Identification of a toxin coding fragment in pBSSB1, a linear plasmid from Salmonella enterica serovar Typhi that can stabilize a multicopy plasmid
Sunjukta Ahsan, David Summers
1Department of Genetics, University of Cambridge, CB2 3EH, UK
2Department of Microbiology, University of Dhaka, Dhaka-1000, Bangladesh
Keywords:Linear plasmid pBSSB2 Salmonella enterica serovar Typhi
ABSTRACT Objective: To identify the region conferring stability to pBSSB2 (a linear plasmid, pBSSB1,containing a kanamycin cassette), which is unique to Indonesian isolates of Salmonella enterica serovar Typhi. Methods: The open reading frame (ORF) 009 was identified as a toxin coding gene in the plasmid through introduction of translational termination codons in the ORF.Results: The stability function was located in a fragment that spanned nucleotides 5 766 to 6 828 in the linear plasmid genetic map. Ectopic expression of ORF009 in pBAD18 vector indicated ORF009 codes for a toxin. This fragment could stabilize plasmid pUC18 previously destabilized through mutation of the pcnB (plasmid copy number control) gene that codes for polyA polymerase. Majority of the cells expressing ORF009 were non-viable according to phase contrast microscopy. Conclusions: This study demonstrated that a linear plasmid fragment that carries a gene encoding a toxin possibly conferred stability to the parent plasmid.It was able to stabilize a multicopy plasmid of Escherichia coli.
1. Introduction
Salmonella entericaserovars Typhi can express several flagellar antigens[1-9]. Indonesian isolates ofSalmonellaTyphi express the novel antigen, H: z66 (z66), encoded by a linear plasmid,pBSSB1[6]. Plasmids are extrachromosomal elements and can replicate independently of the host chromosome, which are usually circular. Linear plasmids were first reported inStreptomyces rocheiin 1979[10]. Linear plasmids ofStreptomycesspp. range in size between 12 and 640 kb[11]. Linear plasmids possessing a different structure have also been reported inBorreliaspp[12]. The linear plasmid pBSSB1 was first reported by Bakeret al.[6]. It is 27 kb containing terminal inverted repeats of 1 230 bp and terminal proteins that make it resistant to 5’ to 3’ exonucleases[6]. Thirty-three open reading frames (ORFs) have been identified, of which ORF031 codes for the flagellar antigen, H: z66[6]. Previously, the protein Fis has been shown to affect the stability of pBSSB1[13]. However, the specific regions involved in multiplication and stable survival of the linear plasmid have not been identified. Here we describe a region of pBSSB1 with a putative toxin-antitoxin function that influences the stability of the plasmid and can also stabilise a circular multicopy plasmid ofEscherichia coli(E. coli). We believe that the findings of this study will help to understand the mechanism of stability of linear plasmids. To our knowledge, this is the first report of the stabilization of a multicopy, circular plasmid by the stability function of a linear plasmid.
2. Materials and methods
2.1. Plasmids
The linear plasmid pBSSB2 (pBSSB1 with a kanamycin resistance cassette integrated at a location of 1 495 bp)[6] was provided by Gordon Dougan, Welcome Trust Sanger Institute, Hinxton, UK.Plasmids pREG531 and pFH450[9] used in stability assays were obtained from Dr. Finbarr Hayes, University of Manchester. The expression vector pBAD18 was described elsewhere[14].
2.2. Bacterial strains
E. coliSP2[15] (pcnB, Δlac,recA) which is a plasmid copy number mutant and DNA polymerase Ⅰ mutant BR825[16] deficient in DNA polymerase Ⅰ was obtained from Finbarr Hayes. Strain BW25141 (lacIqrrnBT14ΔlacZWJ16ΔphoBR580hsdR514ΔaraBADAH33ΔrhaBADLD78galU95 endABT333uidA(ΔMluⅠ)::pir+recA1) was described earlier[14].
2.3. Polymerase chain reaction (PCR)
Oligonucleotide primers are listed in Table 1. Long amplicons were generated by using the Expand Long Template PCR system(www.roche-applied-science.com). Reaction conditions and reaction mixture compositions were prepared according to the manual of the manufacturer.
2.4. DNA digestion and purification
DNA digestion and ligation were carried out according to standard protocols. The Qiagen PCR purification kit was used to purify PCR products and digested DNA following the manufacturer’s instructions. DNA was purified from the agarose gel by using the gel extraction kit (QIAquick, UK).
2.5. Construction of a library of overlapping fragments of pBSSB2
Overlapping 4-6 kb regions of pBSSB2 were amplified using PCR primer pairs designated RP2-RP10 (Table 1).EcoRⅠ andBamHⅠrestriction sites were added as primer tails to allow cloning of fragments.Amplified regions were cloned into pUC18 by usingEcoRⅠandBamHⅠrecognition sites.
2.6. Electroporation
The appropriateE. colistrain was inoculated in ten mL of LB-broth and incubated at 37 ℃ overnight in a shaker incubator. Cells were collected by centrifugation for 10 min (MSE Harrier) at 4 000 rpm and washed using ice-cold sterile distilled water. The centrifugation and cell washing step was repeated thrice. Cells were suspended in 200 µL of ice-cold sterile distilled water. DNA was added to the competent cells (50 µL) and mixed briefly by gentle pipetting. Electroporation was carried out under conditions of pulse at 1.6 kV, 200 Ohms in a Gene Pulser unit (Bio-Rad). Next, 450 µL of SOC (Life technologies,UK) medium was quickly added to the cells, followed by a recovery step of 1 h at 37 ℃ at 120 rpm. Finally, cells were spread on appropriate medium and incubated overnight at 37 ℃.

Table 1 Oligonucleotides used in the present study.
2.7. Chemical transformation
Chemical transformation was used to introduce plasmid DNA intoE. coliBR825 (DNA polymerase Ⅰ mutant). DNA was added to 100 µL of competent cells. Heat shock was applied at 42 ℃ for approximately 1 min, then cells were placed on ice for 2 min. Nine hundred µL of pre-warmed LB-broth was added. Transformed cells were allowed to recover for 3 h at 37 ℃ followed by further recovery for 18-24 h after adding 1 mL culture to 9 mL of LB-broth.Appropriate antibiotics were added to this 10 mL culture. The tube was incubated at 37 ℃ for 18-24 h in a static incubator. Following incubation, harvested cells were re-suspended in LB-broth (100 µL).They were then spread on medium in the presence of the appropriate antibiotic and incubated at 37 ℃ for 18-24 h.
2.8. Stability assay
Stability was tested using the assay described earlier[17] in which strains were streaked twice to single colonies on non-selective agar before being tested for plasmid retention.
2.9. Phase contrast microscopy
Five hundred µL of boiling 1% (w/v) agarose in phosphate buffered saline was spread allowed to solidify on a microscope slide. Between 5 and 10 µL of bacterial culture was spread on top of the agar after it had solidified. The slide was viewed immediately using an oil immersion objective and phase contrast illumination.
2.10. Site-directed mutagenesis
RP4 cloned in pUC18 was mutated by using pre-designed synthetic oligonucleotides following the method described elsewhere[18]. Stop codons were introduced in each ORF (ORF 007-009) by means of site directed mutagenesis[18] to determine its role in stability by using pre-designed oligonucleotides (Table 2).

Table 2 Oligonucleotides to insert stop codon in ORFs in RP4.
2.11. Toxicity assay
The expression vector pBAD18[19] contains the arabinose promoter,PBAD, that is repressed by glucose and induced arabinose in the growth medium. ORF009 was cloned downstream of PBADusingEcoRⅠ andPstⅠ restriction sites and the resulting construct was coined pBAD18-ORF009. For the toxicity assay, cultures ofE. coliXLI-Blue with pBAD18 or pBAD18-ORF009 were grown overnight in LB-broth at 37 ℃. Each overnight culture (250-300 µL) was transferred to fresh LB-broth (50 mL) medium and allowed to reach an OD600≈ 0.3. Each culture was then split into two. In one subculture cells were grown in LB-broth containing glucose (0.2%)while the other contained arabinose (0.2%) to induce ORF009 expression. The culture densities were measured 30 min, 60 min,90 min and 150 min after the culture was split and samples were withdrawn simultaneously for estimation of viable counts on LB-agar plates without glucose or arabinose.
2.12. DNA sequencing
DNA sequencing was carried out by Geneservice Ltd. by applying BigDye Terminator and 3730 capillary sequencer (Applied Biosystems).
3. Results
3.1. Screening for stability functions of pBSSB2
A set of pUC18 derivatives containing overlapping 4-6 kb regions of pBSSB2 (pRP2-pRP10) were screened for increased stability in aE. colipcnB(plasmid copy number control gene) mutant. In the negative control (pUC18, no insert), about 60% of cells retained the plasmid (Figure 1). With the exception of pRP2 and pRP4, all constructs were retained less efficiently than pUC18, presumably because of the increased metabolic load imposed by the 4-6 kb inserts. Construct pRP2 was retained, on average, by 74% of colonies while pRP4 was retained by 94%. The improvement in stability conferred by RP4 was confirmed in an independent stability assay using an unstable bi-replicon plasmid (data not shown).
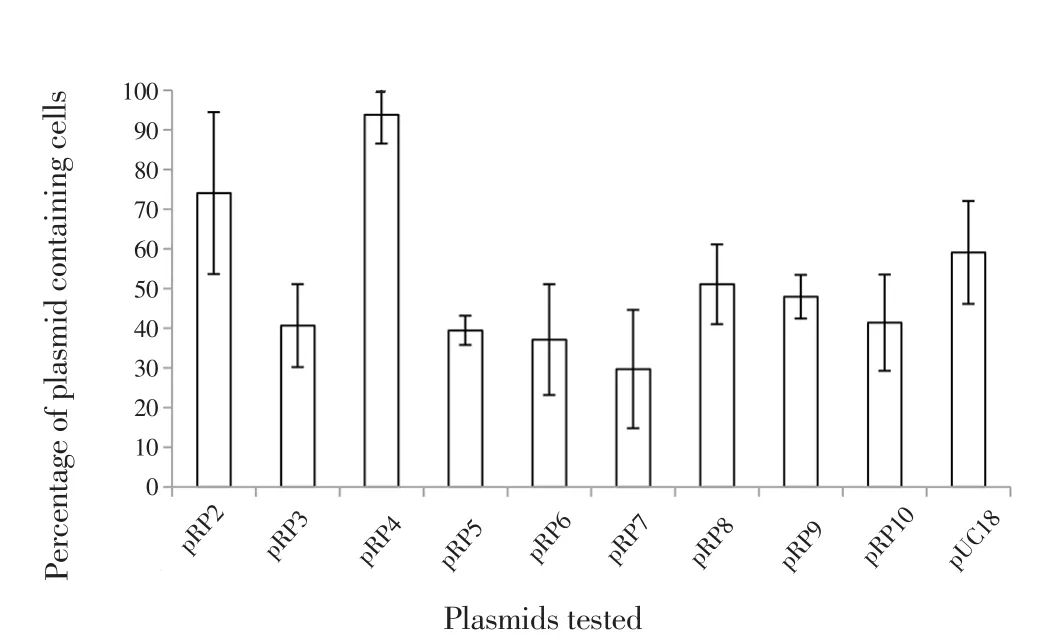
Figure 1. Stability of plasmids containing pBSSB2 fragments, RP2-RP10.
3.2. Deletion analysis of RP4 stability region
Fragment RP4 of pBSSB2 was approximately 4.5 kb (Figure 2a).The construct was digested withHindⅢ to leave approximately 2.2 kb of RP4. The resultant pRP4 derivative that contains three annotated ORFs was named pRP4HindⅢ (Figure 2b). Plasmid stability assays were carried out in apcnBhost to compare the stabilities of pRP4HindⅢ, pRP4 and pUC18. It was found that fragment RP4HindⅢ stabilized pUC18 as effectively as the full RP4 region.
A series of subclones of RP4HindⅢ was amplified by PCR, using primers listed in Table 1. Each PCR product was cloned in pUC18 with the aid ofEcoRⅠ andBamHⅠ restriction sites and amplified a subsection of RP4HindⅢ. Plasmid stability was tested in thepcnBmutant. In all cases where the right end of RP4HindⅢ had been removed, stability of the circular plasmid was reduced (Figure 2).From the left end deletions of 1 173 bp retained the stability function of RP4HindⅢ in a circular derivative (Figure 2c). This defined a 1 068 bp fragment that conferred stability and contained ORFs 008 and 009 [Fragment bounded by arrows (Figure 2c)].

Figure 2. Map of pBSSB2 fragments RP4 and RP4HindⅢ (figure not drawn to scale).
3.3. Mutational analysis of RP4 stability region
RP4HindⅢ contained three open reading frames: ORF007,ORF008 and ORF009 (Figure 2b). Mutated fragments were inserted into pUC18 with the help ofEcoRⅠ andBamHⅠ restriction sites.The presence of the desired mutation was confirmed by sequencing.The effect of individual mutation in ORFs 007-009 in pRP4HindⅢderivatives was investigated in apcnBmutant (Figure 3). The result showed that stability of the plasmid in which a translation termination codon was introduced in ORF009 was reduced, suggesting that only ORF009 was necessary for stabilization of the circular plasmid.

Figure 3. Effect of point mutation of ORFs 007-009 on stability of pUC18 carrying fragment RP4HindⅢ.
3.4. Characterization of the RP4 stability function
The RP4 stability region seems more likely to encode postsegregational killing than active partition since only one ORF is needed. Active partitioning systems normally have two ORFs that code for ATPase and DNA-binding proteins. On the other hand,in a toxin-antitoxin system, while the toxin is normally a protein,the antitoxin can either be protein or RNA. We therefore explored the possibility that ORF009 is the toxin counterpart of a postsegregational killing system.
In presence of pBAD18 only, a similar pattern of growth (OD600)was observed in the presence of glucose and arabinose. Cells containing pBAD18-ORF009 exhibited reduced growth when arabinose was present but grew normally in the presence of glucose.Cells containing pBAD18 or pBAD18-ORF009 exhibited different viable counts in the presence of glucose and arabinose. In the presence of arabinose, viable counts for cells containing pBAD18-ORF009 fell by 104-fold after 30 min. Thus the expression of ORF009 resulted in the death, though not lysis, of 99.99% of the cells when compared with cells containing functional ORF009(Figures 4a & b).
To observe whether expression of ORF009 produced changes in cell morphology,E. coliXLI-Blue with pBAD18 or pBAD18-ORF009 grown in LB-broth supplemented with either glucose or arabinose were observed by phase contrast microscopy.E. coliXL1-Blue transformed with pBAD18 showed no change in cell morphology when grown for 30 min in LB-broth containing glucose or arabinose. However, after induction with arabinose for 30 min,cells became elongated and appeared to be ‘empty’ or ‘ghost’ cells(data not shown). Besides, A psi BLAST search (http://www.sbg.bio.ic.ac.uk/~phyre/) indicated a similarity of ORF009 to RelE like proteins (E-value 2e-27, 41.5% identity).

Figure 4. Growth of cultures containing pBAD18 or pBAD18-ORF009 in presence of glucose (0.2%) or arabinose (0.2%) measured in terms of OD600(a) and viable counts (b).
4. Discussion
The annotation of the sequence of linear plasmid pBSSB1 failed to identify replication or stability functions. A previous study reports that the protein Fis is required to stabilize the linear plasmid,however the study does not continue to identify the mechanism of stability function[13]. Here we have taken a functional approach to identify potential stability functions. In this approach, a SP2 mutant was used in which mutations inpcnBdecreased the copy number of ColE1-like plasmids, causing instability[20]. The gene carries information for a poly (A) polymerase, which polyadenylates the replication repressor RNAi[21,22]. In the absence of polyadenylation,RNAi has a longer half-life and consequently the copy number and stability of ColE1-like plasmids is reduced. By cloning fragments of pBSSB2 into such unstable, circular plasmids, a 1 062 bp fragment covering nucleotides 5 766 to 6 828 in the pBSSB2 genetic map was found to stabilise these plasmids. Only one ORF within this region(ORF009) was required to confer the stability phenotype.
Since a partitioning system usually required two proteins, one an ATPase and the other a DNA-binding protein, it was hypothesized that the stability function was a post-segregational killing system through expression of toxin-antitoxin. This hypothesis was tested by ectopic expression of the putative toxin ORF009. It was found that the great majority of cells expressing ORF009 were non-viable.These cells appeared to be empty when viewed under a phase contrast microscope. Cells with condensed poles with a central clearing as a result of the effect of a toxin have been termed as ‘ghost cells’[23].
ORF009 was found to be similar to RelE like proteins according to psi BLAST search. InE. coli, chromosomal DNA codes for arelEBtoxin-antitoxin locus. The RelE protein toxin which inhibits translation by cleavage of mRNA can be inhibited by binding of the RelB protein antitoxin[24]. A candidate RNA anti-toxin was identified on the opposite strand to ORF009. However, further studies will be required to determine whether the candidate promoter is functional and whether the putative transcript does indeed function as an antitoxin.
A toxin-antitoxin stability determinant has been found previously in a linear plasmid, pCLP, ofMycobacterium celatum[25]. The pCLP system shows similarity to PemI-PemK of plasmid R100 found inE. coli[26]. The pem operon ofMycobacterium celatumexpresses 98 and 84 amino acid peptides that function as PemK (toxin) and PemI(antitoxin), respectively. Upon transformation ofMycobacterium smegmatiswith the shuttle vector pMIP12 carrying the 373 bppemKgene, it was found that transformants were unable to grow on selective medium. A point mutation added into thepemKgene was able to restore growth.
A toxin-antitoxin system is a secondary stability function that kills plasmid-free cells formed when the active partitioning system fails.Sequencing and annotation of the pBSSB2 genome (deposited at the National Centre for Biotechnology Information; accession number AM419040) suggests that ORF017 codes for a nucleotide binding protein that carries structural similarity to ATPase-like ParA protein.A sequence homology to other bacterial plasmid partitioning systems was found including 63% and 51% homology toVibrio caribbenthicusandCorynebacterium glutamicum, respectively[6]. In the present study we did not investigate the function of the ParA homolog in pBSSB2 and neighbouring sequences. However, our data suggest that fragment RP2, which contains ORF017, when inserted into pUC18, may give some improvement in stability in apcnBmutant.Similar to theMycobacterium celatumpCLP linear plasmid ParA homolog[25], fragment RP2 is also located immediately adjacent to the RP7 replicon region. Further studies are required to determine whether RP2 contains a partitioning function in pBSSB2.
The linear plasmid, pBSSB2, fromSalmonella entericaserovar Typhi, harbours a toxin-antitoxin function which is presumed to play a role in plasmid stability. The toxin component is encoded by ORF009. Ectopic expression of the toxin is able to kill host cells.The antitoxin remains unidentified in this study. The identified toxin-antitoxin fragment was demonstrated to be able to stabilize a destabilized multicopy plasmid (pUC18) ofE. coli. This study demonstrates that as with circular plasmids, linear plasmids harbour toxin-antitoxin functions for segregational stability. This constitutes the first report of the stabilization capacity of a linear plasmid function in a circular plasmid.
Conflict of interest statement
We declare no conflict of interest.
 Asian Pacific Journal of Tropical Biomedicine2018年7期
Asian Pacific Journal of Tropical Biomedicine2018年7期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Role of toll like-receptor 2 in inflammatory activity of macrophage infected with a recombinant BCG expressing the C-terminus of merozoite surface protein-1 of Plasmodium falciparum
- Moderating effect of synthesized docosahexaenoic acid-enriched phosphatidylcholine on production of Th1 and Th2 cytokine in lipopolysaccharide-induced inflammation
- Characterization of Cnidoscolus quercifolius Pohl bark root extract and evaluation of cytotoxic effect on human tumor cell lines
- Anti-epileptic effect of morin against experimental pentylenetetrazol-induced seizures via modulating brain monoamines and oxidative stress
- Expression of fluorescent tagged recombinant erythroferrone protein
- Influence of different cultivars of Phoenix dactylifera L-date fruits on blood clotting and wound healing
