Chaihu-Shugan-San exerts an antidepressive effect by downregulating miR-124 and releasing inhibition of the MAPK14 and Gria3 signaling pathways
Qiong Liu, Ning-Ning Sun, Zheng-Zhi Wu, Da-Hua Fan , Mei-Qun Cao,
1 Department of Cardiovascular Medicine, XiangYa Hospital of Central South University, Changsha, Hunan Province, China
2 Shenzhen Institute of Geriatrics, Shenzhen Second People’s Hospital, The First Affiliated Hospital of Shenzhen University, Shenzhen,Guangdong Province, China
3 The Eighth Affiliated Hospital of Sun Yat‐sen University, Shenzhen, Guangdong Province, China
4 Chinese and Wsetern Integrative Medicine, Shcool of Medicine, Jinan University, Guangzhou, Guangdong Province, China.
Introduction
Depression is a clinical mental disorder that severely im‐pairs the quality of life of the patients (Putnam et al., 2017;Riddle et al., 2017). A variety of risk factors contribute to the development of depression (Miller et al., 2015; Duman et al., 2016). Stress‐induced changes in synaptic and structural neuroplasticity in the hippocampus have been implicated in the pathophysiology of depression (de Sousa et al., 2015;Duman et al., 2016; He et al., 2016). The main strategy for the treatment of depression includes increasing the levels of monamine transmitters in the synaptic cleft. However, on average, 50% of patients show poor response to this treat‐ment approach (Juurlink et al., 2006; Papakostas et al., 2010;Jiang et al., 2015). Therefore, there is an urgent need for novel antidepressants with high efficacy and few side effects.Chaihu-Shugan-San(CHSGS), a traditional Chinese herbal preparation, has been widely used clinically to relieve the symptoms caused by liver‐qi stagnation (Kim et al., 2005;Wang et al., 2012; Butler et al., 2013; Qiu et al., 2014). Recent studies have demonstrated that CHSGS has antidepressive activity and that the major components ferulic acid, nar‐ingin, merazin hydrate and neohesperidin are responsible for the antidepressive effect (Kim et al., 2005; Wang et al.,2012; Qiu et al., 2014). However, the molecular mechanisms underlying the therapeutic efficacy of CHSGS in depression remain unclear.
Impaired neuroplasticity is partially caused by aberrant post‐transcriptional regulation of gene expression (Duman et al., 2002; Dwivedi et al., 2009). Accumulating evidence suggests that miRNAs are abundant in the nervous system and are involved in synaptic plasticity (Kosik et al., 2006;Zeng et al., 2009; Baudry et al., 2010). miR‐124, the most abundant miRNA in the brain, plays critical roles in adult neurogenesis, neuronal differentiation and synaptic plasticity (Lagos‐Quintana et al., 2002; Makeyev et al., 2007; Cheng et al., 2009). Increased expression of miR‐124 has been observed in the hippocampus of rats with unpredictable stress‐induced depression (Cao et al., 2013; He et al., 2016).However, the molecular mechanisms by which miR‐124 and its downstream effectors regulate depression remain largely unknown. Recent studies have shown that mitogen‐acti‐vated protein kinase 14 (MAPK14) orchestrates the cellular response to stress and inflammation, which play important roles in the regulation of neuronal cell death (Bruchas et al., 2011; Moretti et al., 2015; Redhead et al., 2015). Keeping MAPK14 at proper levels is essential for neuronal function and survival, which are important for maintaining the health and plasticity of ischemic neurons. However, the factors reg‐ulating MAPK14 expression remain largely unknown.
Glutamate receptor subunit 3 (Gria3), subunit 3 of the ionotropic AMPA receptor, is abundant throughout the brain (Enoch et al., 2014; Di Lorenzo et al., 2015). AMPA receptors have been reported to mediate most excitatory synaptic transmission in the central nervous system, and they play important roles in the induction and maintenance of long‐term potentiation and depression (Bonnet et al.,2009; McCartney et al., 2014). In this study, we investigate the effect of CHSGS on the miR‐124 signaling pathway in rats with depression induced by a combination of solitary condition and chronic unpredictable mild stress (CUMS).
Materials and Methods
Animals
A total of 32 male 8‐week‐old Sprague‐Dawley rats weigh‐ing 180—220 g (license No. SYXK (Yue) 2014‐0140) were obtained from the Guangdong Medical Lab Animal Center,China. The protocols were approved by the Ethics Commit‐tee of Shenzhen Second People’s Hospital, China (Ethical Approval No. 20151211009). Animals were housed (five per cage) under a 12‐hour light/dark cycle (light on from 8:00—20:00), with controlled background noise (40 ± 10 dB)and temperature (20 ± 3°C), with food and water availablead libitum.
Generation of the CUMS model of depression
Rats were randomly divided into four groups (n= 8 per group). The CUMS rat model was produced using a previ‐ously published protocol (Willner et al., 1997; Wang et al.,2014). After a 7‐day habituation period, all rats (except the normal control group) were subjected to CUMS as previ‐ously described. The stress protocol was as follows: food deprivation for 24 hours; water deprivation for 24 hours;a 1‐minute tail pinch; 5‐minute thermal stimulus at 45°C;5‐minute cold swimming at 4°C; 24 hours reversed light/dark cycle; electric foot shock (10 mA, 10‐second duration each, every other minute for 30 minutes); intermittent white noise (85 db); 24‐hour soiled cage and 24‐hour 45° cage tilt.Rats were randomly exposed to one of these stimuli once a day for 28 days. Immediately after each stress exposure, rats were returned to their home cage and maintained under standard conditions.
Drug administration
(1) Control group: Normal control rats were given distilled water, 4.5 mL/kg per day, by gavage once a day. (2) CUMS group:CUMS model rats were given distilled water, 4.5 mL/kg per day, by gavage once a day. (3) CHSGS group: CUMS model rats were given CHSGS extract, 2.835 g/kg per day, by ga‐vage once a day. (4) Fluoxetine group: CUMS model rats were given normal food and fluoxetine, 1.8 mg/kg per day(Sigma Aldrich, St. Louis, MO, USA), by gavage once a day.Food was withdrawn 2 hours prior to drug administration.Drugs or distilled water were administered for 4 consecutive weeks from the third week. Open‐ field test and sucrose consumption test were performed 1 hour after the last drug treatment.
Raw herbal medicines and CHSGS extract
The raw herbal medicine containing Radix Bupleuri (ratio of medicinal materials: 19.05%), Aurantii nobilispericarpium(14.30%), Szechwan Lovage rhizome (19.05%), Nutgrass Galingale rhizome (14.29%), Fructus Aurantii (9.05%),Paeonia (14.29%) and Glycyrrhiza uralensis (4.74%) was purchased from Beijing Tongren Tang (Shenzhen, China).
The composition and the amount of each component were authenticated by Chinese medicine experts in accordance with the requirement of the Chinese pharmacopeia (2015 edition). The phytochemical profiles of CHSGS were identified by high performance liquid chromatography. The air dried herbal pieces of CHSGS were ground and extracted twice with boiling water (0.5 hours of boiling each time; the ratio of herbs to water was 1:10, g/mL). The combined water extract was filtered and concentrated under reduced pres‐sure at 60 °C to yield the dry extracts. The drug was stored at 4°C before administration.
Open field test
The open field test was conducted as previously described,with minor modifications (Zhu et al., 2010). Briefly, the open field device consisted of a square iron enclosure (100 cm × 100 cm × 50 cm) with black inner walls. Each rat was individually placed into the middle of the open‐field apparatus and then allowed to explore freely for 5 minutes. The time and distance in the center and the overall distance were recorded during the test, and the entire process was record‐ed with the Digibehave system (Shanghai Ruanlong Co.,Ltd., Shanghai, China). After each test, the open‐ field apparatus was cleaned with 75% alcohol to avoid contamination.
Sucrose consumption test
The procedure was performed according to a previously published protocol (Jiang et al., 2015). The rats were deprived of food and water for 24 hours prior to the ex‐periments and then exposed to two bottles of 1% sucrose solutions in a 24‐hour period. Followed this step, rats were offered both the test solution (1% sucrose) and distilled drinking water for the next 24 hours. During this period,the positions of the bottles were alternated between the leftand right sides of the cage throughout the experiment. The test was conducted before stress and 2 and 4 weeks after stress. Sucrose consumption was determined by reweighing the pre‐weighed bottle. The sucrose preference value was calculated as follows: Preference value (%) = sucrose intake/(sucrose intake + water intake) × 100%.
Forced swimming test
This test was performed with a transparent plastic cylinder(30 cm diameter × 50 cm height) filled to a depth of 35 cm with water at room temperature. Rats were placed in the cylinder and could not touch the bottom with their tails or hind limbs. Before the experiment, rats were allowed to swim for 15 minutes to acclimate to the new environment.The rats were then forced to swim for 5 minutes. The behaviors during this period, including immobility time and escape, and the amount of climbing and swimming, were recorded with an overhead camera. After the test, rats were dried with a towel and returned to the home cage. The water in the cylinder was changed between trials. The duration of immobility was accurately scored during the last 5 minutes of the total swimming time using a timer. The evaluation was performed by a blinded observer. Rats were judged to be in an immobile posture when they remained in a passively floating state in water without struggling or were swimming just to keep their heads above the water.
Quantitative assessment of hippocampal synaptic density
Cross‐sections of the hippocampus were used for analysis of synapse number. Sections of hippocampus from any given animal were randomly sampled. Synapses were counted in electron microscopic sections in a defined area (40 mm2) and were normalized to the control group. At least 30 such samples from at least 10 sections were examined for each animal.
Real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR)
All rats were sacrificed 24 hours after the last behavioral test. Hippocampal tissues were collected, and total RNA was extracted using the TRIzol reagent (Aidlab Biotechnologies Co., Ltd., Beijing, China). The quality of RNA was evaluated with the NanoDrop ND‐2000 spectrophotometer (Ther‐mo‐Scientific, Rockford, IL, USA). Reverse transcription was performed with 3 μg RNA using Oligo (dT) primers according to the manufacturer’s instructions. RT‐qPCR was performed on the 7900/Viia7 real‐time PCR platform with 2× All‐In‐One qPCR Mix (D01010A; Vazyme, Piscataway,NJ, USA). β‐Actin was used as the normalization control,and the relative expression levels of miR‐124, MAPK14 and Gria3 were calculated with the 2‐ΔΔCtmethod (Livak et al.,2001). The experiment was performed in triplicate. Primer sequences are given below:

MAPK14: Mitogen‐activated protein kinase 14; Gria3: glutamate receptor subunit 3.
miRNA microarray analysis
Specimen labeling and microarray hybridization were per‐formed according to a modified version of the Affymetrix miRNA microarray expression profiling protocol (Capti‐calBio Corporation, Beijing, China). Brie fly, total RNA was extracted using TRIzol Reagent (15596‐018; Invitrogen Life Technologies, Carlsbad, CA, USA), and miRNA was puri‐fied with the MirVana miRNA isolation Kit (AMI1560, Am‐bion). Poly(A) tailing was performed with the PAP enzyme.The miRNAs were then biotin‐labeled with the FlashTag Biotin Ligation Mix (FT30AFYB; Genisphere, Hat field, PA,USA). Hybridization was conducted at 48°C for 16 hours in a hybridization oven. Afterwards, the slides were washed and scanned with the GeneChip Scanner 3000 7G from Applied Biosystems (Foster City, CA, USA). The signals were analyzed with the Affymetrix GeneChip Command Console(v1.1) software from Affymetrix (Santa Clara, CA, USA).
Western blot assay
Hippocampal tissues were homogenized in a 1:10 (w/v) ratio of ice‐cold homogenization buffer (20 mM Tris‐HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100, 1 μg/mL leupeptin, 1 mM EGTA, 1 mM PMSF and 2.5 mM sodium orthovanadate). Samples were centrifuged at 13,400 ×gat 4°C for 15 minutes, and the supernatants were collected.The protein concentration of the lysate was determined with bicinchoninic acid assay kits (Huaxing Bio, Beijing, China).Samples were mixed with 5× sodium dodecyl sulfate (SDS)loading buffer and boiled at 95°C for 10 minutes. Proteins were separated by 15% sodium dodecyl sulfate polyacryl‐amide gel electrophoresis (SDS‐PAGE) and then transferred to a nitrocellulose membrane. The membrane was blocked with 5% milk for 1 hour at room temperature and then in‐cubated with primary antibodies–MAPK14 (mouse mono‐clonal antibody, #9228), Gria3 (rabbit monoclonal antibody,#4676) or GAPDH (rabbit monoclonal antibody, #2118)overnight at 4°C (all primary antibodies were purchased from Cell Signaling Technology, Danvers, MA, USA). After washing the blot extensively in wash buffer (3 × 10 minutes)with gentle agitation, anti‐rabbit secondary antibody (mouse monoclonal antibody, sc‐2357) or anti‐mouse secondary antibody (goat monoclonal antibody, sc‐2005), diluted in wash buffer, was added and incubated for 1 hour at room temperature with gentle agitation (the secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz,CA, USA). The membrane was washed with gentle agitation as follows: 4 × 5 minutes in wash buffer; 3 × 5 minutes in phosphate‐buffered saline with Tween 20; and 2 × 5 minutes in PBS. Protein bands were visualized with enhanced chemi‐luminescence reagents (Millipore, Billerica, MA, USA) and analyzed with Image J software (NIH, Bethesda, MD, USA).
Statistical analysis
All statistical analyses were performed with SPSS 17.0 soft‐ware (SPSS, Chicago, IL, USA). All results were expressed as the mean ± SD from three independent replicates. One‐way analysis of variance was performed, and significant differences among multiple group comparisons were analyzed using least significant difference tests.P‐values < 0.05 were considered statistically significant.
Results
CHSGS exhibited an antidepressant-like effect in the CUMS-induced rat model of depression
To evaluate the antidepressive effect of CHSGS, the sucrose consumption test was performed. As shown in Figure 1A,rats in the CUMS group consumed less sucrose than those in the control group. After a 2‐week treatment with CHSGS or fluoxetine, a significant increase in sucrose consumption was detected in stressed rats compared with normal control rats.
The CUMS group was only given distilled water. In addition, rats with CUMS‐induced depression showed significant decreases in both time and distance in the central area of the open field compared with the control group (Figure 1B, C).Treatment with CHSGS or fluoxetine significantly increased the time and distance in the central area in depressed rats(Figure 1B, C). Consistently, in the forced swimming test,rats in the CUMS group exhibited a substantial increase in immobility time compared with the control group. CHSGS and fluoxetine considerably diminished this increase in immobility time; namely, the treatment increased swimming time and climbing number in rats with CUMS‐induced depression (Figure 1D). Collectively, these results demonstrate that CHSGS exerts an antidepressive effect in the stressed rats, similar to fluoxetine.
CHSGS promoted synapse formation in the hippocampus
It is well documented that the hippocampus undergoes dramatic alterations in depressive disorders (Araki et al.,2015; Constals et al., 2015; Perez et al., 2015). Depression has been proposed to result from defects in synaptic con‐nections and plasticity in the hippocampus. Based on this concept, we speculated that the antidepressant‐like effect of CHSGS might be associated with synaptic remodeling in the hippocampus. To test this hypothesis, hippocampal tissues were collected from the rats of the different groups.Transmission electron microscopy showed that the number of synapses in stressed rats was decreased. Furthermore,the synapses had an enlarged synaptic cleft and a thinner postsynaptic density (Figure 2A, B). CHSGS treatment of depressed rats produced a recovery of synaptogenesis—the number of synapses in the hippocampus was increased,the synaptic cleft was smaller, and the postsynaptic density was thicker (Figure 2C, D). These results demonstrate that CHSGS promotes synapse formation in the hippocampus of stressed rats, highlighting the potential application of CHSGS as an antidepressant.
CHSGS downregulated miR-124 expression
The dramatic antidepressant‐like effect of CHSGS encour‐aged us to examine the underlying molecular mechanisms.Dysregulation of miRNA expression has been found in animal models of depression and postmortem brain tissues of depressed subjects (Lopez et al., 2014). To investigate the molecular mechanisms underlying the antidepressive action of CHSGS, miRNA microarray analysis was performed using hippocampal samples from rats in the control, CUMS and CHSGS groups. This analysis revealed that expression levels of 13 miRNAs were substantially changed (> 2‐fold)in depressed rats compared with the control group (Table 1). CHSGS treatment strikingly suppressed the increase inlevels of miR‐503, miR‐532, miR212, miR‐125a, miR‐182 and miR‐124 in depressed rats (Table 1). Previous studies demonstrated that miR‐124 is the most abundant miRNA in the brain and is specifically expressed there, playing important roles in neuropathological changes. Therefore, we focused on miR‐124 for subsequent experiments.

Table 1 List of candidate miRNAs that may be correlated with the therapeutic action of CHSGS in CUMS rats
RT‐qPCR was performed to quantify the abundance of miR‐124 in the hippocampus of the different groups.Consistent with the microarray data, elevated miR‐124 expression was observed in stressed rats. CHSGS treatment markedly suppressed this upregulation of miR‐124 induced by CUMS (Figure 3), similar to fluoxetine. These results suggest that miR‐124 is a downstream target of CHSGS and that the antidepressive effect of CHSGS might be mediated by the downregulation of miR‐124 expression.
MAPK14 and Gria3 were predicted target genes of miR-124
The post‐transcriptional regulation of gene expression by miRNAs involves cleaving or repressing the translation of target mRNAs (Flynt et al., 2008). To identify the gene tar‐gets of miR‐124, target gene prediction was performed with the TargetScan, miRDB and miRanda databases. As shown in Figure 4A, different miR‐124 target genes were identi‐fied with these databases. Approximately 190 of these genes were found at the intersection of all three databases (data not shown). To fully understand the function of these genes,the KEGG (Kyoto Encyclopedia of Genes and Genomes)pathway annotation was performed using DAVID. Several predicted miR‐124 target genes are associated with dopa‐minergic synapses (Figure 4B). Among these 190 genes,MAPK14 and Gria3 are particularly important for synaptic plasticity and neuronal proliferation in the hippocampus.To examine the regulatory effects of miR‐124 on MAPK14 and Gria3, the 3′‐UTR regions of MAPK14 and Gria3 that contain the presumed miR‐124 binding site were fused to the fire fly luciferase reporter plasmid. As expected, miR‐124 significantly reduced luciferase activity in both the MAPK14 and Gria3 groups compared with the control (Figure 4C).Furthermore, we constructed mutant plasmids by introducing point mutations in the corresponding complementary seed sites in the 3′‐UTRs of MAPK14 and Gria3 to eliminate miR‐124 binding. No significant changes in luciferase activity were observed in the presence of miR‐124 (Figure 4C). These results demonstrated that miR‐124 binds to the 3′‐UTR regions of MAPK14 and Gria3, which represses the expression of these genes.
CHSGS regulated the expression of MAPK14 and Gria3
To further examine the effect of CHSGS, both mRNA and protein levels of MAPK14 and Gria3 were measured. The RT‐qPCR analysis showed that the mRNA expression lev‐els of MAPK14 and Gria3 were dramatically increased by CHSGS treatment, and this effect was correlated with the downregulation of miR‐124 by CHSGS (Figure 5A and B).Consistent with this result, western blot assay demonstrat‐ed that CHSGS treatment markedly increased the protein levels of MAPK14 and Gria3 (Figure 5C and D). The quantitative analysis of the western data for Gria3 and MAPK14 are shown in Figure 5E and F, respectively. These results suggest that the downregulation of miR‐124 and the corre‐sponding upregulation of its target genes may underlie the antidepressive effect of CHSGS in the CUMS‐induced depression model.
Discussion
Chronic stress‐induced dysregulation of gene expression and dysfunctional neuronal plasticity have been implicated in the etiology and pathophysiology of depression (Fisar et al., 2008; Fan et al., 2015). Although remarkable progress has been made in our understanding of the pathophysi‐ological processes that contribute to the development of depression, a large number of patients still respond poorly to commercially available antidepressant therapies (Blier et al., 2016). In this study, we found that CHSGS, a traditional Chinese herbal medicine, alleviated the depression‐like behavioral disorder observed in CUMS rats at least in part by regulating miR‐124 and its downstream targets MAPK14 and Gria3. These findings provide insight into the molecular mechanisms underlying the antidepressive action of CHSGS and highlight the potential application of this medicine in the clinical treatment of depression.

Figure 1 CHSGS exhibited an antidepressive effect on CUMS-induced depression in rats.
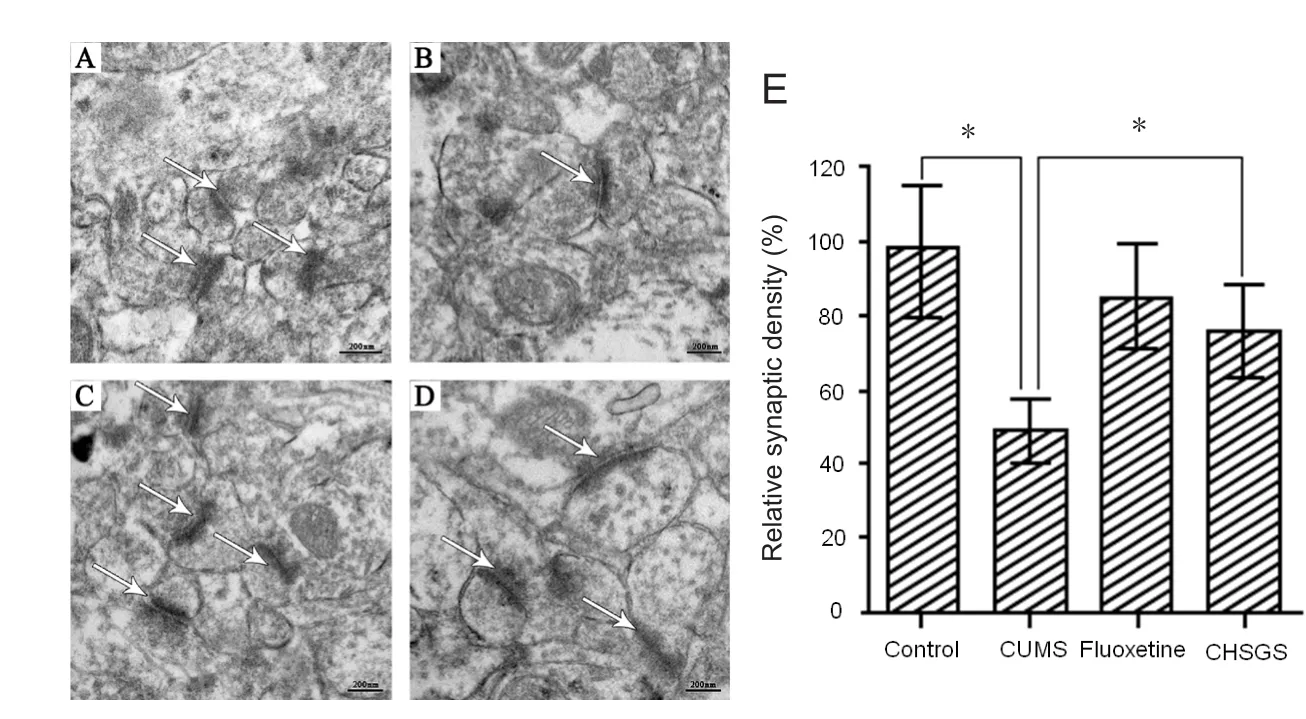
Figure 2 CHSGS promoted synapse formation in the hippocampus.

Figure 3 CHSGS downregulated miR-124 expression.

Figure 4 miR-124 target gene prediction analysis.

Figure 5 CHSGS increased the expression levels of MAPK14 and Gria3.
It is well documented that chronic stress induces depres‐sion‐like behaviors. However, the mechanisms that link the stress signals to the behavioral disorders remain largely un‐known (Kallarackal et al., 2013; Kreisel et al., 2014; Franklin et al., 2018). Previous studies have indicated that miRNA dysregulation strongly contributes to stressed‐induced synaptic plasticity and the pathophysiology of depression(Belzeaux et al., 2012; Fan et al., 2014; Wan et al., 2015).In particular, miR‐124, which is brain‐enriched and neu‐ron‐specific, controls the susceptibility to chronic stress‐in‐duced depression‐like behaviors (Dwivedi et al., 2015; Hi‐guchi et al., 2016; Roy et al., 2016). Overexpression of miR‐124 leads to an increase in the proportion of post‐mitotic neurons, a decrease in dividing precursors, and a significant decrease in astrocytes (Yoo et al., 2009). Diverse molecular mechanisms have been proposed for these effects of miR‐124. Recent studies show that miR‐124 controls the expres‐sion of the histone deacetylases HDAC4/5 and glycogen synthase kinase 3β in the hippocampus, which contributes to stress‐induced dendritic hypotrophy and reduces spine density of granular neurons in the dentate gyrus (Higuchi et al., 2016). Roy et al. (2016) demonstrated that miR‐124 is epigenetically regulated and that its interaction with the RNA‐induced silencing complex is compromised in major depressive disorder. Furthermore, a clinical study showed that miR‐124 expression is increased in major depressive patients (He et al., 2016). In our study, CHSGS treatment decreased levels of the highly expressed miR‐124 in stressed rats and enhanced synaptic plasticity in the hippocampus.These results demonstrate the involvement of miR‐124 in the pathophysiological progression of depression.
miRNAs negatively control gene expression by repressing mRNA translation or mediating cleavage of the target genes(Flynt and Lai, 2008). It has been shown that antidepres‐sants, including selective serotonin reuptake inhibitors and electroconvulsive therapy, modulate the aberrant expression of miRNAs and their targets (Belzeaux et al., 2012; Boc‐chio‐Chiavetto et al., 2013).
In this study, we identified MAPK14 and Gria3 as can‐didate targets of miR‐124. The downregulation of miR‐124 induced by CHSGS treatment was negatively correlated with the increased expression of MAPK14 and Gria3 in the hip‐pocampus of stressed rats. This suggests that CHSGS effec‐tively prevents the stress‐induced dysregulation of MAPK14 and Gria3 levels. Abnormalities in MAPK14 have been ob‐served in the hippocampus of suicide subjects (Pittenger et al., 2008). Glutamate is a major excitatory neurotransmitter in the central nervous system. A component of the AMPA glutamate receptor, Gria3 has been reported to be associated with migraine (Fang et al., 2015).
In summary, we found that CHSGS alleviated the depres‐sive behavior by downregulating miR‐124, which in turn upregulates MAPK14 and Gria3, in the hippocampus of the rat model of CUMS‐induced depression. These results suggest that miR‐124, MAPK14 and Gria3 are promising targets for investigating the pathogenesis of depression and for developing novel therapeutic strategies for the disease.The clinical validation of these findings is ongoing, and the results will be published in the near future.
Author contributions:QL carried out the open- field test, sucrose consumption test and forced swimming test, and drafted the paper. DHF and NNS carried out RT-qPCR, miRNA microarray analysis and western blot assay. ZZW participated in study design and performed statistical analysis. MQC conceived of the study, and participated in study design and coordination and helped to draft the paper. All authors had read and approved the final paper.
Conflicts of interest:The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Financial support:This work was supported by the National Natural Science Foundation of China, No. 81503415, 81574038, 81603671; the China Postdoctoral Science Foundation Grant, No. 2016M600709; a grant from the Science and Technology Planning Project of Guangdong Province of China, No. 2014A020221062; a grant from the Science and Technology Planning Project of Shenzhen City of China, No.JCYJ20150401170235349, JCYJ20160428105749954. The conception,design, execution, and analysis of experiments, as well as the preparation of and decision to publish this manuscript, were made independent of any funding organization.
Institutional review board statement:The protocols were approved by the Ethics Committee of Shenzhen Second People’s Hospital (Ethical Approval No. 20151211009). The experimental procedure followed the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1985).
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak,and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Elizabeth Hernández-Echeagaray, Universidad Nacional Autonoma de Mexico, Mexico.
Additional file:Open peer review report 1.
Araki Y, Zeng M, Zhang M, Huganir RL (2015) Rapid dispersion of SynGAP from synaptic spines triggers AMPA receptor insertion and spine enlargement during LTP. Neuron 85:173‐189.
Baudry A, Mouillet‐Richard S, Schneider B, Launay JM, Kellermann O (2010) miR‐16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329:1537‐1541.
Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano‐Tréziny C, Verrier L, Loundou A, Baumstarck‐Barrau K, Boyer L, Gall V, Gabert J, Nguyen C, Azorin JM, Naudin J, Ibrahim EC (2012)Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry 2:e185.
Blier P (2016) Neurobiology of depression and mechanism of action of depression treatments. J Clin Psychiatry 77:e319.
Bocchio‐Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignot‐ti S, Tardito D, Corrada D, Milanesi L, Gennarelli M (2013) Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol 23:602‐611.
Bonnet C, Leheup B, Béri M, Philippe C, Grégoire MJ, Jonveaux P(2009) Aberrant GRIA3 transcripts with multi‐exon duplications in a family with X‐linked mental retardation. Am J Med Genet A 149A:1280‐1289.
Bruchas MR, Schindler AG, Shankar H, Messinger DI, Miyatake M,Land BB, Lemos JC, Hagan CE, Neumaier JF, Quintana A, Palmiter RD, Chavkin C (2011) Selective p38α MAPK deletion in serotoner‐gic neurons produces stress resilience in models of depression and addiction. Neuron 71:498‐511.
Butler L, Pilkington K (2013) Chinese herbal medicine and depres‐sion: the research evidence. Evid Based Complement Alternat Med 2013:739716.
Cao MQ, Chen DH, Zhang CH, Wu ZZ (2013) Screening of specific microRNA in hippocampus of depression model rats and inter‐vention effect of Chaihu Shugan San. Zhongguo Zhong Yao Za Zhi 38:1585‐1589.
Cheng LC, Pastrana E, Tavazoie M, Doetsch F (2009) miR‐124 regulates adult neurogenesis in the subventricular zone stem cell niche.Nat Neurosci 12:399‐408.
Constals A, Penn AC, Compans B, Toulmé E, Phillipat A, Marais S,Retailleau N, Hafner AS, Coussen F, Hosy E, Choquet D (2015)Glutamate‐induced AMPA receptor desensitization increases their mobility and modulates short‐term plasticity through unbinding from Stargazin. Neuron 85:787‐803.
Di Lorenzo C, Coppola G, Grieco G, Santorelli FM, Pascale E, Pierelli F (2015) O041. GRIA3 (glutamate receptor, ionotropic, ampa 3)gene polymorphism influences cortical response to somatosensory stimulation in medication‐overuse headache (MOH) patients. J Headache Pain Suppl 1:A49.
de Sousa CN, Meneses LN, Vasconcelos GS, Silva MC, da Silva JC,Macêdo D, de Lucena DF, Vasconcelos SM (2015) Reversal of cor‐ticosterone‐induced BDNF alterations by the natural antioxidant alpha‐lipoic acid alone and combined with desvenlafaxine: empha‐sis on the neurotrophic hypothesis of depression. Psychiatry Res 230:211‐219.
Duman RS (2002) Pathophysiology of depression: the concept of syn‐aptic plasticity. Eur Psychiatry Suppl 3:306‐310.
Duman RS, Aghajanian GK, Sanacora G, Krystal JH (2016) Synaptic plasticity and depression: new insights from stress and rapid‐acting antidepressants. Nat Med 22:238‐249.
Dwivedi Y (2009) Brain‐derived neurotrophic factor: role in depres‐sion and suicide. Neuropsychiatr Dis Treat 5:433‐449.
Dwivedi Y, Roy B, Lugli G, Rizavi H, Zhang H, Smalheiser NR (2015)Chronic corticosterone‐mediated dysregulation of microRNA network in prefrontal cortex of rats: relevance to depression patho‐physiology. Transl Psychiatry 5:e682.
Enoch MA, Rosser AA, Zhou Z, Mash DC, Yuan Q, Goldman D (2014)Expression of glutamatergic genes in healthy humans across 16 brain regions; altered expression in the hippocampus after chronic exposure to alcohol or cocaine. Genes Brain Behav 13:758‐768.
Fan HM, Sun XY, Guo W, Zhong AF, Niu W, Zhao L, Dai YH, Guo ZM, Zhang LY, Lu J (2014) Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J Psychiatr Res 59:45‐52.
Fan HM, Sun XY, Niu W, Zhao L, Zhang QL, Li WS, Zhong AF,Zhang LY, Lu J (2015) Altered microRNA expression in peripheral blood mononuclear cells from young patients with schizophrenia. J Mol Neurosci 56:562‐571.
Fang J, An X, Chen S, Yu Z, Ma Q, Qu H (2015) Case‐control study of GRIA1 and GRIA3 gene variants in migraine. J Headache Pain 17:2.
Fisar Z, Raboch J (2008) Depression, antidepressants, and peripheral blood components. Neuro Endocrinol Lett 29:17‐28.
Flynt AS, Lai EC (2008) Biological principles of microRNA‐mediated regulation: shared themes amid diversity. Nat Rev Genet 9:831‐842.
Franklin TC, Wohleb ES, Zhang Y, Fogaça M, Hare B, Duman RS(2018) Persistent increase in microglial RAGE contributes to chronic stress‐induced priming of depressive‐like behavior. Biol Psychia‐try 83:50‐60.
He S, Liu X, Jiang K, Peng D, Hong W, Fang Y, Qian Y, Yu S, Li H(2016) Alterations of microRNA‐124 expression in peripheral blood mononuclear cells in pre‐and post‐treatment patients with major depressive disorder. J Psychiatr Res 78:65‐71.
Higuchi F, Uchida S, Yamagata H, Abe‐Higuchi N, Hobara T, Hara K, Kobayashi A, Shintaku T, Itoh Y, Suzuki T, Watanabe Y (2016)Hippocampal microRNA‐124 enhances chronic stress resilience in mice. J Neurosci 36:7253‐7267.
Jiang Y, Zhu J (2015) Effects of sleep deprivation on behaviors and abnormal hippocampal BDNF/miR‐10B expression in rats with chronic stress depression. Int J Clin Exp Pathol 8:586‐593.
Juurlink DN, Mamdani MM, Kopp A, Redelmeier DA (2006) The risk of suicide with selective serotonin reuptake inhibitors in the elderly.Am J Psychiatry 163:813‐821.
Kallarackal AJ, Kvarta MD, Cammarata E, Jaberi L, Cai X, Bailey AM,Thompson SM (2013) Chronic stress induces a selective decrease in AMPA receptor‐mediated synaptic excitation at hippocampal tem‐poroammonic‐CA1 synapses. J Neurosci 33:15669‐15674.
Kim SH, Han J, Seog DH, Chung JY, Kim N, Hong Park Y, Lee SK(2005) Antidepressant effect of Chaihu‐Shugan‐San extract and its constituents in rat models of depression. Life Sci 76:1297‐1306.
Kosik KS (2006) The neuronal microRNA system. Nat Rev Neurosci 7:911‐920.
Kreisel T, Frank MG, Licht T, Reshef R, Ben‐Menachem‐Zidon O,Baratta MV, Maier SF, Yirmiya R (2014) Dynamic microglial alter‐ations underlie stress‐induced depressive‐like behavior and sup‐pressed neurogenesis. Mol Psychiatry 19:699‐709.
Lagos‐Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue‐specific microRNAs from mouse.Curr Biol 12:735‐739.
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCtmethod. Meth‐ods 25:402‐408.
Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, Mauss‐ion G, Yang JP, Yerko V, Vigneault E, El Mestikawy S, Mechawar N, Pavlidis P, Turecki G (2014) miR‐1202 is a primate‐specific and brain‐enriched microRNA involved in major depression and anti‐depressant treatment. Nat Med 20:764‐768.
Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The Mi‐croRNA miR‐124 promotes neuronal differentiation by triggering brain‐specific alternative pre‐mRNA splicing. Mol Cell 27:435‐448.
McCartney AJ, Zolov SN, Kauffman EJ, Zhang Y, Strunk BS, Weisman LS, Sutton MA (2014) Activity‐dependent PI (3,5) P2 synthesis controls AMPA receptor trafficking during synaptic depression.Proc Natl Acad Sci U S A 111:E4896‐E4905.
Miller BR, Hen R (2015) The current state of the neurogenic theory of depression and anxiety. Curr Opin Neurobiol 30:51‐58.
Moretti M, Budni J, Freitas AE, Neis VB, Ribeiro CM, de Oliveira Balen G, Rieger DK, Leal RB, Rodrigues AL (2015) TNF‐α‐in‐duced depressive‐like phenotype and p38 (MAPK) activation are abolished by ascorbic acid treatment. Eur Neuropsychopharmacol 25:902‐912.
Papakostas GI (2010) The efficacy, tolerability, and safety of contemporary antidepressants. J Clin Psychiatry Suppl E1:e03.
Perez de Arce K, Schrod N, Metzbower SW, Allgeyer E, Kong GK,Tang AH, Krupp AJ, Stein V, Liu X, Bewersdorf J, Blanpied TA, Lu‐cić V, Biederer T (2015) Topographic mapping of the synaptic cleftinto adhesive nanodomains. Neuron 88:1165‐1172.
Pittenger C, Duman RS (2008) Stress, depression, and neuroplasticity:a convergence of mechanisms. Neuropsychopharmacology 33:88‐109.
Putnam KT, Wilcox M, Robertson‐Blackmore E, Sharkey K, Bergink V,Munk‐Olsen T, Deligiannidis KM, Payne J, Altemus M, Newport J,Apter G, Devouche E, Viktorin A, Magnusson P, Penninx B, Buist A,Bilszta J, O’Hara M, Stuart S, Brock R et al. (2017) Clinical pheno‐types of perinatal depression and time of symptom onset: analysis of data from an international consortium. Lancet Psychiatry 4:477‐485.
Qiu J, Hu SY, Zhang CH, Shi GQ, Wang SE, Xiang T (2014) The effect of Chaihu‐Shugan‐San and its components on the expression of ERK5 in the hippocampus of depressed rats. J Ethnopharmacol 152:320‐326.
Redhead M, Satchell R, Morkūnaitė V, Swift D, Petrauskas V, Golding E, Onions S, Matulis D, Unitt J (2015) A combinatorial biophysical approach; FTSA and SPR for identifying small molecule ligands and PAINs. Anal Biochem 479:63‐73.
Riddle M, Potter GG, McQuoid DR, Steffens DC, Beyer JL, Taylor WD (2017) Longitudinal cognitive outcomes of clinical phenotypes of late‐life depression. Am J Geriatr Psychiatry 25:1123‐1134.
Roy B, Dunbar M, Shelton RC, Dwivedi Y (2016) Identification of Mi‐croRNA‐124‐3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology 42:864‐875.
Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J, Liu L, Zhang C (2015)Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS One 10:e0121975.
Wang S, Hu S, Zhang C, Qiu J, Li Y (2014) Antidepressant‐like activity of Chaihu‐Shugan‐San aqueous extract in rats and its possible mechanism. Pharmacogn Mag Suppl 1:S50‐S56.
Wang Y, Fan R, Huang X (2012) Meta‐analysis of the clinical effectiveness of traditional Chinese medicine formula Chaihu‐Shugan‐San in depression. J Ethnopharmacol 141:571‐577.
Willner P (1997) Validity, reliability and utility of the chronic mild stress model of depression: a 10‐year review and evaluation. Psy‐chopharmacology 134:319‐329.
Yoo AS, Staahl BT, Chen L, Crabtree GR (2009) MicroRNA‐mediated switching of chromatin‐remodelling complexes in neural develop‐ment. Nature 460:642‐646.
Zeng Y (2009) Regulation of the mammalian nervous system by mi‐croRNAs. Mol Pharmacol 75:259‐264.
Zhu X, Li T, Peng S, Ma X, Chen X, Zhang X (2010) Maternal depri‐vation‐caused behavioral abnormalities in adult rats relate to a non‐methylation‐regulated D2 receptor levels in the nucleus ac‐cumbens. Behav Brain Res 209:281‐288.
- 中国神经再生研究(英文版)的其它文章
- Weak phonation due to unknown injury of the corticobulbar tract in a patient with mild traumatic brain injury: a diffusion tensor tractography study
- Exosomes: a novel therapeutic target for Alzheimer’s disease?
- Inhibition of retinal ganglion cell apoptosis:regulation of mitochondrial function by PACAP
- Trillium tschonoskii maxim extract attenuates abnormal Tau phosphorylation
- Association between Alzheimer’s disease pathogenesis and early demyelination and oligodendrocyte dysfunction
- Nogo receptor expression in microglia/macrophages during experimental autoimmune encephalomyelitis progression