展示样品间的关系,而样品间基因表达水平相关性是检验试验可靠性和样品选择是否合理的重要指标。根据RPKM定量结果,计算出所有样品(每个样品3个重复)两两间的相关系数(图7),发现相关系数均在0.5900以上,说明样品检测的重复性较好,结果可信度较高。
3 讨论
近年来,高通量测序技术已广泛应用于模式和非模式生物的相关研究领域,如在兴国红鲤、白鲫、红鲫及红白鲫杂交子代中均有利用高通量测序技术进行转录组学分析的研究报道(岳华梅等,2016;Zhang et al.,2017c;Liu et al.,2018)。在转录组学研究中,为获取较多转录本以避免个体变异而产生的负面影响,通常从不同组织或个体中提取RNA,等体积混合后进行转录组测序分析(Huang et al.,2012;Li et al.,2012;Liao et al.,2013;李冰冰等,2017)。本研究利用RNA-seq高通量测序技术对3种白鲫杂交子代进行转录组学测序分析,共得到225858条unigenes,平均長度为668 bp,N50为938 bp。与红白鲫杂交子代转录组学分析结果(Liu et al.,2018)相比,本研究中的unigene数目较多,且质量较好,可能与本研究利用多个样本多个组织的总RNA进行测序有关,所获得的转录组数据相比单个组织或单个样品测序更全面。
目前,有关水产动物杂交分子机理的研究主要集中在差异表达基因与杂种优势的相关性方面(Zhai et al.,2013;Zhang et al.,2017a)。孙颖(2016)通过对棕点石斑鱼(E. fuscogutatus ♀)×鞍带石斑鱼(E. lanceolatus ♂)杂交F1代(虎龙斑)生长优势的转录组学进行研究,结果在虎龙斑中找到大量的差异表达基因,且这些差异表达基因主要分布在GH/IGF轴及其下游涉及蛋白与糖原合成的信号通路上,对虎龙斑生长优势有重要贡献。本研究结果表明,在白鲫(♀)×黑龙江野鲤(♂)与白鲫(♀)×兴国红鲤(♂)的杂交子代中差异表达基因最多,为6448个;其次是白鲫(♀)×散鳞镜鲤(♂)与白鲫(♀)×兴国红鲤(♂)的杂交子代,为4249个;差异表达基因最少的是白鲫(♀)×散鳞镜鲤(♂)与白鲫(♀)×黑龙江野鲤(♂)的杂交子代,仅有1615个;3个转录组的共有基因为43个。差异表达基因的KEGG分析结果显示,大多数差异表达基因注释到免疫相关通路中,包括内吞作用、Jak-STAT信号通路、溶酶体、吞噬体和Wnt信号通路等;也有差异表达基因注释到Toll样受体信号通路和NOD样受体信号通路中。此外,从差异表达基因中鉴定出过氧化氢酶(unigene071729)、铁蛋白(unigene045233)、peroxiredoxin-6(unigene058224)及α-2M(unigene079502)等抗氧化和免疫相关的基因,在3个白鲫杂交子代中这些基因的表达量均呈不同程度上调趋势。免疫系统可保护生物体免受自然环境中寄生虫或致病菌的感染,是机体产生抗病性的生理基础,也是育种过程中进行人工选择的重要指标之一(Rauw,2012)。免疫相关基因或通路的鉴定,既可为鲫鲤杂交的人工选育提供依据,又能佐证这些基因或通路对3个白鲫杂交子代的生长优势具有重要贡献。
生长速度和体重一直是选育过程中的关键指标。本课题组的前期研究结果表明,3种白鲫杂交子代间的体重存在显著差异。本研究也鉴定出与生长和发育相关的差异表达基因通路,如MAPK信号通路(ko04010)、Hippo信号通路(ko04390)和背腹轴形成(ko04320)等。Zhang等(2017b)对中华鳖(Pelodiscus sinensis)的杂交子代进行转录组学分析,也曾鉴定出这3条通路,说明这3条通路在促进杂交子代生长和发育的过程中发挥重要作用。其中,MAPK信号通路(ko04010)和Hippo信号通路(ko04390)在调节器官发育及器官发育过程中发挥重要作用(Schaeffer and Weber,1999;Yu et al.,2012)。此外,与生长相关的表皮生长因子受体(unigene078218)、微管相关蛋白(unigene024234)、血小板源生长因子受体(unigene038197和unigene068685)等基因均明显上调,而生长抑制蛋白(unigene067656)明显下调。这些基因的差异表达有助于开展杂交子代的生长性状比较分析。
分子标记是进行种群遗传学、生物地理学及进化关系研究的关键(Li and Gui,2007;Jia et al.,2008;Wang et al.,2011;陈春林等,2018)。通过高通量测序技术获得的转录组序列,可为分子标记的挖掘和开发提供一种更直观、高效的方法(Davey et al.,2011;Fu and He,2012;Ji et al.,2012)。至今,利用转录组测序技术进行SSR分子标记挖掘在国际上已有很多成功的报道(Tian et al.,2014;Chen et al.,2015;Xiao et al.,2015)。Liao等(2013)从鲫鱼的转录组数据库中挖掘到11295个SSR位点,且大多数SSR位点为二核苷酸重复基元。岳华梅等(2016)从兴国红鲤的转录组中鉴定出13652个SSR位点,并从中随机筛选出30个进行PCR验证,发现有20对SSR分子标记可扩增出清晰稳定的条带。本研究从3个白鲫杂交子代中鉴定出20272个SSR位点,其中有62.15%的SSR位点为二核苷酸重复基元,与Liao等(2013)的研究结果相似。这些挖掘获得的SSR位点可为后续的遗传多样性、遗传图谱构建及QTL定位等研究打下基础。
4 结论
不同白鲫杂交子代间存在较多的差异表达基因,从中获得参与抗氧化、免疫和生长发育相关的通路和基因序列,且挖掘出20272个SSR位点,有助于选择性育种、分子标记开发及开展遗传多样性、遗传图谱构建和QTL定位等研究。
参考文献:
陈春林,田易萍,陈林波,邓少春,徐丕忠,李朝云. 2018. 基于荧光标记的紫娟茶树转录组EST-SSR标记开发[J]. 江苏农业学报,34(4):747-753. [Chen C L,Tian Y P,Chen L B,Deng S C,Xu P Z,Li C Y. 2018. EST-SSR marker development of Zijuan tea tree transcriptome based on the fluorescent labeling[J]. Jiangsu Journal of Agricultural Sciences,34(4):747-753.]
李冰冰,赵振利,邓敏捷,曹亚兵,董焱鹏,范国强. 2017. 盐胁迫对南方泡桐基因表达的影响[J]. 河南农业大学学报,51(4):471-480. [Li B B,Zhao Z L,Deng M J,Cao Y B,Dong Y P,Fan G Q. 2017. Effect of salt stress on gene expressions of different genotypes of Paulownia australis[J]. Journal of Henan Agricultural University,51(4):471-480.]
李盛文,賈智英,柏盈盈,李池陶,石连玉. 2014. 散鳞镜鲤两个保种群体的遗传多样性[J]. 水产学杂志,27(2):1-8. [Li S W,Jia Z Y,Bai Y Y,Li C T,Shi L Y. 2014. Microsatellite marker analysis of genetic diversity in two sca-ttered scale mirror carp populations[J]. Chinese Journal of Fisheries,27(2):1-8.]
刘义新,肖祖国,徐振秋. 2007. 几种杂交鲤的形态学特征及主要经济性状[J]. 水产科学,26(11):619-621. [Liu Y X,Xiao Z G,Xu Z Q. 2007. Morphological and main economic characteristics of several common carp(Cyprinus carpio L.) hybrids[J]. Fisheries Science,26(11):619-621.]
楼允东. 1999. 我国鱼类育种研究五十年回顾[J]. 淡水渔业,29(9):1-3. [Lou Y D. 1999. Review of fish breeding research in China in the past fifty years[J]. Freshwater Fishe-ries,29(9):1-3.]
孙颖. 2016. 棕点石斑鱼(♀)×鞍带石斑鱼(♂)杂交F1代生长优势的转录组学研究[D]. 广州:中山大学. [Sun Y. 2016. Transcriptomic studies on the growth superiorities in grouper hybrid(Epinephelus fuscogutatus ♀)×(Epinephelus lanceolatus ♂)[D]. Guangzhou:Sun Yat-sen University.]
唐玉娟,黄国弟,罗世杏,周俊岸,莫永龙,李日旺,赵英,张宇,宋恩亮,宁琳. 2018. 芒果2个不同花芽分化时期转录组分析[J]. 南方农业学报,49(7):1257-1264. [Tang Y J,Huang G D,Luo S X,Zhou J A,Mo Y L,Li R W,Zhao Y,Zhang Y,Song E L,Ning L. 2018. Transcriptome of Mangifera indica L. in two different flower bud differentiation stages[J]. Journal of Southern Agriculture,49(7):1257-1264.]
王静,肖军,曾鸣,徐康,陶敏,张纯,段巍,刘文彬,罗凯坤,刘筠,刘少军. 2015. 白鲫×红鲫杂交后代的遗传变异[J]. 中国科学:生命科学,45(4):371-380. [Wang J,Xiao J,Zeng M,Xu K,Tao M,Zhang C,Duan W,Liu W B,Luo K K,Liu Y,Liu S J. 2015. Genomic variation in the hybrids of white crucian carp and red crucian carp[J]. Scien-tia Sinica(Vitae),45(4):371-380.]
武耀,賈智英,李池陶,葛会争,石连玉. 2012. 筛选杂交鲤亲子鉴定的微卫星标记(英文)[J]. 农业生物技术学报,20(5):549-559. [Wu Y,Jia Z Y,Li C T,Ge H Z,Shi L Y. 2012. Microsatellite markers for parentage identification of cross-breeding carp(Cyprinus carpio) in a selective breeding programme[J]. Journal of Agricultural Biotechnology,20(5):549-559.]
岳华梅,翟晴,宋明月,叶欢,杨晓鸽,李创举. 2016. 基于转录组测序的兴国红鲤微卫星标记筛选[J]. 淡水渔业,46(1):24-28. [Yue H M,Zhai Q,Song M Y,Ye H,Yang X G,Li C J. 2016. Development of microsatellite mar-kers in Cyprinus carpio var. singuonensis using next-ge-neration sequencing[J]. Freshwater Fisheries,46(1):24-28.]
朱健,柴学森,李冰,张成锋. 2014. 建鲤和黑龙江野鲤自交以及正反交子代生长比较[J]. 渔业科学进展,35(2):35-41. [Zhu J,Chai X S,Li B,Zhang C F. 2014. Growth comparison of inbreeds of Cyprinus carpio var. Jian,Cyprinus carpio haermatopterus and the reciprocal F1 hybrid[J]. Progress in Fishery Sciences,35(2):35-41.]
朱健,王建新,龚永生,郁桐炳. 2000. 我国鲤鱼遗传改良研究概况[J]. 浙江海洋学院学报(自然科学版),19(3):266-271. [Zhu J,Wang J X,Gong Y S,Yu T B. 2000. An introduction to the studies on common carp genetic improvement in China[J]. Journal of Zhejiang Ocean University(Natural Science),19(3):266-271.]
Arcella T E,Perry W L,Lodge D M,Feder J L. 2014. The role of hybridization in a species invasion and extirpation of resident fauna:Hybrid vigor and breakdown in the rusty crayfish,Orconectes Rusticus[J]. Journal of Crustacean Biology,34(2):157-164.
Bougas B,Granier S,Audet C,Bernatchez L. 2010. The transcriptional landscape of cross-specific hybrids and its possible link with growth in brook charr(Salvelinus fontinalis Mitchill)[J]. Genetics,186(1):97-107.
Chen X,Mei J,Wu J,Jing J,Ma W,Zhang J,Dan C,Wang W,Gui J. 2015. A comprehensive transcriptome provides candidate genes for sex determination/differentiation and SSR/SNP markers in yellow catfish[J]. Marine Biotechnology,17(2):190-198.
Davey J W,Hohenlohe P A,Etter P D,Boone J Q,Catchen J M,Blaxter M L. 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing[J]. Nature Reviews Genetics,12:499-510.
Firdaus R F,S L L,Kawamura G, Shapawi R. 2016. Assessment on the acceptability of hybrid grouper,Epinephelus fuscoguttatus ♀×Epinephelus lanceolatus ♂ to soybean meal-based diets[J]. AACL Bioflux,9(2):284-290.
Fu B D,He S P. 2012. Transcriptome analysis of silver carp (Hypophthalmichthys molitrix) by paired-end RNA sequencing[J]. DNA Research,19(2):131-142.
Gao B Q,Liu P,Li J,Wang Q Y,Li X P. 2014. Genetic diversity of different populations and improved growth in the F1 hybrids in the swimming crab(Portunus trituberculatus)[J]. Genetics & Molecular Research,13(4):10454-10463.
Gao Y,Zhang H,Gao Q,Wang L L,Zhang F C,Siva V S,Zhou Z,Song L S,Zhang S. 2013. Transcriptome analysis of artificial hybrid pufferfish Jiyan-1 and its parental species:Implications for pufferfish heterosis[J]. PLoS One,8(3):e58453.
Huang Q,Dong S,Fang C,Wu X,Ye T,Lin Y. 2012. Deep sequencing-based transcriptome profiling analysis of Oryzias melastigma exposed to PFOS[J]. Aquatic Toxicology,120-121:54-58. doi: 10.1016/j.aquatox.2012.04.013.
Ji P F,Liu G M,Xu J,Wang X M,Li J T,Zhao Z X,Zhang X F,Zhang Y,Xu P,Sun X W. 2012. Characterization of common carp transcriptome:Sequencing,de Novo assembly,annotation and comparative genomics[J]. PLoS One,7(4):e35152.
Jia Z Y,Shi L Y,Liu X F,Sun X W. 2008. The genetic diversity of diploid and triploid crucian carp from six populations in Heilongjiang River System[J]. Hereditas,30(11):1459-1465.
Jin J J,Sun Y W,Qu J,Syah R,Lim C H,Alfiko Y,Rahman N,Suwanto A,Yue G,Wong L,Chua N H,Ye J. 2017. Transcriptome and functional analysis reveals hybrid vi-gor for oil biosynthesis in oil palm[J]. Scientific Reports,7(1):439. doi: 10.1038/s41598-017-00438-8.
Li B,Dewey C N. 2011. RSEM:Accurate transcript quantification from RNA-seq data with or without a reference genome[J]. BMC Bioinformatics,12:323. doi:10.1186/1471-2105-12-323.
Li C,Zhang Y,Wang R,Lu J,Nandi S,Mohanty S,Terhune J,Liu Z,Peatman E. 2012. RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish,Ictalurus punctatus[J]. Fish & Shellfish Immunology,32(5):816-827.
Li F B,Gui J F. 2007. Clonal diversity and genealogical relationships of gibel carp in four hatcheries[J]. Animal Genetics,39(1):28-33.
Li J Q,Wang M L,Fang J G,Liu X,Mao Y Z,Liu G M,Bian D P. 2017. Reproductive performance of one-year-old Pacific abalone(Haliotis discus hannai) and its crossbreeding effect on offspring growth and survival[J]. Aquaculture,473:110-114.
Liao X,Cheng L,Xu P,Lu G,Wachholtz M,Sun X,Chen S. 2013. Transcriptome analysis of crucian carp (Carassius auratus),an important aquaculture and hypoxia-tolerant species[J]. PLoS One,8(4):e62308.
Liu Q,Qi Y,Liang Q,Xu X,Hu F,Wang J,Xiao J,Wang S,Li W,Tao M,Qin Q,Zhao R,Yao Z,Liu S. 2018. The chimeric genes in the hybrid lineage of Carassius auratus cuvieri(♀)×Carassius auratus red var.(♂)[J]. Scien-ce China. Life Sciences,61(9):1079-1089.
Liu X J,Liang H W,Li Z,Liang Y J,Lu C,Li C,Chang Y,Zou G,Hu G. 2017. Performances of the hybrid between CyCa nucleocytplasmic hybrid fish and scattered mirror carp in different culture environments[J]. Scientific Reports,7:46329. doi: 10.1038/srep46329.
Liu Y F,Ma D Y,Zhao C Y,Wang W Q,Zhang X L,Liu X,Liu Y,Xiao Z Z,Xu S H,Xiao Y S,Liu Q H,Li J. 2014. Histological and enzymatic responses of Japanese flounder(Paralichthys olivaceus) and its hybrids(P. olivaceus♀×P. dentatus ♂) to chronic heat stress[J]. Fish Physiology and Biochemistry,40(4):1031-1041.
Mao X Z,Cai T,Olyarchuk J G,Wei L. 2005. Automated genome annotation and pathway identification using the KEGG Orthology(KO) as a controlled vocabulary[J]. Bioinformatics,21(19):3787-3793.
Rauw W. 2012. Immune response from a resource allocation perspective[J]. Frontiers in Genetics,3:267. doi: 10.3389/fgene.2012.00267.
Schaeffer H J,Weber M J. 1999. Mitogen-activated protein kinases:Specific messages from ubiquitous messengers[J]. Molecular and Cellular Biology,19(4):2435-2444.
Sinhorin V D G,Sinhorin A P,Teixeira J M D S,Miléski K M L,Hansen P C,Moreira P S A,Kawashita N H,Baviera A M,Loro V L. 2014. Effects of the acute exposition to glyphosate-based herbicide on oxidative stress parameters and antioxidant responses in a hybrid Amazon fish surubim(Pseudoplatystoma sp)[J]. Ecotoxicology and Environmental Safety,106:181-187.
Tian C X,Liang X F,Yang M,Dou Y Q,Zheng H Z,Cao L,Yuan Y C,Zhao C. 2014. New microsatellite loci for the mandarin fish Siniperca chuatsi and their application in population genetic analysis[J]. Genetics and Molecular Research,13(1):546-558.
Wang Z W,Zhu H P,Wang D,Jiang F F,Guo W,Zhou L,Gui J F. 2011. A novel nucleo-cytoplasmic hybrid clone formed via androgenesis in polyploid gibel carp[J]. BMC Research Notes,4(1):82. doi: 10.1186/1756-0500-4-82.
Xiao T Q,Lu C Y,Xu Y L,Li C,Zheng X H,Cao D C,Cheng L,Mahboob S,Sun X. 2015. Screening of SSR markers associated with scale cover pattern and mapped to a genetic linkage map of common carp(Cyprinus carpio L.)[J]. Journal of Applied Genetics,56(2):261-269.
Yan L L,Su J Q,Wang Z P,Yan X W,Yu R H,Ma P,Li Y,Du J. 2017. Transcriptomic analysis of Crassostrea sikamea× Crassostrea angulata hybrids in response to low salinity stress[J]. PLoS One,12(2):e0171483.
Young M D,Wakefield M J,Smyth G K,Oshlack A. 2010. Gene ontology analysis for RNA-seq: Accounting for selection bias[J]. Genome Biology,11(2):R14. doi:10.1186/ gb-2010-11-2-r14.
Yu F X,Zhao B,Panupinthu N,Jewell J L,Lian I,Wang L H,Zhao J,Yuan H,Tumaneng K,Li H,Fu X D,Mills G B,Guan K L. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling[J]. Cell,150(4):780-791.
Zhai R R,Feng Y,Wang H M,Zhan X D,Shen X,Wu W,Zhang Y,Chen D,Dai G,Yang Z,Cao L,Cheng S. 2013. Transcriptome analysis of rice root heterosis by RNA-seq[J]. BMC Genomics,14:19. doi:10.1186/1471-2164-14-19.
Zhang C B,Lin C J,Fu F Y,Zhong X,Peng B,Yan H,Zhang J,Zhang W,Wang P,Ding X,Zhang W,Zhao L. 2017a. Comparative transcriptome analysis of flower heterosis in two soybean F1 hybrids by RNA-seq[J]. PLoS One,12(7):e0181061.
Zhang H,Xu X,He Z,Zheng T,Shao J. 2017b. De novo transcriptome analysis reveals insights into different mechanisms of growth and immunity in a Chinese soft-shelled turtle hybrid and the parental varieties[J]. Gene,605:54-62.
Zhang Y Q,Liu J H,Fu W,Xu W T,Zhang H,Chen S,Liu W,Peng L,Xiao Y. 2017c. Comparative transcriptome and DNA methylation analyses of the molecular mechanisms underlying skin color variations in crucian carp (Carassius carassius L.)[J]. BMC Genetics,18(1):95. doi: 10.1186/s12863-017-0564-9.
Zhang Z H,Chen J,Ling L I,Tao M,Zhang C,Qin Q B,Xiao J,Liu Y,Liu S J. 2014. Research advances in animal distant hybridization[J]. Science China. Life Scien-ces,57(9):889-902.
Zhou Z C,Dong Y,Sun H J,Yang A F,Chen Z,Gao S,Jiang J W,Guan X Y,Jiang B,Wang B. 2013. Transcriptome sequencing of sea cucumber(Apostichopus japonicus) and the identification of gene-associated markers[J]. Molecular Ecology Resources,14(1):127-138.
(責任编辑 兰宗宝)

