一种高通量提取桃DNA方法的建立与应用
张南南,牛良,崔国朝,潘磊,曾文芳,王志强,鲁振华
一种高通量提取桃DNA方法的建立与应用
张南南,牛良,崔国朝,潘磊,曾文芳,王志强,鲁振华
(中国农业科学院郑州果树研究所/国家桃葡萄品种改良中心/农业部果树育种技术重点实验室,郑州 450009)
【目的】DNA制备是大规模基因型筛选和分子标记辅助选种的重要前提,本研究采用一种1.2 mL八联排管代替单个离心管,探索一种操作简便、节约时间、成本低的桃DNA快速提取方法,以满足高通量遗传研究的需求,提高工作效率。【方法】以普通生长型桃(standard type,ST)‘中油桃8号’为母本,温度敏感半矮生型桃(temperature-sensitive semi-dwarf in,)‘09-1-112’为父本,杂交获得F1代分离群体500株实生苗为载体,包括温度敏感半矮生型246株,普通生长型254株,建立一种高通量、低成本桃DNA提取方法。利用1.2 mL八联排管结合八通道移液器,简化提取步骤,提取桃幼嫩叶片中的基因组DNA;通过紫外分光光度计和琼脂糖凝胶电泳对所提取的DNA浓度、纯度和完整性进行检测。基于高分辨率熔解曲线(high resolution melting,HRM),采用96孔板对温度敏感半矮生型桃和普通生长型桃共500个F1分离后代单株进行PCR扩增和基因分型,区分基因型与基因型。基于双亲表型与基因型一致,开发InDel位点,PCR扩增后,采用SDS-PAGE在F1群体中进行验证,确定利用分离的DNA是否正确区分不同基因型。【结果】分离的DNA经紫外分光光度计检测,浓度范围约为25—200 ng·μL-1,OD260nm/OD280nm约为1.81—1.98,DNA纯度较高;琼脂糖凝胶电泳条带清晰、单一,DNA完整度较高。参考桃基因组(Version 2.0),根据双亲深度测序数据,开发获得SNP标记SNP_Pp03_3758620,应用于高分辨率熔解曲线基因分型,发现温度敏感半矮生型和普通生长型呈现明显不同的峰型,证明提取的DNA模板可应用于HRM基因分型。基于双亲基因型与表型一致,开发InDel标记InDel_Pp03_3829009,聚丙烯酰胺凝胶电泳的验证结果显示PCR扩增具有较高的强度,获得与目的片段大小一致的特异性条带,且在两种不同生长型单株中具有明显的多态性,表明PCR扩增稳定,提取的DNA可用于基于InDel标记的多态性分析。使用该提取方法,每人每天可以完成1 000个样品的DNA提取,成本较低,且不会对幼苗早期生长造成影响。【结论】建立了一种简便、有效、低成本的桃基因组DNA提取方法,可以满足基因分型、品种鉴定及遗传分析等分子生物学研究,实现了大批量不同样本基因组DNA的同时提取,具有较高应用价值。
桃;高通量;DNA提取方法
0 引言
【研究意义】桃()原产中国,是我国栽培最为广泛的落叶果树之一,仅次于苹果、梨和葡萄[1]。桃基因组较小,染色体数量少,单基因控制性状较多,被认为是多年生果树研究的模式植物[2]。随着分子标记技术在桃育种工作中的应用,大规模样本DNA提取操作过程复杂、耗时长、成本高等问题突出,而基因分型、分子标记辅助选择、品种鉴定等工作对DNA的浓度和产量要求不高[3],不需要DNA样品长时间保存。因此,寻找一种简单、快速、高通量、成本低的DNA提取方法非常必要。【前人研究进展】全基因组测序的完成为桃优良性状基因定位、遗传多样性分析、资源评价及分子辅助选种体系的建立等奠定了基础[4-7]。随着植物科学向分子水平进一步发展,利用分子标记进行定位和克隆控制桃优良性状的基因、分子标记选择育种、大规模杂交后代单株基因型鉴定等成为桃分子育种的重要环节[8],而该类分子生物学和遗传学研究都需大批量DNA以应用于下游生物学反应[9-11]。目前,快速高通量提取DNA的方法在作物中的应用已有许多报道,Randhawa等[12]将小麦DNA高通量提取方法成功应用于分子标记辅助选择育种;Xin等[13]结合CTAB与DNA提取试剂盒的方法提取高粱叶片与种子中的基因组DNA;Devi等[14]报道了适用于PCR扩增等下游生物学研究的生姜基因组DNA快速提取方法。在木本果树中高通量提取DNA的研究与应用较少,Kim等[15]最先通过改良已有的4种DNA提取方法,报道了苹果、梨、葡萄及柿子的DNA快速提取方法,可获得高质量的DNA;Cheng等[16]以柑橘为研究材料,报道了可应用于20多种热带和亚热带果树作物基因组DNA快速提取的方法。关于桃DNA快速DNA提取的研究虽已有报道,但仅提高DNA提取过程中的研磨速度,操作过程复杂,无法满足高通量的需求[17]。【本研究切入点】实验室中常用的传统DNA提取方法,如CTAB或SDS等,操作步骤复杂,耗时耗力,难以适应科研快速发展的需求[18]。虽然已经有专用的设备可以实现高通量大规模的DNA提取,但是对于普通实验室而言,构建一种简单、快速且低成本的中小通量的植物基因组DNA提取方法很有必要。【拟解决的关键问题】采用改良CTAB法,以1.2 mL八联排管代替2 mL离心管,建立一种高通量提取桃基因组DNA的有效方法,成功应用于基因分型,提高桃大群体基因型筛选和分子标记辅助选种的效率。
1 材料与方法
试验于2017年在农业部果树育种技术重点实验室进行。
1.1 材料
以‘中油桃8号’(standard type,ST)为母本,基因型为;‘09-1-112’(temperature-sensitive semi-dwarf in,)为父本,来源于‘SD9238’,基因型为。手工去雄后人工授粉,获得F1杂交后代500株实生苗,其中型246株,普通生长型254株,=0.800,2=0.064,比例约为1﹕1,符合孟德尔遗传规律,采集各单株幼嫩叶片用于DNA提取。
1.2 基因组DNA提取
采用CTAB[19]法提取桃叶片基因组DNA,略作修改(图1),具体如下:(1)取桃幼嫩叶片约30 mg,用镊子放入1.2 mL八联排离心管底部,每孔各加入1个5 mm钢珠(图1-A),放入96孔底座,为防止液氮进入离心管,确保离心管盖高于液氮液面,并充分冷冻;(2)手动摇晃数次,保证钢珠充分打碎样品,DNA自动研磨仪(上海领成生物科技有限公司)进行研磨,频率30 Hz,时间90 s(图1-B);(3)立即用镊子轻轻敲打离心管盖,使附着在管盖内壁的样品掉落,待离心管盖充分解冻后,于通风橱中开盖,各离心管盖朝上分别放于干净实验纸上;(4)用300 mL量程8通道移液器加入配制好的CTAB液600 μL(图1-C),放入60℃电热恒温鼓风干燥箱加热30 min,其间约每10 min轻摇匀;(4)称量配平,放入冷冻离心机(Eppendorf 5810 R)4℃条件下,4 000 r/min瞬时离心,将管盖及内壁液体离心至底部;加入氯仿和异戊醇混合液,体积比为24﹕1,直至1.2 mL的八联排离心管满载线,后缓慢颠倒混匀5 min,配平后,放入冷冻离心机(Eppendorf 5810 R)4℃条件下,4 000 r/min,离心10 min;(5)分别吸取上清液100 µL于两个200 µL PCR板中,其中一板加入等体积的无水乙醇,于-20℃冰箱1 h,称量配平,4℃条件下,4 000 r/min,离心10 min,板孔朝下,迅速弃上清液;另一板放入4℃冰箱,保存备用;(6)在带有沉淀的96孔PCR板中加入150 μL的70%的乙醇,4 000 r/min瞬时离心,洗涤沉淀两次,加入无水乙醇洗涤沉淀一次,用100 μL的8通道移液器(Eppendorf)吸除离心管底部剩余无水乙醇,更换吸头,自然晾干;(7)在室温下自然风干沉淀后,加入150 μL的0.1×TE溶解,同时加入0.5 μL的RNase(图1-D),37℃放置1 h,祛除RNA污染(长期保存在-20℃冰箱,常用则存于4℃冰箱),以用于后续研究。
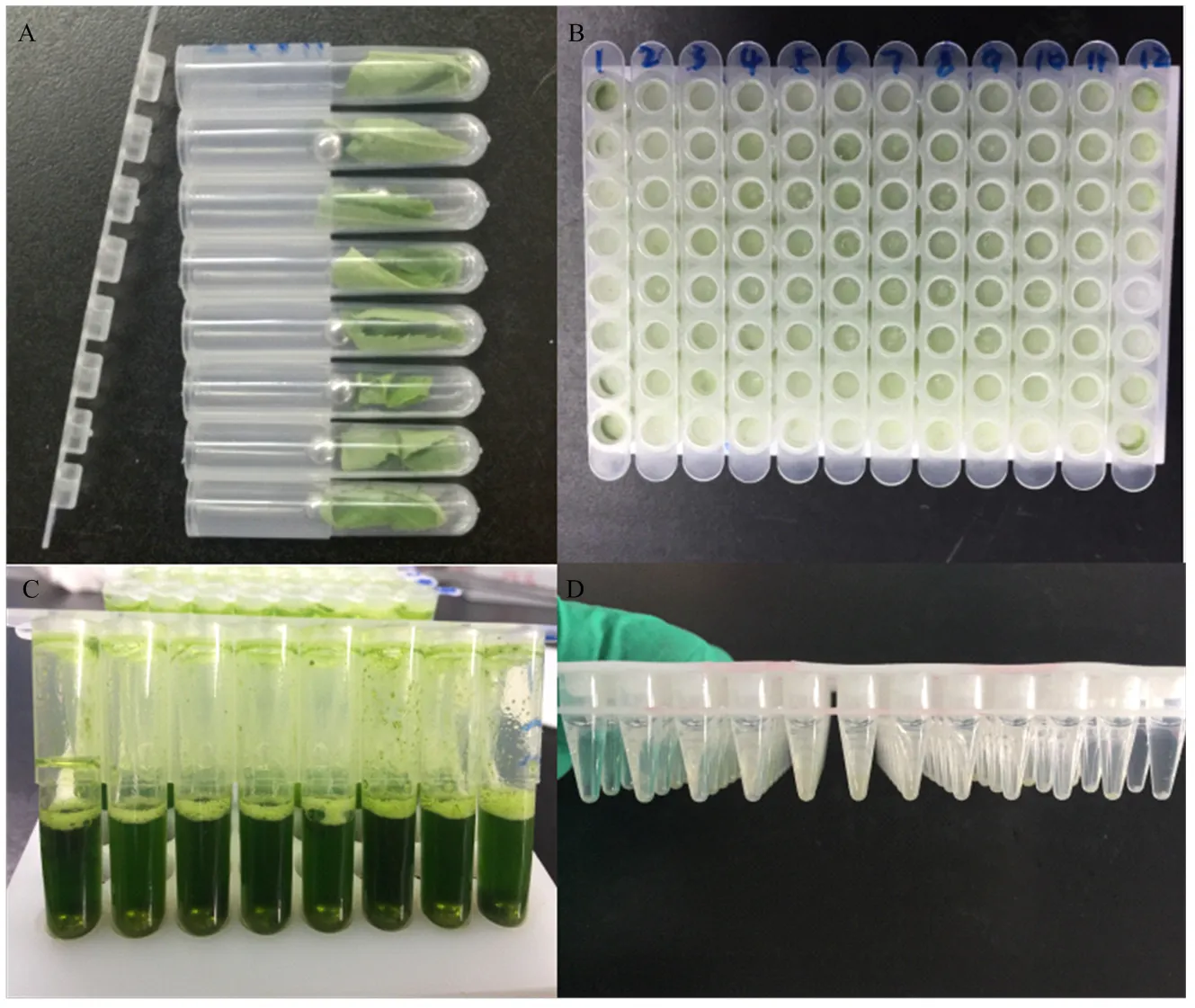
A:取幼嫩叶片放入1.2 ml八联排管Young leaves sampled into 1.2 mL thin-wall 8 strip polypropylene PCR tubes;B:研磨后的DNA样品Samples after grinding;C:加入预热的CTAB DNA samples with preheated extraction buffer;D:150 μL TE溶解DNA沉淀DNA dissolved in 150 μL TE
1.3 DNA质量和浓度检测
用1%琼脂糖凝胶电泳对提取的DNA片段大小和完整度进行检测,在紫外凝胶成像仪上观察并拍照。吸取1 μL DNA样品,采用NanoDrop 1000 spectrophotometer(Themo)紫外分光光度计检测DNA浓度和纯度(OD260mm/OD280mm),稀释成工作液浓度(约25 ng·μL-1),以用于后续研究。
1.4 基于高分辨率熔解曲线(high resolution melting,HRM)的SNP基因分型
基于重测序数据开发与双亲基因型和表型紧密连锁的SNP标记,采用Primer 3.0软件(http://primer3. ut.ee/)设计引物,设置引物的退火温度60—63℃,扩增片段长度100—200 bp。采用HRM master mix(Roche)进行PCR扩增,反应总体积为15 μL,利用LightCycler 480II定量PCR仪(Roche)进行PCR扩增和HRM分析[20]。
1.5 PCR扩增验证
参考桃基因组(Version 2.0)数据在亲本(‘中油桃8号’ב09-1-112’)中开发InDel标记。采用Primer 3.0软件(http://primer3. ut.ee/)设计引物,引物的退火温度60℃左右,扩增片段长度150—200 bp。采用TaKaRa EX在F1群体中进行PCR扩增,反应总体积为15 μL,其中含DNA模板1 μL、酶0.075 μL、dNTP 1.2 μL、10×buffer(含Mg2+)1.5 μL、正/反向引物和ddH2O。扩增程序为95℃ 2 min;94℃ 15 s,56.5℃ 15 s,72℃ 20 s,30个循环;72℃ 4 min,4℃冷却10 min。取1.2 μL PCR产物进行8%聚丙烯酰胺凝胶电泳检测,硝酸银染色,氢氧化钠脱色后观察。
2 结果
2.1 DNA浓度和纯度
经紫外分光光度计检测提取的DNA浓度和纯度,部分结果见表1。由表1 可知,OD260mm/OD280mm介于1.81—1.98,浓度介于25—200 ng·μL-1。RNA酶处理后,经琼脂糖凝胶电泳检测(图2),结果表明条带明亮清晰,整齐单一,DNA较完整。说明所提取的DNA 较纯,符合进一步研究分析要求。
2.2 SNP基因分型检测
参考桃基因组(Version 2.0),基于双亲重测序数据获得引物SNP_Pp03_3758620,正向引物5′-GTGAA GTCCCACCAGTGCAG-3′,反向引物5′-TGGAGTCA GAGAGGATCGTCAA-3′,扩增片段长度171 bp;型在该位点的基因型为A/G,普通生长型的基因型为G/G。基于HRM的基因分型结果发现,96个样品全部扩增成功,经计算后,型与普通生长型呈现明显不同的峰型曲线(图3)。表明该方法提取的桃基因组DNA质量好、纯度高,适用于HRM基因分型。

M: DNA Marker

表1 部分样品DNA纯度和浓度
2.3 PCR电泳检测
基于双亲‘中油桃8号’ב09-1-112’基因型开发InDel标记InDel_Pp03_3829009,缺失大小4 bp,采用Primer 3.0软件(http://primer3. ut.ee/)设计引物,正向引物:5′-AGCCCTGTATTGGTTCCATCCT-3′, 反向引物5′-AGAAGGTAGCGACTCCTTTTCCT-3′,扩增片段大小201 bp。从聚丙烯凝胶电泳分析图谱结果显示(图4),目的条带整齐清晰、分辨率高,PCR扩增结果良好,型与ST型具有明显的多态性,基因型与表型相符,证明PCR扩增稳定,提取的DNA可用于基于InDel标记的基因型分析。
3 讨论
目前,已报道的植物DNA提取方法有几十种,如CTAB法、碱裂解法、高低盐pH法、苯酚法、试剂盒提取DNA法等。传统的DNA提取方法经过改良后,分离的DNA质量与纯度较高,满足了一般分子生物学实验所需DNA的质量要求[21],但其操作步骤繁琐,且需消耗大量人力和时间,达不到高通量的要求。一些简易DNA提取方法,虽简化了提取步骤,提取速度快,但分离DNA纯度不高,且扩增效果不稳定[22-24];许多DNA提取试剂盒虽然操作简单,但其成本较高,产量低,不适宜大规模高通量DNA制备[25-28]。本研究采用1.2 mL的八联排管结合改良CTAB法快速提取大批量桃叶片基因组DNA的方法,可以在3 min之内研磨192个样品,八联排管与8通道移液枪相结合,操作简便,每人每天可提取1 000个样品,节约时间,每个样品提取成本约为DNA提取试剂盒的1/10,极大提高了工作效率;分离的DNA产率大、纯度高,经检测可以满足InDel分析、PCR扩增及SNP基因分型等大规模高通量分子生物学研究。
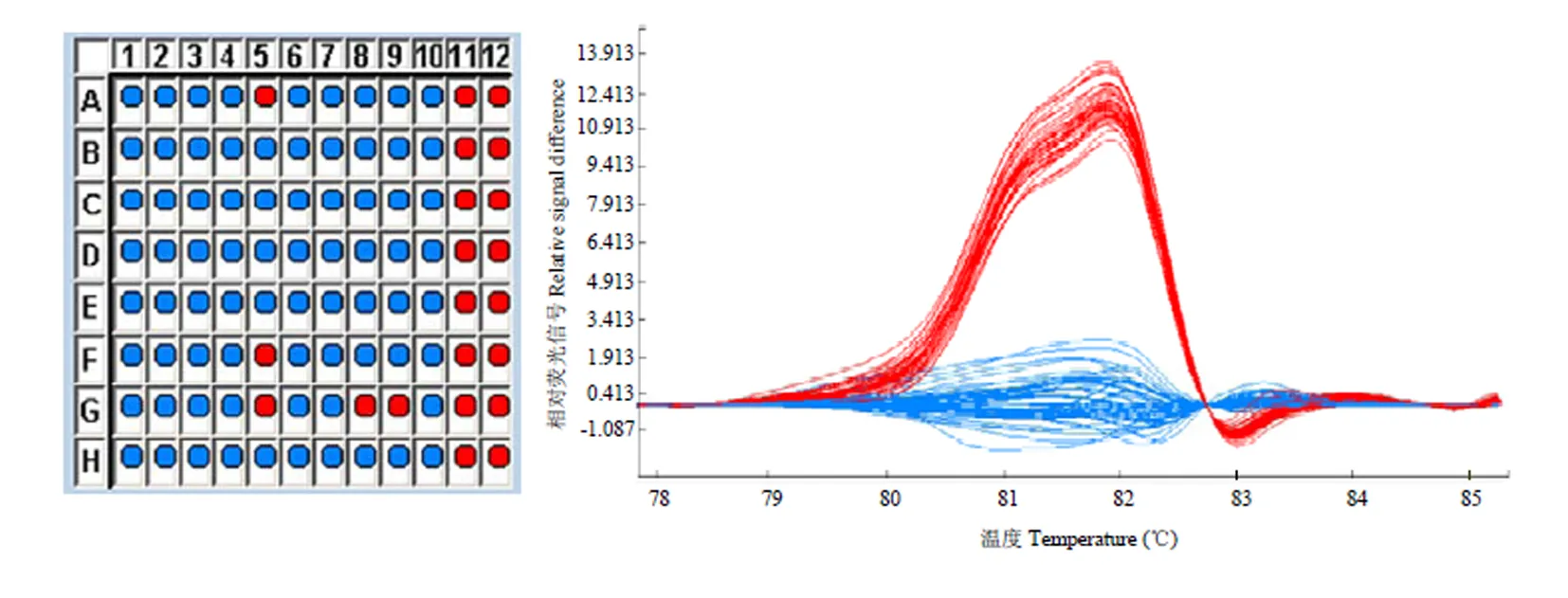
图3 基于HRM分析PpTssd型A/G(蓝色)和普通型G/G(红色)位点的SNP鉴定

图4 PCR扩增片段聚丙烯酰胺凝胶电泳图
DNA提取过程中,首先应对1.2 mL的八联排离心管和管盖进行位置和方向标记,防止打乱样品顺序。用20 cm枪状镊子取幼嫩叶片,利于将叶片放入离心管底部,取样完成后,每个1.2 ml八联排深孔管中各放一个5 mm钢珠,液氮冷冻后,研磨前手工摇动96孔板,保证钢珠充分晃动,研磨时间仅需90 s,避免样品研磨不充分或在研磨过程中组织褐变;研磨后立即用镊子轻轻敲打离心管盖,使附着在内壁的样品掉落,待管盖充分解冻后,再开盖,防止因液氮挥发造成样品溅飞,造成交叉污染;另外,实验过程中每次开盖前,称量配平,瞬时离心,八通道移液枪悬空加样,吸头污染时及时更换可有效防止样品间交叉污染。传统CTAB法所需的样品量约为0.5 g,且需要研钵手动研磨,采用磨样仪磨样时一次只能磨几十个样品,且磨样时间较长,本研究所需样品组织量仅为30 mg,不会影响幼苗的生长及试验材料的其他研究工作,且幼嫩叶片DNA含量多,糖类、酚类、蛋白质等次生代谢产物积累少[11,29],有利于高质量DNA的分离。实验中采用氯仿、异戊醇混合液仅抽提一次,简化了实验步骤,但获得的DNA样品OD260mm/OD280mm仍介于1.81—1.98,与普通提取方法所获得的DNA纯度几乎无差异。DNA沉淀采用150 μL的0.1×TE溶解,样品浓度最低约为25 ng·μL-1,虽无法达到CTAB法或高成本试剂盒高浓度要求,但类似于普通DNA提取试剂盒浓度,产量和浓度均可充分保证PCR检测、HRM分型、聚丙烯酰胺凝胶电泳等基础研究的进行。
利用提取的DNA进行HRM基因分型和聚丙烯酰胺凝胶电泳分析,两种研究方法灵敏度高,能够准确直观反应DNA模板的质量,有效对不同基因型进行分析;且所需模板量约为1 μL,一次提取的DNA量可进行100次SNP基因分型、InDel标记分析等生物学研究,可保证后续研究进行。
本研究提出的DNA提取方法快速高效,节约时间和成本,应用于大规模群体样本DNA的制备,PCR效果和重复性好,虽简化了提取步骤,但兼具CTAB法提取DNA的高质量[30]和简易提取DNA高通量的优点,克服了传统DNA提取方法程序复杂、耗时长的缺点,适用于大规模的基因型检测、自交系和品种纯度检测等研究工作。
4 结论
建立了一种简便、有效、低成本的桃基因组DNA提取方法,可以满足基因分型、品种鉴定及遗传分析等分子生物学研究,实现了大批量不同样本基因组DNA的同时提取,具有较高的应用价值。
[1] 俞明亮, 马瑞娟, 沈志军, 蔡志翔.中国桃种质资源研究进展. 江苏农业学报, 2010, 26(6): 1418-1423.
Yu m l, ma r j, shen z j, cai z x. Research advances in peach germplasm in china., 2010, 26(6): 1418-1423. (in Chinese)
[2] Aranzana M J, Abbassi E K, Howad W, Arús P. Genetic variation, population structure and linkage disequilibrium in peach commercial varieties., 2010, 11(1): 69.
[3] 陈平华, 王恒波, 许莉萍, 陈由强, 陈如凯.碱裂解叶片两步快速制备PCR模板技术研究. 热带作物学报, 2010, 31(3): 422-429.
CHEN H P, WANG H B, XU L P, CHEN Y Q, CHEN R K. Alkali lysis of leaves by two-step handling for preparation of PCR templates.,2010, 31(3): 422-429. (in Chinese)
[4] Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori M T, Sansavini S. Microsatellite DNA in peach (L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars., 2000, 43(3): 512-520.
[5] Li X W, Meng X Q, Jia H J, Yu M L, Ma R J, Wang L R, Cao K, Shen Z J, Niu L, Tian J B, Chen M J, Xie M, Arus P, Gao Z S, Aranzana M J. Peach genetic resources: diversity, population structure and linkage disequilibrium., 2013, 14(1): 84.
[6] Cao K, Zheng Z, Wang L, Liu X, Zhu G, Fang W, ChengS, ZengP, ChenC, WangX, XieM, ZhongX, WangX, ZhaoP, BianC, ZhuY, ZhangJ, MaG, ChenC, LiY, HaoF, LiY, HuangG, LiY, LiH, GuoJ, XuX, WangJ. Comparative population genomics reveals the domestication history of the peach,, and human influences on perennial fruit crops., 2014, 15(7): 415.
[7] Aranzana M J, Illa E, Howad W, Arús P. A first insight into peach [(L.) Batsch] SNP variability., 2012, 8(6): 1359-1369.
[8] Martínez-García P J, Peace C P, Parfitt D E, OGUNDIWIN E A, FRESNEDO-RAMíREZ J, DANDEKAR A M, GRADZIEL T M, CRISOSTO C H. Influence of year and genetic factors on chilling injury susceptibility in peach ((L.) Batsch)., 2012, 185(2): 267-280.
[9] Irfan M, ZHANG T T, Wang Y, ZHANG C y, MIAO Q, ZHANG L j, LIN F. Modification of CTAB protocol for maize genomic DNA extraction., 2013, 8(1): 41-45.
[10] Gupta P K, Varshney R K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat., 2000, 113(3): 163-185.
[11] Springer N M. Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB., 2010, 2010(11): pdb.prot5515.
[12] Randhawa H S, Mutti J S, Kidwell K, Morris C F, Chen X, Gill K S. Rapid and targeted introgression of genes into popular wheat cultivars using marker-assisted background selection., 2009, 4(6): e5752.
[13] Xin Z, Chen J. A high throughput DNA extraction method with high yield and quality., 2012, 8(1): 26.
[14] Devi K D, Punyarani K, Singh N S, devi h s. An efficient protocol for total DNA extraction from the members of order Zingiberales-suitable for diverse PCR based downstream applications., 2013, 2: 669.
[15] Kim C S, Lee C H, Shin J S, Chung Y S, Hyung N I. A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP., 1997, 25(5): 1085-1086.
[16] CHENG Y J, GUO W W, YI H L, PANG X M, DENG X x. An efficient protocol for genomic DNA extraction fromspecies., 2003, 21(2): 177-178.
[17] 刘航空, 王安柱, 赵彩平, 韩明玉, 李金金, 李芳. 高通量提取桃树叶片组织基因组DNA的研究. 北方园艺, 2015(13): 120-125.
LIU H K, WANG A Z, ZHAO C P, HAN M Y, LI J J, LI F. Study on extracting DNA from peach leaves efficiently., 2015(13): 120-125. (in Chinese)
[18] DILWORTH E, FREY J E. A rapid method for high throughput DNA extraction from plant material for PCR amplification., 2000, 18(1): 61-64.
[19] DOYLE J J, DOYLE J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue., 1987, 19: 11-15.
[20] 鲁振华, 牛良, 张南南, 崔国朝, 潘磊, 曾文芳, 王志强. 基于HRM获得与桃紧密连锁的SNP标记. 中国农业科学, 2017, 50(8): 1505-1513.
LU Z H, NIU L, ZHANG N N, CUI G C, PAN L, ZENG W F, WANG Z Q. SNP marker tightly linked tofor peach using high resolution melting analysis., 2017, 50(8): 1505-1513. (in Chinese)
[21] TEL-ZUR N, ABBO S, MYSLABODSKI D, MIZRAHI Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of the generaand(Cactaceae)., 1999, 17(3): 249-254.
[22] MINAS K, MCEWAN N R, NEWBOLD C J, SCOTT K P. Optimization of a high-throughput CTAB based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures., 2011, 325(2): 162-169.
[23] HANDAYANI F, WULANDARI R A, MURTI R H. Genomic DNA extraction method from mature leaf of lai (Becc.)., 2016, 38(1): 73-79.
[24] ABDEL-LATIF A, OSMAN G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize., 2017, 13: 1.
[25] XIN Z, CHEN J. A high throughput DNA extraction method with high yield and quality., 2012, 8(1): 26.
[26] VON POST R, VON POST L, DAYTEG C, NILSSON M, FORSTER B P, TUVESSON S. A high-throughput DNA extraction method for barley seed., 2003, 130(2): 255-260.
[27] NAEEM R, MIRZA B. High-throughput DNA extraction and optimization of PCR efficiency for barley SSRs genotyping., 2018, 43(1): 143-154.
[28] WANG S, KNOX R E, DEPAUW R M, CLARKE J M, WANG B L. A simple DNA preparation method for PCR amplifications in marker-assisted selection of wheat., 2005, 4(7): 481-485.
[29] HEALEY A, FURTADO A, COOPER T, HENRY R J. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species., 2014, 10(1): 21.
[30] FANG G, HAMMAR S, GRUMET R. A quick and inexpensive method for removing polysaccharides from plant genomic DNA., 1992, 13(1): 52-54, 56.
(责任编辑 岳梅)
Establishment and application of a high-throughout protocol for Peach () DNA extraction
ZHANG NanNan, NIU Liang, CUI GuoChao, PAN Lei, ZENG WenFang, WANG ZhiQiang, LU ZhenHua
(Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences/National Peach and Grape Improvement Center/Key Laboratory of Fruit Breeding Technology of Ministry of Agriculture, Zhengzhou 450009)
【Objective】Preparation of large quantity and high-quality DNA is an important prerequisite for large-scale genotypic screening and molecular marker-assisted of plant breeding. The objective of this study is to present a low-cost, high-throughput peach (L. Batsch) genomic DNA extraction method, meet the needs of high-throughput genetic researches and improve the working efficiency.【Method】The population were obtained from a cross between female parent ‘CN8’ (standard type, ST) and male parent ‘09-1-112’ (temperature-sensitive semi-dwarf in,type) to establish a high-throughout protocol for peach DNA extraction. The F1segregating population were generated to assess the phenotype characteristics, resulting in observed 1﹕1 (254 standard type and 246 semi-dwarf type individuals). Subsequently, DNA extraction was carried out on the young leaves of two parents and 500 progenies by procedure using 1.2 mL thin-wall 8 strip polypropylene PCR tubes instead of a single centrifuge tube. After extraction, the quality of DNA samples was examined with ultraviolet spectrophotometry and 1% agarose gel electrophoresis, respectively. Referencing the peach genome (version 2.0) and using re-sequencing data, single nucleotide polymorphism (SNP) markers were developed and the HRM analysis was employed on F1population to conduct SNP genotyping. Ultimately, the extracted DNA samples were validated by using an InDel marker to verify the genotype of 500 individuals.【Result】The concentrations of DNA were in a range between 25 to 200 ng·μL-1and the UV absorbance ratios values (1.81-1.98) to determine DNA quality were acceptable and with high-purity. The result of agarose gel electrophoresis proved that DNA bands were clear, single with a high degree of DNA integrity.Referencing the peach genome and using whole genome re-sequencing data of two parents, SNP_Pp03_3758620 was developed in female and male parents, and the HRM analysis was employed to conduct SNP genotyping and divided temperature-sensitive semi-dwarf and standard type individuals into two groups, respectively, which proved DNA templates extracted from this DNA isolation procedure could be employed for HRM genotyping. Based on the genotype and phenotype of two parents, InDel_Pp03_3829009 was developed and the results of polyacrylamide gel electrophoresis showed that PCR amplification products showed desired fragment size, and were polymorphic intype and ST type with bright and clear target fragment. It concluded that the extracted DNA samples could be used for indel analysis. Using this method, 1 000 samples of DNA could be extracted per day with low cost and no effect on the early growth of seedlings.【Conclusion】A simple, effective and low-cost method for extracting genomic DNA from peach was established, which can be used for molecular biology, such as genotyping, variety identification and genetic analysis. Simultaneous extraction of genomic DNA from large quantities of different samples was realized. It has high application value.
peach (); high-throughout; DNA extractionmethod
2018-01-19;
2018-04-02
国家自然科学基金(31500558)、河南省重点研发专项(182102110134)、中国农业科学院科技创新工程专项经费(CAAS-ASTIP-2018-ZFRI)
张南南,E-mail:18763895031@163.com。
鲁振华,E-mail:luzhenhua@caas.cn。通信作者王志强,E-mail:wangzhiqiang@caas.cn
10.3864/j.issn.0578-1752.2018.13.016

