Bioelemental patterns in the cerebrospinal fluid as potential biomarkers for neurodegenerative disorders
Neurodegenerative disorders like Parkinson’s disease (PD) or atypi‐cal Parkinsonian syndromes including the different synucleinopa‐thies and tauopathies are an important burden for patients, rela‐tives, care providers and incur mounting costs for the health care system in our aging society. The lack of disease modifying strategies and the failure in translating promising molecules from bench to bedside is also attributable to a relatively late diagnosis: when pa‐tients become symptomatic and seek medical advice, neurodegen‐eration has already widely spread through the central nervous sys‐tem and thus represents a major obstacle for disease‐modifying and/or regenerative therapies. The detection of neurodegenerative processes at an earlier stage by biomarkers is urgently needed to de fine state and trait of a disease condition, which ideally would in‐crease precision of diagnosis, allow for strati fied therapeutic inter‐ventions and monitoring of treatment effects. The mechanisms causing neurodegeneration in aggregation‐related disorders like PD are not completely understood but most likely are multifactorial,including factors like oxidative stress, autophagic‐lysosomal dys‐function, mitochondrial dysfunction and prion‐like spreading of misfolded proteins (Maiti et al., 2017). Importantly, biometals and other bioelements were shown to modify these mechanisms under multiple circumstances. For example, aggregation of alpha‐synucle‐in, a hallmark of PD and other synucleinopathies, can be enhanced by different biometals like iron, copper, aluminium and magnesium(Uversky et al., 2001). Pathological protein aggregation triggered by bioelements has not only been shown for PD but also for other neurodegenerative disorders like Amyotrophic lateral sclerosis(ALS) and Alzheimer’s disease (Aizenman and Mastroberardino,2015), suggesting a common disease mechanism. In addition to ef‐fects on protein aggregation, biometals can be involved in neurode‐generative pathogenesis by catalysis of oxidative reactions. One ex‐ample is the contribution of iron to the Haber‐Weiss‐ and Fenton‐reactions, which results in increased oxidative stress in do‐paminergic neurons, their dysfunction and consequent demise(Carboni and Lingor, 2015). Because disease pathology is unlikely to be in fluenced by one single element alone, the characterization of bioelemental patterns may be more promising for the identi fica‐tion of a biomarker. Advanced bioinformatics methods extending beyond descriptive statistics are helpful to detect changes in such multivariable patterns. A number of computational machine learn‐ing algorithms allowing for predictive pattern recognition are avail‐able and can be used for analysis of complex data sets in biomedical research applications. Recently, our group used machine learning techniques to analyse patterns in the elemental composition of ce‐rebrospinal fluid (CSF) samples of PD patients and age‐matched healthy controls. CSF has a close spatial relation to the affected brain regions in neurodegenerative pathology and thus has the po‐tential to re flect pathophysiological changes of the disease process‐es. Although the acquisition of CSF is more invasive in comparison to blood samples, CSF may represent a more faithful biomarker source when it comes to neurodegenerative disorders like PD. Ele‐mental serum levels may be variable for example in correlation to specific food and supplement intake while CSF levels are more tightly regulated and thus may be more meaningful in regard to their diagnostic value. In our discovery trial (Maass et al., 2018),inductively‐coupled plasma mass spectrometry (ICP‐OES and ICP‐sf‐MS) was used for element determination and a total of 28 ele‐ments were quantified in CSF samples of 36 PD patients and 42 age‐matched controls, including some elements in trace concentra‐tions down to nanogram levels. Nineteen out of those initial 28 ele‐ments were stably detectable and used for further analysis. In the single element comparison, arsenic, magnesium and selenium showed signi ficantly higher mean CSF levels in the PD group com‐pared with the control group after multiple adjustments. However,previous trials showed contradictory results on the levels of single elements, which may be due to their high variability and limited sample size (Jiménez‐Jiménez et al., 2014). These limitations also applied to our trial and we therefore aimed at the identi fication of patterns instead of individual elemental levels. In order to establish differences in the bioelemental pattern, machine‐learning was ap‐plied to this dataset, showing the best performance for the discrim‐ination of PD patients and controls for the gradient tree boosting algorithm, yielding a good area under the receiver‐operated curve(AUROC) of 0.83. The term “boosting” belongs to an ensemble method in the field of machine learning, which creates a strong classi fier from multiple weak classi fiers, in this case based on differ‐ent decision trees (Figure 1). The classi fication decision tree itself is a predictive model, based on a binary tree‐like system with nodes representing the input variable (abundance of bioelements) used for prediction, branches representing the test outcome and leaves representing the output class label (PDvs. control). Since the analy‐sis of many elements is not economical, we aimed to reduce the number of required elements to a minimum using the AUC results from the 10 times repeated 10‐fold cross validations as a feature se‐lection criterion to determine the optimal set of elements for a good classi fier. This resulted in the identi fication of a cluster of six single elements (Se, Fe, As, Ni, Mg, Sr), which most importantly contributed to the sample discrimination. Based on the six remain‐ing elements from the feature selection analysis, the feature impor‐tance for each of these elements was quanti fiedvia10 times 10‐fold cross validation of a classi fier trained on these elements only. It can be suspected that the elemental pattern identi fied in this analysis is not a mere epiphenomenon but re flects the role of these elements in disease pathogenesis. Changes in endogenous elemental pattern may represent a disease speci fic dysregulation, which could be in flu‐enced by external (e.g., environmental) factors. Across the contigu‐ous 48 states of the USA, PD mortality rates have a signi ficant posi‐tive correlation with soil strontium concentrations, and an inverse correlation with soil selenium concentrations (Sun, 2018). Further investigation in other industrial areas is needed to validate this po‐tential relationship,e.g., in Scandinavian countries, where epidemio‐logical registers of high quality are available. While the role of iron in the pathogenesis of PD has been widely described, our analysis ar‐gues for an important contribution of selenium, which had the high‐est impact in this model. There is growing evidence for the impor‐tance of selenium and selenoproteins in neurodegenerative disorders like PD but there is also evidence for an involvement in Alzheimer’s disease, Multiple sclerosis, ALS and Huntington’s disease (Cardoso et al., 2015). In PD, one function of selenoproteins seems to be the pro‐tection of dopaminergic neurons against oxidative stress and cell death. A more detailed characterization of selenoproteins could have the potential to enhance the sensitivity of the bioelemental finger‐print, likely yielding an even better sensitivity. Our analysis with a proof‐of‐concept approach showed that changes in elemental signa‐tures in the CSF detected by machine learning techniques may have the potential to be used as a biomarker signature for the diagnosis of neurodegenerative diseases like PD. This approach using the CSF composition of elements as a biomarker for neurodegeneration seems also to be promising in ALS. Alterations in heavy metals, such as lead, cadmium and mercury, have been described in this moto‐neuron disease (Vinceti et al., 2017a). Changes in elemental levels were also recently detected in serum, urine and hair samples of ALS patients, again showing dysregulated selenium levels as a marker for neurodegeneration (Oggiano et al., 2018). In Alzheimer’s disease,higher levels of inorganic selenium in the CSF may predict the con‐version from mild cognitive impairment (Vinceti et al., 2017b). In the field of Parkinsonian syndromes, such bioelemental patterns may also be useful for an earlier and precise differential diagnosis be‐tween idiopathic PD and atypical Parkinsonian syndromes (e.g.,Multisystem atrophy, Dementia with Lewy bodies, Progressive su‐pranuclear palsy or Corticobasal degeneration), which is a more fre‐quent challenge in the clinical setting than the mere diagnosis of PD.Biomarker patterns could also facilitate the design of disease‐modi‐fying trials, particularly when it comes to the identi fication of sub‐groups with different progression rates.
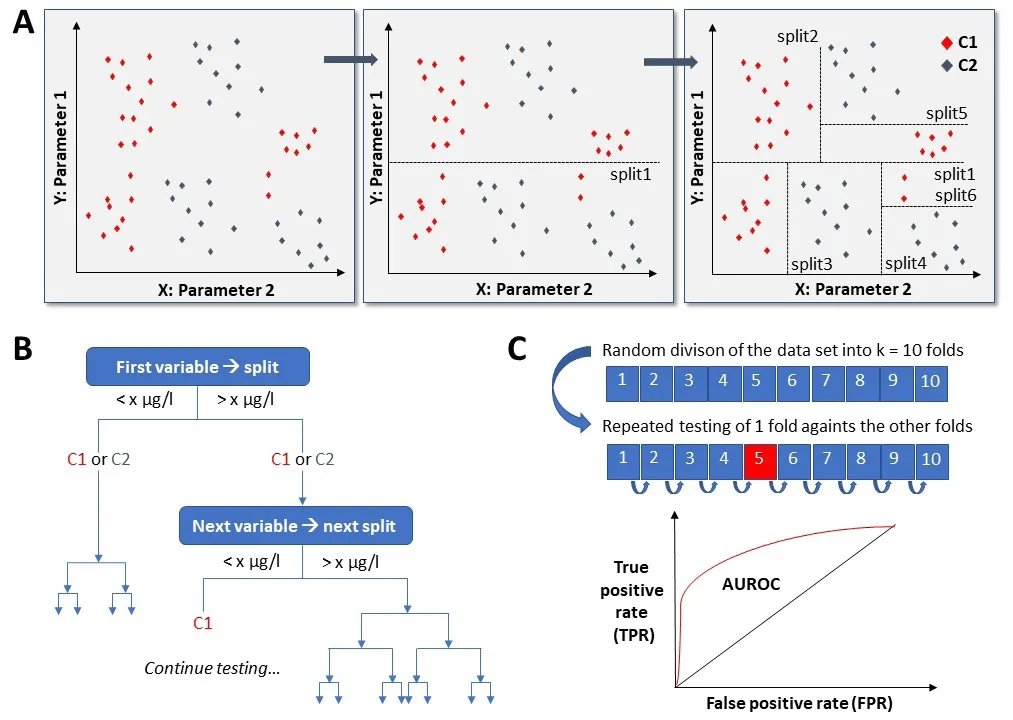
Figure 1 Scheme showing a classification task by “decision tree” machine learning.
In a currently ongoing analysis, we are now validating the ele‐mental pattern identified by the gradient tree boosting algorithm in a prospective multicentre validation cohort including more PD patients and controls as well as patients from different centres to ex‐clude a center bias. Increasing the number of patients may also yield a sufficient power to detect correlations of disease severity (assessed by the Hoehn & Yahr stage or the Uni fied PD rating scale, UPDRS)and the bioelemental pattern, which was not possible in the original discovery cohort. Since our patient cohort is followed‐up for at least one year, we will also be able to include longitudinal data on individ‐ual patients that will permit to correlate bioelemental patterns with disease progression, which could reflect disease progression more faithfully than commonly used clinical scales.
We expect that the analysis of elemental pro file patterns, includ‐ing biometals, as well as associated metalloproteins will contribute to a better accuracy in the diagnosis of PD. Ongoing analyses
including patients with other neurodegenerative disorders will show whether this also holds true for phenotypic mimics. A better differentiation of patients particularly in the early disease stages will permit to choose the most appropriate therapy, to better in‐form patients and caregivers about the individual prognosis and to stratify patients for clinical trials. Data from trials analyzing single elements in other neurodegenerative disorders suggests that the identi fication of bioelemental patterns has also the potential to be translated to other disease entities.
FM was funded by the TRANSMED Kolleg Göttingen, which was supported by the Ministerium für Wissenschaft und Kultur, Niedersachsen, Germany. PL was funded by the DFG-Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB), Göttingen, Germany.
Fabian Maass, Paul Lingor*Department of Neurology, University Medical Center Goettingen,Germany (Maass F, Lingor P)Cluster of Excellence Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB), Goettingen, Germany(Lingor P)
*Correspondence to:Paul Lingor, M.D., plingor@gwdg.de.
orcid:0000-0001-9362-7096 (Paul Lingor)
Accepted:2018-06-05
doi:10.4103/1673-5374.235239
Copyright transfer agreement:The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Chengbiao Wu, University of California San Diego, USA.
Additional file:Open peer review report 1.
- 中国神经再生研究(英文版)的其它文章
- In Memoriam: Ray Grill (1966–2018)
- Reorganization of injured anterior cingulums in a hemorrhagic stroke patient
- A novel chronic nerve compression model in the rat
- Analgesic effect of AG490, a Janus kinase inhibitor, on oxaliplatin-induced acute neuropathic pain
- Three-dimensional visualization of the functional fascicular groups of a long-segment peripheral nerve
- Novel conductive polypyrrole/silk fibroin scaffold for neural tissue repair