甘蓝型油菜皖油20号种子不同部位油脂合成的转录调控分析
张宇婷 鲁少平 金 诚 郭 亮
甘蓝型油菜皖油20号种子不同部位油脂合成的转录调控分析
张宇婷 鲁少平 金 诚 郭 亮*
华中农业大学作物遗传改良国家重点实验室, 湖北武汉 430070
甘蓝型油菜是主要的油料作物之一, 种子含油量一般在35%~50%。油脂主要储存于油菜种子胚中, 胚主要由子叶[包括外子叶(OC)和内子叶(IC)和胚轴(EA)]组成。低芥酸油菜品种皖油20号(WY20)种子不同部位的含油量存在显著差异。WY20的胚中, OC含油量最高, EA含油量最低。同时, 脂肪酸组成在种子不同部位也存在差异, EA中棕榈酸(C16:0)、亚油酸(C18:2)及二十碳酸(C20:0)的比例均显著高于子叶, 特别是C16:0在EA中的比例约为子叶的2倍。而油酸(C18:1)及二十碳烯酸(C20:1)在子叶中的比例均显著高于EA。硬脂酸(C18:0)在OC中含量最低, 在IC和EA中无差别。亚麻酸(C18:3)则在OC中含量最高, 在IC和EA中无差异。对发育34d种子的IC、OC和EA进行转录组分析, 将三个部位中基因表达定量分析的结果两两比较后共发掘出7192个差异表达基因, 其中OC和IC之间差异表达基因数目较少, 子叶和EA间有较多的差异表达基因。子叶和胚轴中的差异表达基因富集在光合作用、脂肪酸代谢和叶绿素合成等生物学过程。基因功能注释显示, 差异表达基因中有355个和脂质代谢相关, 且多集中在质体中脂肪酸从头合成途径。本研究表明油脂合成途径关键基因的差异调控是造成油菜种子不同部位含油量和脂肪酸组成差异的主要因素。
甘蓝型油菜; 种子不同部位; 含油量; 脂肪酸组成; 转录调控
油菜是世界第三大油料作物, 也是我国第一大油料作物, 菜籽油是我国最主要的食用植物油来源之一[1]。油料作物的含油量是一项重要的生产指标, 是油料作物产量的重要因素之一。油菜种子含油量每提高1个百分点, 单位面积油菜的产油量就能提高2.3~2.5个百分点[2]。近年来, 我国油菜单产已达到世界平均水平, 但油菜种子中的油脂含量偏低, 导致国产菜籽油在国际市场缺乏竞争力且受到进口植物油的冲击。因此, 提高油菜籽含油量对我国油菜产业发展极其重要[3-5]。油菜种子中各类脂肪酸的组成影响菜籽油的品质及营养价值。脂肪酸根据结构分为饱和脂肪酸和不饱和脂肪酸, 其中饱和脂肪酸(C16:0和C18:0等)熔点较高, 人体不易消化吸收, 易凝固在血管壁上。不饱和脂肪酸(C18:1、C18:2、C18:3等)熔点较低, 易被人体消化和吸收, 不易凝固或沉淀在血管壁上[6]。我国市场上销售的食用植物油有很多种, 其中菜籽油的饱和脂肪酸的含量是所有植物油中最低的, 其油酸、亚油酸和亚麻酸含量比例合理, 是最健康的食用植物油之一[7]。
植物细胞中, 质体和内质网是脂肪酸合成的主要场所。光合作用产生的糖类在细胞质中经糖酵解转化为丙酮酸进入质体, 丙酮酸在丙酮酸脱氢酶的作用下生成乙酰辅酶A, 此过程为脂肪酸从头合成的第一步[8]。乙酰辅酶A再经过一系列的缩合、还原及脱水反应生成丁酰-酰基转运蛋白, 完成脂肪酸链的第一次延伸[9]。接着, 酰基转运蛋白继续与乙酰辅酶A反应并重复上述过程, 每反应一次碳链增加2个碳原子, 直至碳链上碳原子数为16时停止延伸[10]。此时, 合成的16:0-ACP在酮脂酰基ACP合酶(KASII)和硬脂酰ACP脱氢酶(SAD)的作用下生成18:1-ACP。然后, 在酰基-ACP硫脂酶(FATA和FATB)催化下, 脂肪酸从脂酰ACP脱离并在长链脂酰-CoA合成酶(LACS)作用下合成酰基CoA[11]。酰基CoA被转运到内质网或胞质中的酰基CoA池中, 一部分酰基CoA经3-磷酸甘油酰基转移酶(GPAT)、溶血性磷脂酸酰基转移酶(LPAAT)、磷脂酸磷酸酶(PAP)和二酰甘油转酰酶(DGAT)途径最后合成三酰基甘油(TAG), 另一部分酰基CoA经溶血卵磷脂酰基转移酶(LPCAT)作用将脂肪酸转移到卵磷脂(PC)上, 经过脂肪酸脱氢酶FAD2和FAD3对脂肪酸链修饰产生带有多不饱和脂肪酸酸链的PC[12]。此时, PC在磷脂二酯酰甘油酰基转移酶(PDAT)的催化下与二酰基甘油(DAG)反应生成TAG, 或者在磷脂酰胆碱: 二酰甘油磷酸胆碱转移酶(PDCT)和磷酸胆碱转移酶(CPT)的作用下生成DAG, 然后再合成TAG[13]。近年来研究表明, TAG和其他脂类分子在棉花、亚麻芥、油菜和拟南芥等种子中的含量是不均一的, 暗示脂质代谢途径在不同种子中存在着差异[14-17]。推测转录水平或酶活水平的调控是造成种子不同部位中脂质代谢差异的原因, 然而具体的机制并不明确[18]。
随着测序技术的发展, 转录组测序已广泛应用于生命科学的各个领域[19-20]。RNA测序(RNA-seq)能揭示在特定时间内或部位中RNA的存在和数量, 是一种高通量的分析基因表达的手段[21-23]。Manuel等[24]基于深度表达序列标记测序对4种油料作物种子的4个发展阶段的基因表达进行转录组分析, 揭示了参与合成油脂的基因具有保守性和物种特异性。Stéphane等[25]对油棕果实和种子的3个不同部位的转录组进行了比较, 发现EgWRI1-1和EgWRI1-2转录因子在果皮和胚乳的油脂积累过程中被大量转录。Lu等[26]比较高、低含油量油菜种子不同部位中脂质代谢物与油脂合成相关基因表达, 结果表明种子不同部位的脂质含量存在明显差异, 这些差异主要由种子不同部位中参与油脂合成相关基因表达量的不同造成。但该研究没有对造成种子不同部位脂质含量差异的相关差异表达基因进行深入挖掘和分析。本研究从转录组水平分析并鉴定调控油菜种子不同部位含油量和脂肪酸组成的代谢网络和关键基因, 从转录水平解析油菜种子不同部位中油脂合成的调控机制。
1 材料与方法
1.1 材料及处理方法
供试材料为甘蓝型低芥酸油菜皖油20 (WY20)种子。显微镜下分离WY20发育34 d种子的内子叶(IC)、外子叶(OC)、胚轴(EA)和种皮(SC), 设置5个生物学重复。参考Lu等[26]的方法用脂肪酸甲酯化和气质联用仪(GC-MS)分析种子各部位的油脂含量和脂肪酸组成。提取IC、OC和EA总RNA进行转录组测序, 设每组3个生物学重复。
1.2 文库构建与测序
使用植物RNA提取试剂盒(DP432, http://www. tiangen.com/)提取种子各个部位的总RNA, 将提取的RNA样品送GenoSeq公司(http://www.genoseq. cn/), 用Illumina Hiseq进行转录组测序, 每个样品6G数据量。
1.3 转录组数据分析
用fastp软件去除接头序列和低质量读数, 测序数据质量控制和过滤后, 通过hisat2软件将9组转录组数据比对到油菜参考基因组(http://www.genoscope. cns.fr/brassicanapus/)。利用featureCounts软件定量得到TPM值(transcripts per kilobase of exon model per million mapped reads)来计算基因的表达量[27-28]。再用DESeq2软件包进行差异表达基因的筛选, 筛选条件为-value <0.01, |log2fold change|> 1, 并对所筛选出的差异表达基因进行GO富集分析[29-30]。
1.4 候选基因的鉴定及筛选
利用blastp工具将油菜蛋白序列与数据库中拟南芥蛋白序列对比, 设置E-value为1e–5, Coverage> 50%, 找到油菜在拟南芥中对应的同源基因及基因ID。利用拟南芥约700个与脂质代谢相关的基因和甘蓝型油菜全基因组测序得到的1000余个与油脂相关的基因作为参考, 进行油菜基因同源性分析从而构建本研究中油菜油脂合成基因数据库。将差异表达分析的结果与数据库相结合, 筛选出与油脂合成相关的差异表达基因, 进一步对油脂合成相关的差异表达基因进行功能注释和表达数据的分析。最后将与油脂合成相关的差异表达基因比对到油脂合成的代谢途径中, 并在代谢途径中标记出差异基因在IC、OC和EA两两之间表达量差异的倍数。
2 结果与分析
2.1 WY20种子不同部位脂肪酸组成和含油量分析
WY20种子切片在显微镜下可以明显地看到OC、IC、EA和SC四个不同部位(图1-A)。脂肪酸分析结果表明, 不同部位之间含油量存在显著差异, OC的含油量最高, SC的含油量最低(图1-B)。比较种子不同部位所占种子重量的百分比, 含油量最高的OC的比例远大于其他3个部位, 含油量最少的SC比例高于EA (图1-C)。WY20种子不同部位的脂肪酸组成存在显著差异, 其中C18脂肪酸在整颗种子中的脂肪酸含量比较高(图1-D, E)。C16:0、C18:2和C20:0在胚轴中的含量均显著高于子叶, 特别是C16:0在EA中的含量约为子叶的2倍。C18:1和C20:1在子叶中的含量则均显著高于EA, C18:0在OC中含量显著低于IC和EA, 而在IC和EA中无差别。C18:3则在OC中含量最高, 在IC和EA中无差异(图1-D)。
2.2 转录组质量控制及reads的比对
数据统计约有3.08亿条原始数据, 平均每个部位有1.02亿条。经质量控制和过滤后, 得到质量较好的读段(reads), reads和全基因组比对有93.87%被映射到油菜参考基因组。总读数中约有6.13%的reads可能由于筛选参数设定较严格、测序组装错误或者参考基因组不完整而匹配不上。
2.3 基因差异表达分析
调查3个部位中基因表达量的分布并对样品3个生物学重复之间的相关性进行检测和主成分分析(PCA)(图2-A)表明, IC和OC三个生物学重复之间的皮尔森系数均高于0.98, EA三个生物学重复之间的皮尔森系数高于0.92, 但生物学重复之间差异均不显著, 证明3个生物学重复的重复性较好。对3个部位的基因表达量进行两两间的差异分析表明, IC和OC间比较得到了525个差异基因, 其中有233个基因表达上调, 292个表达下调。IC和EA间比较得到了5436个差异基因, 表达上调的有2520个, 下调的有2916个。OC和EA间比较得到了5749个差异基因, 表达上调的有2535个, 下调的有3214个(图2-B)。3个组合去重复后的差异表达基因有7192个, 有116个基因在EA与IC, EA与OC, IC与OC 3个比较组合中均有表达上的差异(图2-C)。此结果也表明内子叶和外子叶间差异基因较少, 暗示着OC和IC的基因表达模式比较一致。
2.4 GO富集分析
GO富集分析发现, OC与EA和IC与EA之间的差异基因主要富集在光合作用、一元羧酸生物合成过程和脂肪酸生物合成过程。子叶与EA间存在较多参与脂质合成相关的差异表达基因, 这些基因将成为我们后续分析的重点。而OC与IC中的差异表达基因很少, 且主要富集在胞外区和核仁等与油脂合成不直接相关的生物学过程。我们列出了每组比较差异表达基因GO富集到的前10个代谢通路(图3)。
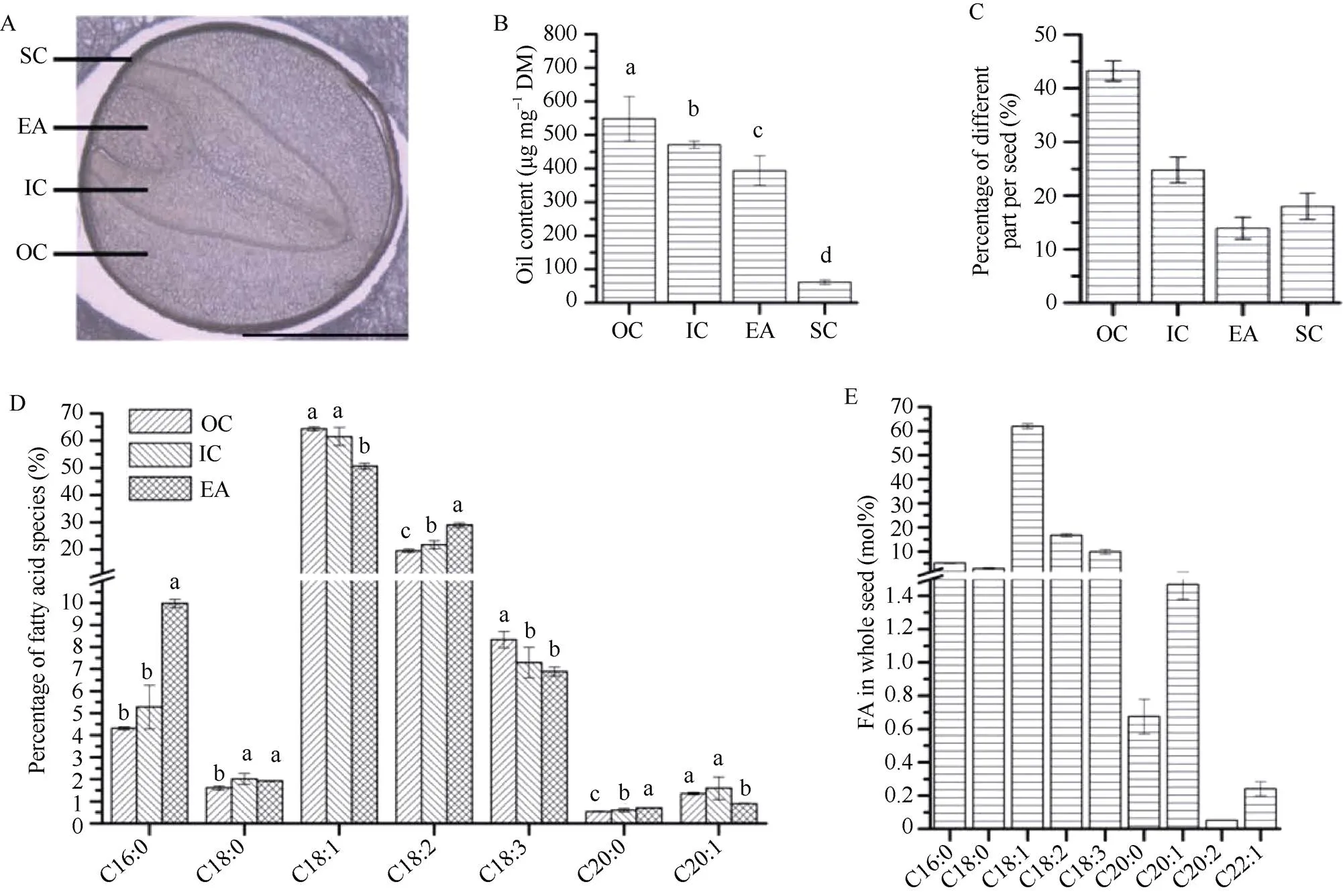
图1 甘蓝型油菜WY20种子不同部位含油量和脂肪酸组成
A: 显微镜明视野下WY20种子部位; B: 种子不同部位含油量; C: 种子不同部位重量百分比; D: 种子不同部位脂肪酸组成; E: 整颗种子脂肪酸组成。
A: bright-field image in different parts of WY20 seed under microscope; B: oil content in different parts of seed; C: weight percentage in different parts of seed; D: fatty acid composition in different parts of seed; E: whole seed fatty acid composition.

图2 3个种子部位差异表达基因分布情况
A: 样本间PCA分析; B: IC、OC和EA差异表达基因数目及百分比; C:差异表达基因维恩图。
A: PCA analysis of different samples; B: differentially expressed genes in IC, OC, and EA; C: Venn diagram of differentially expressed genes.
2.5 候选基因的鉴定与筛选
在油菜种子不同部位之间筛选的7192个差异表达基因中, IC与OC之间的差异表达基因中有18个油脂合成相关基因, IC与EA之间的差异表达基因中有268个油脂合成相关基因, OC与EA之间的差异表达基因中有286个油脂合成相关基因。去除相互之间重复基因, 一共筛选出355个和油脂合成相关的差异基因。为了确保差异基因在种子中有表达, 去除这些基因的表达量(TPM值)在3个部位中均小于1的基因, 最终剩余336个基因。再将336个基因匹配到油菜种子油脂合成途径中, 共得到53个可以匹配到代谢通路上的基因, 我们将同一基因的不同拷贝算做1类基因。IC与OC间得到了4类基因, IC与EA间得到了49类基因, OC与EA间得到了46类基因。其中BnaA02g11570D、BnaA03g23490D、BnaA03g13590D和BnaC03g27860D为3种组合对比中共有的差异基因。而IC与EA、OC与EA两种对比组合间共有匹配到通路上的和油脂合成相关的差异表达基因有38个, IC与EA间特有7个, OC与EA间特有4个。我们将得到的53个基因根据不同对比组合进行了注释和分析(图4和表1)。油脂合成途径中IC和OC中的基因表达量都普遍高于EA, 而IC和OC之间的基因表达水平差别不显著(图4)。

图3 油菜胚的3个部位差异表达基因GO功能分布
A: IC和OC差异表达基因功能分布; B: IC和EA差异表达基因功能分布; C: OC和EA差异表达基因功能分布。
A: GO analysis of differentially expressed genes between IC and OC; B: GO analysis of differentially expressed genes between IC and EA; C: GO analysis of differentially expressed genes between OC and OC.
3 讨论
植物种子中的油脂合成过程是一个复杂的网络, 发生在细胞的多个细胞器中, 受多种酶、代谢物转运、转录因子和能量代谢等影响[31]。近年来研究表明, 脂质代谢途径在油料植物种子胚的不同部位存在着差异[14-17]。本研究对低芥酸油菜WY20种子不同部位的分析结果表明, 含油量在IC、OC和EA中存在着显著差异, OC含油量最高, IC含油量次之, EA含油量最低。C16:0和C18:2在EA中的比例显著高于IC和OC, 而C18:3在EA中比例最低(图1-B, C)。本研究结果与前人利用不同的油菜材料研究结果基本一致[26], 表明油菜种子不同部位中油脂合成和脂肪酸脱饱和过程受到不同的调控。Borisjuk等[32]研究表明油菜种子不同部位能量代谢有明显差异, 这可能与油菜种子结构有关, 不同种子部位获得的空间和光不同, 导致光合作用等存在着差别。
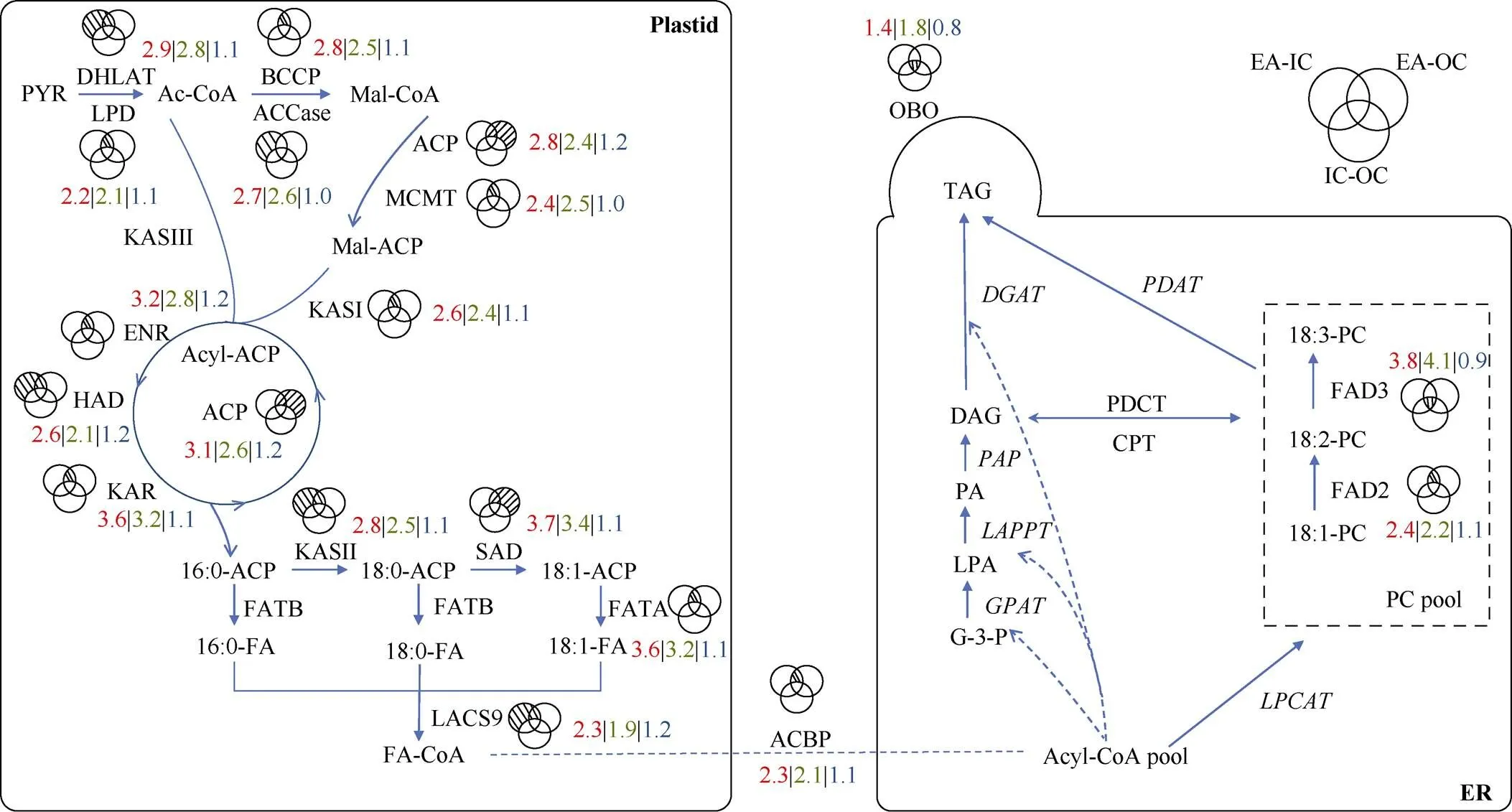
图4 油脂合成相关差异表达基因在油脂代谢通路中的比较分析
维恩图中阴影代表差异表达基因的分布。红色数字表示蛋白家族对应基因TPM值的平均值在IC和EA间的比值, 绿色数字表示蛋白家族对应基因TPM值的平均值在OC和EA间的比值, 蓝色数字表示蛋白家族对应基因TPM值的平均值在IC和OC间的比值。
编码蛋白质的基因: DHLAT: 二氢硫辛酰胺乙酰转移酶; LPD: 二氢硫辛酰胺脱氢酶; BCCP: 生物素羧基载体蛋白; ACCase: 乙酰辅酶a羧化酶; MCMT: 酰基载体蛋白; KASI: 3-酮酰基-酰基载体蛋白合酶I; KASII: 3-酮酰基-酰基载体蛋白合酶II; KASIII: 3-酮酰基-酰基载体蛋白合酶III; KAR: 酮脂酰还原酶; HAD: 硫酯酶蛋白; ENR: 烯酰ACP还原酶; ACP: 酰基载体蛋白; SAD: 硬脂酰-酰基载体蛋白脱饱和酶蛋白; FATA: FATA硫酯酶; FATB: FATB硫酯酶; LACS9: 长链酰基辅酶9; FAD2: 脂肪酸去饱和酶2; FAD3: 脂肪酸去饱和酶3; CPT: CDP胆碱-甘油二酯胆碱酯酶; PDCT: 磷脂酰胆碱-甘油二酯胆碱酯酶; GPAT: 磷酸甘油脂酰转移酶; PDAT: 磷脂-二酰甘油酰基转移酶; LPAAT: 溶血磷脂酸酰基转移酶; PAP: 磷脂酸磷酸酶; DGAT: 二酰甘油酰基转移酶; OBO: 油体蛋白; LTP: 脂质转运蛋白; ER: 内质网。
The shadow in venn diagram represents where the differentially expressed genes are located. Number in red indicates the ratio of genes’ average TPM of between IC and EA, number in green indicates the ratio of genes’ average TPM of between OC and EA, number in blue indicates the ratio of genes’ average TPM of between IC and OC.
Abbreviation of genes that encode proteins: DHLAT: dihydrolipoamide acetyltransferase; LPD: dihydrolipoamide dehydrogenase; BCCP: biotin carboxyl carrier protein; ACCase: acetyl-CoA carboxylase; MCMT: malonyl-CoA: ACP malonyltransferase; KASI: 3-ketoacyl-acyl carrier protein synthase I; KASII: 3-ketoacyl-acyl carrier protein synthase II; KASIII: 3-ketoacyl-acyl carrier protein synthase III; KAR: ketoacyl-ACP reductase; HAD: hydroxyacyl-ACP dehydrase; ENR: enoyl-ACP reductase; ACP: acyl carrier protein; SAD: stearoyl-acyl carrier protein desaturase; FATA: acyl-ACP thioesterase A; FATB: acyl-ACP thioesterase B; LACS: long-chain acyl-CoA synthetase; FAD2: FA desaturase 2; FAD3: FA desaturase 3; CPT: CDP-choline: diacylglycerol cholinephosphotransferase; PDCT: phosphatidylcholine:diacylglycerol cholinephosphotransferase; GPAT: glycerol-3-phosphate acyltransferase; PDAT: phospholipid:diacylglycerol acyltransferase; LPAAT: lysophosphatidic acid acyltransferase; PAP: phosphatidic acid phosphatase; DGAT: diacylglycerol acyltransferase; OBO: oil body oleosin; LTP: lipid transfer protein; ER: endoplasmic reticulum.

表1 甘蓝型油菜皖油20种子不同部位油脂合成差异表达基因
(续表1)

基因名Gene ID注释描述Annotation description蛋白家族缩写Protein family abbreviations分组Group BnaC03g27860DHydroxysteroid dehydrogenase 1OBOEA-IC-OC BnaCnng57830DHydroxysteroid dehydrogenase 1OBOEA-IC BnaA09g02110DOleosinOBOEA-OC BnaA03g20420DStearoyl-acyl-carrier-protein desaturase proteinSADEA-IC/EA-OC BnaA01g32860DStearoyl-acyl-carrier-protein desaturase proteinSADEA-IC/EA-OC BnaC03g24420DStearoyl-acyl-carrier-protein desaturase proteinSADEA-IC/EA-OC BnaC09g41580DStearoyl-acyl-carrier-protein desaturase proteinSADEA-IC/EA-OC BnaA10g18080DStearoyl-acyl-carrier-protein desaturase proteinSADEA-IC/EA-OC BnaA05g03490DStearoyl-acyl-carrier-protein desaturaseSADEA-OC BnaC04g03030DStearoyl-acyl-carrier-protein desaturaseSADEA-OC BnaC09g19280D3-ketoacyl-acyl carrier protein synthase IKASIEA-IC/EA-OC BnaA02g24400D3-ketoacyl-acyl carrier protein synthase IKASIEA-IC/EA-OC BnaA06g36060D3-ketoacyl-acyl carrier protein synthase IKASIEA-IC/EA-OC BnaC06g35760D3-ketoacyl-acyl carrier protein synthase IIKAS IIEA-IC/EA-OC BnaA07g31890D3-ketoacyl-acyl carrier protein synthase IIKAS IIEA-IC/EA-OC BnaA07g21940D3-ketoacyl-acyl carrier protein synthase IIKAS IIEA-IC/EA-OC BnaC06g22680D3-ketoacyl-acyl carrier protein synthase IIKAS IIEA-IC BnaA04g07120DAcyl-ACP thioesteraseFATAEA-IC/EA-OC BnaCnng41490DAcyl-ACP thioesteraseFATAEA-IC/EA-OC BnaCnng00070DFATA acyl-ACP thioesterase FATAFATAEA-IC/EA-OC BnaA07g05070DFATA acyl-ACP thioesterase FATAFATAEA-IC/EA-OC BnaC03g75820DKetoacyl-ACP ReductaseKAREA-IC/EA-OC BnaA02g13310DBeta-ketoacyl reductaseKAREA-IC/EA-OC BnaA07g26670DBeta-ketoacyl reductaseKAREA-IC/EA-OC BnaC06g28830DBeta-ketoacyl reductaseKAREA-IC/EA-OC BnaC09g16320DAcyl carrier proteinACPEA-IC/EA-OC BnaC09g03000DAcyl carrier proteinACPEA-IC/EA-OC BnaA09g03610DAcyl carrier proteinACPEA-IC/EA-OC BnaAnng23710DAcyl carrier proteinACPEA-OC BnaC03g45040DEnoyl-ACP ReductaseENREA-IC/EA-OC BnaC07g04330DEnoyl-ACP ReductaseENREA-IC/EA-OC BnaA03g38220DEnoyl-ACP ReductaseENREA-IC/EA-OC BnaAnng02240DE2 component of pyruvate dehydrogenase complexDHLATEA-IC/EA-OC BnaC06g08280DE2 component of pyruvate dehydrogenase complexDHLATEA-IC/EA-OC BnaC07g23030DE2 component of pyruvate dehydrogenase complexDHLATEA-IC BnaA06g33300DE2 component of pyruvate dehydrogenase complexDHLATEA-IC BnaAnng22560DChloroplasticacetyl coenzyme A carboxylaseBCCPEA-IC/EA-OC BnaC09g42420DBiotin carboxyl carrier protein 2BCCPEA-IC/EA-OC BnaA03g02830DThioesterase superfamily proteinHADEA-IC/EA-OC BnaA02g00390DThioesterase superfamily proteinHADEA-IC BnaA07g20920DLong chain acyl-CoA synthetase 9LACS9EA-IC/EA-OC BnaC06g20910DLong chain acyl-CoA synthetase 9LACS9EA-IC BnaA01g17630DE3 component of pyruvate dehydrogenase complexLPDEA-IC/EA-OC BnaCnng75250DAcetyl-CoA carboxylaseACCaseEA-IC BnaA05g12180DAcyl-carrier-proteinMCMTEA-IC/EA-OC
Lu等[26]对ZS11和WH5557两个油菜材料的种子不同部位进行分析表明, IC、OC和EA中脂质代谢物含量存在显著差异, IC、OC和EA中糖酵解途径和油脂合成途径的基因表达水平普遍存在着显著差异。并进一步鉴定了编码LPAAT、PAP、DGAT、Oleosin等调控种子不同部位脂质含量差异的关键基因。本研究在前期工作基础上, 对低芥酸油菜WY20种子不同部位进行转录组分析, 进一步从转录水平解析油菜种子不同部位含油量和脂肪酸差异的调控机制。WY20种子3个部位转录组分析结果显示, EA与IC和OC之间的差异表达基因主要富集在光合作用、一元羧酸合成和脂肪酸合成与代谢过程, 这些差异表达基因可能是造成不同部位含油量和脂肪酸差异的主要因素(图3-B, C)。IC和OC之间差异表达基因仅有18个基因与脂质代谢相关, 而EA与IC和OC之间与脂质代谢相关的差异表达基因分别为268个和286个, 与EA含油量和脂肪酸组成与IC和OC差异比较一致(图1-B, D)。对油脂合成途径进行深入分析显示, IC和OC中与油脂合成相关的基因的表达量普遍是EA的2~3倍, 例如在IC和OC中的表达量分别是EA中的2.8倍和2.5倍(图4)。这些差异表达基因主要集中在质体中脂肪酸合成途径, 因此我们推断这些关键基因在转录水平的调控是造成EA含油量比IC和OC低的主要原因(图4)。
脂肪酸组成主要由脂肪酸脱饱和酶和脂肪酸延长酶调控[33]。植物质体中由SAD催化合成18:1-ACP, 而其余不饱和脂肪酸合成的反应主要发生在内质网中。从质体转运出来的18:1-CoA被合成PC, 接着在油酸去饱和酶(FAD2)和亚油酸去饱和酶(FAD3)作用下分别生成18:2-PC和18:3-PC。本研究结果表明在IC和OC中表达量分别为EA的2.4倍和2.2倍, 与EA中C18:1比例最低而C18:2比例最高结果一致。在IC和OC中表达量分别为EA的3.8倍和4.1倍, 与EA中C18:3比例最低结果一致。而EA中C16:0的含量约为IC和OC的2倍, 从代谢途径看,、和在IC和OC中的表达量均显著高于EA (2.5~3.7倍), 因此C16:0在IC和OC中被更高效地转化为C18:1, 这与IC和OC中C16:0比例比EA低一致(图1-D和图4)。说明、、、和等关键基因在WY20种子的表达有部位特异性, 这些基因的表达量决定了种子不同部位中脂肪酸的组成。
4 结论
WY20油菜种胚不同部位的含油量和脂肪酸组成存在着差异, 尤其是EA的含油量最低并且脂肪酸组成与子叶存在较大差异。EA与子叶之间参与油脂合成的差异表达基因较多。EA中参与质体中脂肪酸合成的基因表达量普遍低于子叶, 造成EA含油量比子叶低。EA中、、、和等关键基因表达量显著低于子叶, 造成EA中C16:0和C18:2高于子叶。转录调控是WY20油菜种子不同部位油脂合成差异的主要机制, 该研究对于理解油菜种子不同部位含油量和脂肪酸组成差异具有一定的科学意义, 对油菜和其他油料作物高含油量育种和品质改良具有重要的指导意义。
[1] 沈金雄, 傅廷栋. 我国油菜生产、改良与食用油供给安全. 中国农业科技导报, 2011, 13(1): 1–8 Shen J X, Fu T D. Rapeseed production improvement and edible oil supply in China., 2011, 13(1): 1–8 (in Chinese with English abstract)
[2] 沈琼. 中国油菜产业竞争优势与劣势分析. 农业产品加工, 2008, (8): 57–59 Shen Q. Analysis on competitive advantages and disadvantages of Chinese rapeseed industry., 2008, (8): 57–59 (in Chinese with English abstract)
[3] 李殿荣, 田建华, 陈文杰, 张文学, 李永红, 王灏. 甘蓝型油菜特高含油量育种技术与资源创新. 西北农业学报, 2011, 20(12): 83–87 Li D R, Tian J H, Chen W J, Zhang W X, Li Y H, Wang H. Breeding technologies and germplasm innovation on extra- high-oil content in., 2011, 20(12): 83–87 (in Chinese with English abstract)
[4] 张永霞, 赵锋, 张红玲.中国油菜产业发展现状、问题及对策分析. 世界农业, 2015, (4): 96–99 Zhang Y X, Zhao F, Zhang H L. Analysis on the development status, problems and countermeasures of Chinese rapeseed industry., 2015, (4): 96–99 (in Chinese with English abstract)
[5] 王汉中, 殷艳. 我国油料产业形势分析与发展对策建议. 中国油料作物学报, 2014, 36: 414–421 Wang H Z, Yin Y. Analysis and strategy for oil crop industry in China., 2014, 36: 414–421 (in Chinese with English abstract)
[6] 熊秋芳, 张效明, 文静, 李兴华, 傅廷栋, 沈金雄. 菜籽油与不同食用植物油营养品质的比较——兼论油菜品质的遗传改良. 中国粮油学报, 2014, 29: 122–128 Xiong Q F, Zhang X M, Wen J, Li X H, Fu T D, Shen J X. Comparison of nutritive quality between rapeseed oil and different edible vegetable oil—on the genetic improvement of rapeseed quality., 2014, 29: 122–128 (in Chinese with English abstract)
[7] 熊源. 植物油的种类与营养价值. 中国粮食经济, 2014, (6): 72 Xiong Y. Types and nutritional value of vegetable oil., 2014, (6): 72 (in Chinese with English abstract)
[8] Saha S, Enugutti B, Rajakumari S. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic. diacylglycerol acyltransferase., 2006, 141: 1533–1543
[9] Thelen J J, Ohlrogge J B. Metabolic engineering of fatty acid biosynthesis in Plants., 2002, 4: 12–21
[10] Dahlqvist A, Stahl U, Lenman M. Phospholipid: diacylglycerol acyltransferase: An enzyme that catalyzes the Acyl-CoA- Independent formation of triacylglycerol in yeast and plants., 2000, 97: 6487–6492
[11] 周奕华, 陈正华. 植物种子中脂肪酸代谢途径的遗传调控与基因工程. 植物学通报, 1998, 15(5): 16–23 Zhou Y H, Chen Z H. Genetic manipulation and gene engineering of fatty acid metabolism in plant seeds., 1998, 15(5): 16–23 (in Chinese with English abstract)
[12] 周丹, 赵江哲, 柏杨, 张群, 井文, 章文华. 植物油脂合成代谢及调控的研究进展. 南京农业大学学报, 2012, 35(5): 81–90 Zhou D, Zhao J Z, Bai Y, Zhang Q, Jing W, Zhang W H. Research advance in triacylglycerol synthesis, metabolism and regulation in plants. 2012, 35(5): 81–90 (in Chinese with English abstract)
[13] Bates P D, Stymne S, Ohlrogge J B. Biochemical pathways in seed oil synthesis., 2013, 16: 358–364
[14] Horn P J, Korte A R, Neogi P B, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee Y J, Chapman K D. Spatial mapping of lipids at cellular resolution in embryos of cotton., 2012, 24: 622–636
[15] Horn P J, Silva J E, Anderson D, Fuchs J, Borisjuk L, Nazarenus T J, Shulaev V, Cahoon E B, Chapman K D. Imaging heterogeneity of membrane and storage lipids in transgenicseeds with altered fatty acid profiles., 2013, 76: 138–150
[16] Sturtevant D, Dueñas M E, Lee Y J, Chapman K D. Three- dimensional visualization of membrane phospholipid distributions inseeds: a spatial perspective of molecular heterogeneity., 2017, 1862: 268 –281
[17] Woodfield H K, Sturtevant D, Borisjuk L, Munz E, Guschina I A, Chapman K, Harwood J L. Spatial and temporal mapping of key lipid species inseeds., 1998, 173: 1998–2009
[18] Sturtevant D, Lee Y J, Chapman K D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ., 2015, 37: 53–60
[19] Lu X, Chen D, Shu D. The differential transcription network between embryo and endosperm in the early developing maize seed., 2013, 162: 440–455
[20] He R, Salvato F, Park J J. A systems-wide comparison of red rice () tissues identifies rhizome specific genes and proteins that are targets for cultivated rice improvement., 2014, 14: 46–66
[21] Schuster S C. Next-generation sequencing transforms today’s biology., 2008, 5: 16–18
[22] Metzker M L. Sequencing technologies: the next generation., 2010, 11: 31–46
[23] Louisa F L. RNA-Seq: a revolutionary tool for transcriptomics., 2008, 9: 568–574
[24] Troncoso-Ponce M A, Kilaru A, Cao X, Durrett T P, Fan J, Jensen J K, Thrower N A, Pauly M, Wilkerson C, Ohlrogge J B. Comparative deep transcriptional profiling of four developing oil seeds., 2011, 68: 1014–1027
[25] Dussert S, Morcillo F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition., 2013, 162: 1337–1358
[26] Lu S P, Sturtevant D, Aziz M, Jin C, Li Q, Chapman K D, Guo L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oilL. seeds., 2018, 94: 915–932
[27] Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D R, Pimentel H, Salzberg S L, Rinn J L, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks., 2012, 7: 562–578
[28] Anders S, Pyl P T, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data., 2015, 31: 166–169
[29] Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data., 2010, 26: 136–138
[30] Love M I, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2., 2014, 15: 550–575
[31] Baud S, Lepiniec L. Physiological and developmental regulation of seed oil production., 2010, 49: 235–249
[32] Borisjuk L, Neuberger T, Schwender J, Heinzel N, Sunderhaus S, Fuchs J, Hay J O, Tschiersch H, Braun H P, Denolf P, Lambert B, Jakob P M, Rolletschek H. Seed architecture shapes embryo metabolism in oilseed rape., 2013, 25: 113–128
[33] Napier J A, Haslam R P, Beaudoin F. Understanding and manipulating plant lipid composition: Metabolic engineering leads the way., 2014, 19: 68–75
Transcriptional regulation of oil biosynthesis in different parts of Wanyou 20 () seeds
ZHANG Yu-Ting, LU Shao-Ping, JIN Cheng, and GUO Liang*
National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China
is one of the main oil crops and the seed oil content is generally between 35% and 50%. Oil is mainly stored in the seed embryo. Embryo is composed of cotyledons (including outer and inner cotyledons) and embryonic axis. The oil content and fatty acid composition of different parts of low erucicWY20’s seed were analyzed. There was a significant difference in oil content in different parts of the seed. The oil content in the outer cotyledon was the highest while embryonic axis had the lowest oil content. At the same time, the fatty acid composition also showed significant difference in different parts of the seed. The ratio of C16:0, C18:2, and C20:0 in embryonic axis was significantly higher than that in cotyledon. The ratio of C16:0 in the embryonic axis was about twice more than that of the cotyledons. The ratio of C18:1 and C20:1 in cotyledons was significantly higher than that in embryonic axis. C18:0 had the lowest content in the outer cotyledon and no difference in the inner cotyledons and embryonic axis. C18:3 had the highest content in the outer cotyledons and no difference between inner cotyledons and embryonic axis. Transcriptome analysis was performed for the inner cotyledon, outer cotyledon and embryonic axis of the 34-day-old seed. A total of 7192 differentially expressed genes (DEGs) were identified after pairwise comparison of gene expression of the three parts. There were much fewer DEGs between cotyledons and more DEGs between cotyledon and embryonic axis. These DEGs were enriched in biological processes such as photosynthesis, fatty acid metabolism and chlorophyll metabolism. Gene function annotations revealed that there were 355 genes involved in lipid metabolism, especially in the de novo fatty acid biosynthesis in plastid. This study suggests that transcriptional regulation of key genes involved in oil biosynthesis results in different oil contents and fatty acid compositions in different parts of seed in.
; different seed parts; oil content; fatty acid composition; transcriptional regulation
2018-07-28;
2018-12-24;
2019-01-03.
10.3724/SP.J.1006.2019.84105
郭亮, E-mail: guoliang@mail.hzau.edu.cn
E-mail: 249749481@qq.com
本研究由国家自然科学基金青年科学基金项目(31701458)和中央高校基本科研业务费专项资金资助项目(2662015PY090)资助。
This study was supported by the National Science Foundation for Young Scientists of China (31701458) and Fundamental Research Funds for the Central Universities (2662015PY090).
URL: http://kns.cnki.net/kcms/detail/11.1809.S.20190101.1031.002.html

