Biotransformation and degradation of 2,4,6-trinitrotoluene by microbial metabolism and their interaction
Mónica Y.Serrano-González,Rashmi Chandra*,,Carlos Castillo-Zacarias,Felipe Robledo-Padilla,Magdalena de J.Rostro-Alanis,Roberto Parra-Saldivar**,
Tecnologico de Monterrey,School of Engineering and Science,Campus Monterrey,Ave.Eugenio Garza Sada 2501,Monterrey,N.L.,CP 64849,Mexico
1.Introduction
Military operations often result in dissemination of nitro aromatic explosives such as 2,4,6-trinitrotoluene(TNT)into the environment,causing contamination of soil,surface,and ground water[1-4].Some of the explosives contain aromatic compounds with nitro groups,which are noxious and recalcitrant,and their microbial degradation is very slow[3,5].Although the TNT production has ceased,the contamination persists[6].TNT is toxic to aquatic flora and other organisms and animals,including reptiles.TNT is classified as mutagenic,poisonous,and carcinogenic.Even at a very low concentration of 50 mg/L,TNT is lethal for microorganisms including Gram-positive bacteria and yeasts[7].These explosives contain nitrogen aromatic compounds and have a potential of selfoxidation to derivatives and small gaseous molecules[8].These derivatives are just as toxic as TNT.Their presence in the environment poses an ecological and health hazard.Hence,a method to clean the contaminated sites and ensure environmental safety is urgently needed[4].Investigations have focused on the removal of nitro aromatic compounds from the ecosystem,and various remediation technologies have been developed,such as adsorption,oxidation processes,phytoremediation,and microorganism remediation.All of the above offer some possibilities for reducing toxicity and TNT's derivative pollutants from the contaminated sites[1,9,10].
TNT is a relatively complex chemical due to the presence of a stable aromatic ring and nitro groups that makes its degradation challenging.Its three symmetrically arranged nitro groups bring a high electron de ficit at the aromatic ring[11].Some microorganisms have a metabolism capable of reducing nitro aromatic compounds and their metabolites.Transforming TNT requires an oxygenic microenvironment at the beginning of the process and then,a high electronegative reducing environment.The combination of the electron flux resulting from the biocatalyst metabolism facilitates and increases the rate of TNT reduction.The reduction of the third nitro group requires anaerobic bacteria,since aerobic microorganisms are capable of reducing only 2 out of the 3 nitro groups of TNT[12].The main microorganisms capable of the biodegradation of the nitro groups are Pseudomonas,Enterobacter cloacae,Rhodococcus erythropolis,and Mycobacterium which can metabolize TNT in anoxic conditions.Clostridium and Desulfovibrio species and some fungi can completely mineralize TNT[11].The bacterial metabolism might not necessarily open the ring,and that could explain why TNT is usually transformed,but not mineralized.An important alternative to achieve mineralization of TNT is degradation by fungi.For example,several studies report that white-rot fungi,such as the litter degrading Phanerochaete chrysosporium and Stropharia species can mineralize TNT under aerobic conditions[13-15].After the fungus-mediated TNT degradation,an enzymatic process involving nitroreductases,can degrade it further[16-19].This review aims to examine the state of TNT degradation.We havesummarized variouspossible methodsforTNT degradation from chemical and physical treatments to several biological methods.We focus our review on the several kinds of biological treatment:anaerobic,aerobic,combined,and enzymatic processes.
2.Worldwide deposits
Every year 1000 tons of TNT are produced globally,and nearly 2 million L of TNT-contaminated wastewater,along with other nitro aromatic compounds are discarded into the environment every day[20].More than 2000 sites,covering 15 million acres in the United States are expected to have TNT contamination from military activities.About 87%of those sites present groundwater contamination and a total of 30 sites have been listed in the US Environment Protection Agency's National Priority List(NPL)[8,21,22].As per the Environmental Protection Agency,TNT was widely used by the military sector in the USA,and the production was restricted to military activities.Claus reported that 3200 sites in Germany contaminated with TNT require environmental renovation and restoration(Claus 2014a).In 2000,a total of 103 defense training sites in Canada were connected with TNT contamination,3 out of those 103 sites were even found burning and exploding[13](where TNT contamination by the use of weapons was found at levels of about 200g/kg and 100 mg/L in soil and water,respectively[23]).TNT contaminates groundwater by the discharge of contaminated effluent,also known as red water,which is generated during the manufacture,packing,storage,and handling of nitro aromatic compounds used for weapons.As well as TNT,red waters contain more than 30 other aromatic compounds,which may affect thousands of people within three miles of a contaminated site[8].
Countries that have suffered serious armed con flicts(those affected by wars in Africa,Middle East,Australia and Eastern Europe)might have critical environmental problems from TNT contamination.During World War I,TNT was one of the major ingredients for explosives used by the US,which was also adopted by otherarmies.Nitramine,a class of nitratedorganic compounds,was used as an explosive during World War II.(Singh 2012)(Boileau et al.,1987;Gilbert,1980).Large-scale production resulted in the disposal of huge wastes containing nitrated organic by-products of explosives into the environment.The US and Germany ware largest producers of TNT in 1945,with about approximately 50 tons of TNT per production line[5,24,25].About 1.2 million tons of soil have been contaminated with explosives in the US[26].Man-made chemicals from common military explosives and their conversion products cause toxic and mutagenic effects[26].Cleanup of explosive-contaminated areas is now a public health concern,and massive efforts are undergoing to find an economical remediation technology.The least expensive means of destroying organic pollution is biological treatment,which can achieve complete mineralization of TNTand reduce the total downstream waste.This review will examine the most important bioremediation efforts to treat contaminated sites by explosives.We report on the chemical and biological transformations TNTand its derivatives undergo and list the commercial processes developed to perform these transformations for the treatment of contaminated soil.
3.Toxicity of TNT
TNT is used in the manufacture of grenades,bombs,shells,and in some other military and industrial applications.As a result of manufacturing processes and the destruction of military arsenal,nitro aromatic compounds are introduced to the environment as solid waste or as wastewater.TNT exposure occurs through inhaling,eating,drinking,or touching contaminated water,soil,air,or food.Reported cases of people who have been exposed to TNT point to anemia,abnormal liver function,skin irritation,and cataracts.Animal studies indicate that TNT is a carcinogen and TNT is also toxic to aquatic organisms,algae,and invertebrates.Besides,it inhibits the activity of soil microbial processes,such as the dehydrogenase activity and the nitrogen- fixation process even at low levels(10mg/L)of TNT contamination[27,28].The toxicity of TNT depends on the length of time and exposure conditions.The US EPA(United States Environmental Protection Agency)recommended limit for TNT is 0.001mg per liter of drinking water[1].Because of the negative impact of TNTon the ecosystem and on human health,research has to expand from cleanup operations to the development of sustainable military weapons,and environmental-friendly military activities[29].
4.Remediation methods
4.1.Biological process for TNT bioremediation
Biological treatments use living organisms to degrade environmental pollutants into less toxic or non-toxic substances.These methods use bacteria,fungi,algae,or superior organisms such as plants to detoxify and provide effective remediation of contaminated soil and water.The microorganisms used in biological treatment methods could be indigenous from the contaminated site,or they could be previously isolated elsewhere and applied to the contaminated site.Pollutant substances are transformed by microorganisms through a variety of reactions such as oxidation/reduction reactions that occur as part of their natural metabolism.In-situ bioremediation of pollutants results from the action of various microbial species.The biotreatment of explosive compounds has been investigated to find microorganisms that may be applied ex-situ(in a bio-reactor)or in-situ.Different microbial strains capable of degrading TNT,entirely or partially,have been isolated and identified from contaminated sites[30].Additionally,a variety of processes such as bio-reactors inoculated with sludge,composting,and land farming have been developed[31].Snellinx et al.reported that aerobic bacteria transform the TNT molecule by a reduction of one or two nitro groups to hydroxyl amino or amino acid groups while generating different isomers of amino nitroaromatic compounds.
During microbial degradation of 2,4,6-TNT,oxidized metabolites might occur if degradation is not completed.Since these metabolites are highly water soluble,it must be assumed that they represent a much greater threat to groundwater than 2,4,6-TNT itself.Many of the potential risks associated with microbial bioremediation are shared by bacteria used for TNT degradation.Several microorganisms(Pseudomonas,Burkholderia,Sphingomonas,Ralstonia,Comamonas,Achromobacter,Alcaligenes,Rhodococcus,Dehalococcoides)are known to degrade TNT or to accumulate or detoxify heavy metals and,for more than 30 years,it has been hoped that these bacteria might be usedin the clean-up of toxic like phenol,trichloroethylene,trinitrotoluene,dioxins etc.
4.1.1.Bacterial TNT degradation
Depending on the type of microorganism and the culture conditions,the main products and the degree of the TNT degradation reactions vary.However,the de-nitration pathway is pro fitable to microorganisms due to the decrease of the electrophilic nature of TNT,which allows them to use dioxygenases and mono-or dinitrotoluenes as substrates[32,33].The explosive compounds serve as a carbon or nitrogen source to many aerobic or anaerobic microorganisms[13].
The main dif ficulty in microbial TNT degradation is the symmetrical arrangement of three nitro-groups that creates a high electron de ficit condition at the aromatic ring and mostly the mesomeric effects along the whole structure.The use of TNT as a carbon and energy source is very challenging for microbes.The oxidative degradation results in transformation or destruction of TNT to other forms,but not to complete mineralization.The initial metabolites in the biotransformation of TNT are Hydroxyl-aminodi-nitro-toluenes(HADNTs),Amino-di-nitro-toluenes(ADNTs),di-Amino-mono-nitrotoluenes(DANTs)and Tetranitroazoxytoluenes(AZTs)[9,13].Due to the electron de ficiency of the ring,the initial degradation of TNT by microorganisms is characterized by reductive reactions[34].The nitro-moieties of TNT(-NO2)can be successively reduced to nitroso(-NO),hydroxylamino(-NHOH)and finally amino(-NH2)groups.Obligate anaerobic bacteria,such as Clostridium sp.,Desulfovibrio sp.and Archaea as Methanococcus sp.,can reduce TNTcompletely to 2,4,6-triaminotoluene(TAT)[35-37].Depending upon redoxconditions,these compounds can be further converted by biotic and abiotic mechanisms to azo-,azoxy-,hydrazone-,and phenol-acetyl derivatives[13].These compounds can be easily degraded by the application of enzymes like laccases,which we discuss later.
4.1.1.1.Aerobic TNT degradation.Pseudomonas can growon TNT as solitary nitrogen source.P.savastanoi,are able to convert TNT into 2,4-DNT.If the medium is enriched with glucose,TNT degradation enhances the production of 4-Amino-2,6-dinitrotoluene(4-A-2,6-DNT)and 2-Amino-4,6-dinitrotoluene(2-ADNT)(Fig.1)and decreases the de-nitration of TNT[11].It has been reported that the 4-Amino-2-nitroso-6-nitrotoluene(4A-2NOC-6-NT),which is one of the final compounds resulting of the aerobic degradation pathway,has been detected in cultures of Bacillus sp.,Staphylococcus sp.,and Pseudomonas aeruginosa[11].P.Aeruginosa also metabolized amino-di-nitro-toluenes to extremely polar compounds.Table 1 provides a comprehensive list of various aerobic microorganisms involved in TNT degradation.
The aerobic degradation of TNT initiates with the reduction of one of the three nitro groups.The nitro group is easier to reduce when the nitrogen atom combines its high electronegativity and its partially positive charged[7].Anaerobic degradation starts here.The first compounds to be transformed by anaerobic reductive reaction are 2-ADNT and 4-A-2,6-DNT(Fig.1).The final conversions of the anaerobic route lead to the production of diverse isomers of amino nitro aromatic compounds,which tend to accumulate in the growth medium without further metabolism.Anaerobic degradation of TNT also produces 2,6-diamino-4-nitrotoluene(2,6-DA-4-NT),2-amino-4-nitrotoluene(2A-4-NT),and 4A-2NOC-6-NT followed by 2-ADNT and 4-A-2,6-DNT.Aerobic bacteria usually transform the TNT molecule by reducing one or two nitro groups to hydroxylamine via non-specific NAD(P)H-dependent nitroreductase(Fig.1).Hydroxylamine is then reduced to amino groups by nitroreductases and formation of 2,4 amino-2-hydroxylamine-6-nitrotoluene or 2 amino-4,6-dinitrotoluene from the hydroxylamine,though nitrobenzene reductases.During the reduction process more compounds are produced other than the 2,4 amino-2 hydroxylamino-6-nitrotoluene.However in some cases,partially reduced forms of TNT can react in the presence of oxygen to make recalcitrant azoxytetranitrotoluenes[11].
As stated above,the three symmetrically arranged nitro groups in TNT induce a high electron de ficiency at the aromatic ring[11].Because a high electronegative-reducing environment is necessary to transform TNT;the addition of oxygen combined with the electron flux from biocatalyst metabolism can facilitate the rate of reduction of TNT.The reduction of the third nitro group requires anaerobic bacteria,since aerobic bacteria can only reduce 2 out of the 3 nitro groups of TNT[12].When the electrons conforming the TNTaromatic ring are removed by the electronegative nitro groups,the nucleus becomes electrophilic[38]and in some cases this results in the formation of non-aromatic structures such as a Meisenheimer complex,followed by the production of dinitrotoluenes[11].Bacterial strains,including Rhodococcus erythropolis and Mycobacterium sp,are able to form the Meisenheimer complexes from TNT[12].In addition,oxygen is not necessary for the formation of those complexes,hence aerobic TNT degradation is an alternative metabolism when it is not possible to remove oxygen from the environment[11].
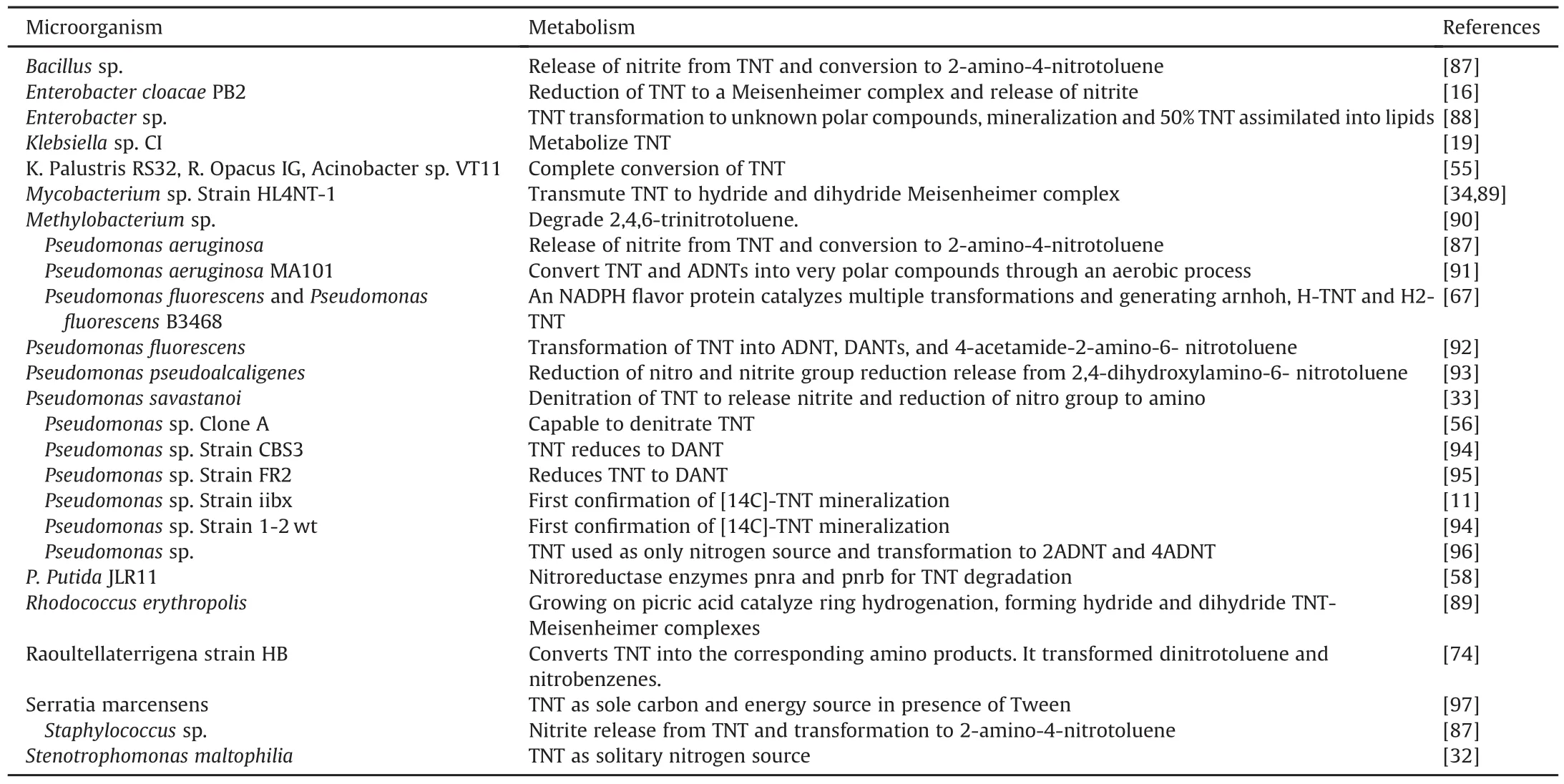
Table 1Microorganisms and mechanisms reported for the TNT degradation and transformation under aerobic conditions.

Table 2Microorganisms and mechanisms reported for TNT degradation and transformation under anaerobic conditions.
4.1.1.2.Anaerobic degradation.TNT is entirely reduced to TAT by strictly anaerobic bacteria,such as Desulfovibrio sp.,Clostridium sp.,and Archaea such as Methanococcus sp[36].Clostridium species are generally capable to reduce TNT anaerobically[39].Desulfovibrio species can metabolize TNT in anoxic conditions and use it as the exclusive nitrogen source.Clostridium and Desulfovibrio species are ableto catalyzethecompletereduction ofTNT to 2,4,6-triaminotoluene,which is one of the final steps of the degradation.Several intermediate reactions occur before the transformation to TAT,such as the reduction of DANTs to TAT,which needs pyruvate and H2to proceed.This is the step that determines the rate of the overall process,according to Preuss[40].Table 2 provides a list of microorganisms and mechanisms reported for anaerobic TNT degradation.
Microbial anaerobic processes are capable of rapid TNT degradation at low redox potential,and minimization of oxidative polymerization reaction[41].As it occurs during aerobic degradation of TNT,the nitro-groups can be consecutively reduced to nitroso,then hydroxylamino,and finally amino groups.At this point the final compounds are the same as the aerobic degradation main products:2-ADNT and 4-A-2,6-DNT.However and only by anaerobic degradation,C.acetobutylicum might transform 4-hydroxylamine-2,4-dinitrotoluene to 2-amino-5-hydroxyl-4-hydroxylamino-6-nitrotoluene.Both,4-A-2,6-DNT and 2-ADNT arereducedto2,4-diamino-6-nitrotoluene(DANT)vianonspecific NAD(P)H nitroreductase(Fig.2).Besides and after TAT is produced bydissimilatorysul fite reductase,DANT is transformed to 2,4-Diamino-6-hydroxylaminotoluene(DAHAT)by hydrogenase or carbon monoxide dehydrogenase,.Moreover,DANTcan be reduced directly to TAT with the aid of C.sordelli,C.bifermentans,and C.sporogenes.TAT can besuccessively converted to 2,4,6-trihydroxytoluene,then 4-hydroxytoluene(p-Cresol).Thep-Cresol is one of the intermediate metabolites in the typical toluene pathway degradation,and precedes the final transformations of the route to produce succinyl CoA and Acetyl-CoA.
4.1.1.3.Combined aerobic and anaerobic TNT degradation.Mineralization of TNT has been found in mixed or unde fined culture systems;however the literature on mineralization under aerobic oranaerobic conditionsbypure bacterial systems is missing[42].In comparisonwith differentelectron acceptors such as sulfate and carbon dioxide,nitrate gives the best TNT degradation rate in soil bacteria[43].Under mutually occurring aerobic and anaerobic conditions,TNT is transformed to amino derivatives via nonspecific NAD(P)H-dependentnitro-reductase[42].MostoftheTNT-degrading bacteria use only one initial metabolic pathway for its degradation;however,some bacteria can use simultaneously two different pathways[34,44].One of the metabolic pathways is a consecutive reduction of two electrons in the nitro groups of the aromatic ring,which produces reduced intermediates such as DANT,4-hydroxyl-2,6-aminodinitrotoluene,2- hydroxyl-4,6-aminodinitrotoluene,2-ADNT,4-A-2,6-DNT,isomers,and azo and azoxy dimers[41].The second pathway is a direct two-electrons reduction of the aromatic ring via hydride addition,the products of this reaction include hydride Meisenheimer complex,dihydride Meisenheimer complex,and protonated tautomers[11,34,44].Several protonated tautomers can be re-aromatized after nitrite release with the formation of dinitrotoluenes(dnts).
Disaggregating the two initial biochemical routes in aerobic degradation,the first step to reduce TNT is its transformation to 4-Hydroxylamino-2,6-dinitrotoluene (4-HDNAT) or 2-Hydroxylamino-4,6-dinitrotoluene(2-HDNT),depending on the microorganism this is mediated by nitroreductase/dihydropteridinereductase,N-ethylmaleimidereductase,and nitrobenzene nitroreductase.Those reactions are followed by the formation of 4-A-2,6-DNT or 2-ADNT via nitroreductases(Fig.3).In addition to this,during the aerobic degradation of 4-HDNATand 2-HDNT more compounds are formed such as 4,4′-dimethyl-3,3′,5,5′-tetranitroazoxybenzene, 2,2′-dimethyl-3,3′,5,5′-tetranitro-azoxybenzene and 2,4′-dimethyl-3,3′,5,5′-tetranitro-azoxybenzene.The final transformations of the aerobic route leads to the production of 2A-4-NT,4A-2NOC-6-NT,and 2,6-DA-4-NT then 4-A-2,6-DNT and 2-ADNT are produced by anaerobic degradation to finalize the process.With anaerobic bacteria,both,4-A-2,6-DNT and 2-ADNT are reduced to DANT via non-specific NAD(P)H nitroreductase.Further on DANT is transformed to DAHAT by hydrogenase or carbon monoxide dehydrogenase,which is followed by the formation of the TAT by dissimilatory sul fite reductase.Alternatively,DANT can also be directly reduced to TAT by C.sordelli,C.bifermentans and C.sporogenes.TAT can be successively reduced to 2,4,6-trihydroxytoluene and then to p-Cresol.The p-Cresol is an intermediate metabolite inside the toluene degradation pathway;its formation during TNT degradation is the result of the reduction of two of the hydroxide bonds of the aromatic ring from 2,4,6 Trihydroxytoluene.The action of the 4-cresol dehydrogenase,p-Cresol is transformed to 4-hydroxybenzaldehyde,followed by the oxidation ofthemethylgroup toform 4-hydroxybenzoate[45].4-hydroxybenzoate can be successively oxidized to Benzoyl-CoA,which suffers a series of reactions: first it is converted into 2,3 epoxybenzoyl- CoA, followed by transformation to 3,4 Dehydroadipyl-CoA-Semialdehyde,and then 3,4 Dehydroadipyl-CoA to finally produce succinyl-CoA and Acetyl-CoA[12].Succinyl CoA and Acetyl-CoA are important intermediates in the citric acid cycle(TCA).
4.1.2.TNT degradation by fungi
Thebacterialmetabolismpreviouslymentioneddoesn't necessarily open the ring in the TNT structure[13].Even though TNT is usually transformed by bacteria,it is not mineralized.A viable alternative is degradation by fungi.Some reports state that white-rot fungi,such as the litter degrading fungus Phanerochaetechryso sporium and Stropharia species can mineralize TNT under aerobic conditions[11,15].Saprophytic micromycetes and basidiomycetes have the ability to metabolize and mineralize TNT[46]and(C14)TNT mineralization can certainly occur by wood and litter decaying basidiomycetes.This study also reported that during a period of 64 days,Clitocybuladusenii tmb12 mineralized 42%of the initial(C14)TNT to(C14)CO2,and Strophariarugosa annulata DSM11372 mineralized 36%.The white-rot fungi mechanism of tntis the ligninolytic enzyme system,which consists of a number of extracellular enzymes(mostly peroxidases)and oxido-reductases(laccases)which catalyze the degradation of the wood constituent lignin[38,47].Another wood white-rot fungus,Phanerochaete chrysosporium can degrade xenobiotics,Also,this fungus can be exposed to wide range of conditions:limitation of carbon,sulfur,or nitrogen sources to favor TNT degradation[11].The reactions involved for TNT transformation by fungi are mostly lignindegrading enzymes.For example,fungi can utilize manganese peroxidases,lignin peroxidases,reductases,oxidases,hydrogen peroxidases,and quinol oxidases[11,32].The first steps of TNT transformation normally imply a reduction of the molecule.For example,underligninolyticconditions,Phanerochaetechrysosporium reduces TNT to nitroso-toluene,4-ADNT,2-ADNT,4-HDNAT,AZTs,phenolic,and acylated(acetylated and formylated)derivatives(Fig.4).Lignin peroxidase and MnO2oxidize the 4-HDNAT part,and cause a futile cycle of reduction-oxidation and the formation of AZTs.Once 4-A-2,6-DNT is produced,an additional derivatization and reduction reaction,and then oxidation by ligninolytic systems occurred[41,48].The 4-A-2,6-DNT is made to produce 4-formamido-2,6-dinitrotoluene,then this last one is reduced to 2-amino-4-formamido-6-nitrotoluene,which can be transformed to DANT and accumulated under non-ligninolytic conditions.But if ligninolytic conditions are present,mineralization happens[37,41].However,the mechanism employed by fungi to mineralize is not completely understood[49].One important aspect related to TNT mineralization is that low concentrations of TNT inhibit the growth of P.Chrysosporium,and then its catabolic potential[50].Table 3 lists several kinds of fungi responsible for TNT degradation under aerobic condition.
Study by Ziganshin et al.shows reactive oxygen and nitrogen species generated during TNT transformation by Y.Lipolytica participate in the abiotic conversion of TNT[51,52].The most promising and efficient pathwayof nitrogen release from TNT is the transformation of parent compound through the addition of hydride ions resulting in the formation of Meisenheimer hydride complexes followed by their subsequent destruction.Among the aromatic ring hydrogenation products, eight hydride-Meisenheimer complexes of TNT were detected,which were previously described in detail[51,53,54].The ability of Y.Lipolytica ANL15 to reduce the TNT aromatic ring to form TNT-hydride complexes,followed by their denitration,makes this strain a potential candidate for bioremediation ofsites contaminated with explosives.
4.1.3.TNT as a source of nitrogen or electron acceptor
Even though TNT biodegradation can be done by a co-metabolic process,it is not mineralized by an individual bacterial strain.The co-metabolism is accomplished in the presence of a reduction equivalent donating substrate,such the Acinetobacter sp.VT11 strain,which use TNT as sole growth substrate[55].On the other hand,some reports show that TNT can be used as a source of nitrogen and also as external electron acceptor by many microorganisms[35,56].One of these microorganisms is Pseudomonas sp.JLR 11,which releases nitrite from the aromatic ring before reducing it to ammonium.Through this mechanism,approximately 85%of the nitrogen present in TNT is integrated as organic nitrogen into the cells[11].The TNT degradation can be done by destruction of the TNT-monohydride complex or by destruction of oneprotonated TNT-dihydride complex.The best known way to release the nitrogen from TNT is by abiotic condensation of hydroxylamino-dinitroluene-(HADNT)isomers and protonated dihydride-meisenheimer-complexes to form secondary diarylamines with the concomitant release of nitrite[57].The released nitrite presumably participates in a nitrite reductase process and then is converted to ammonium.After this,it is assimilated via the glutamine-glutamate synthase pathway as the last step[58].TNT can also be used as an alternative electron acceptor in the respiratory chain forming ATP by some Pseudomonas species[11].In this sense,TNT can be used by anaerobic Clostridiae for reoxidation of reduced electron carriers to maintain the fermentative metabolism[5].
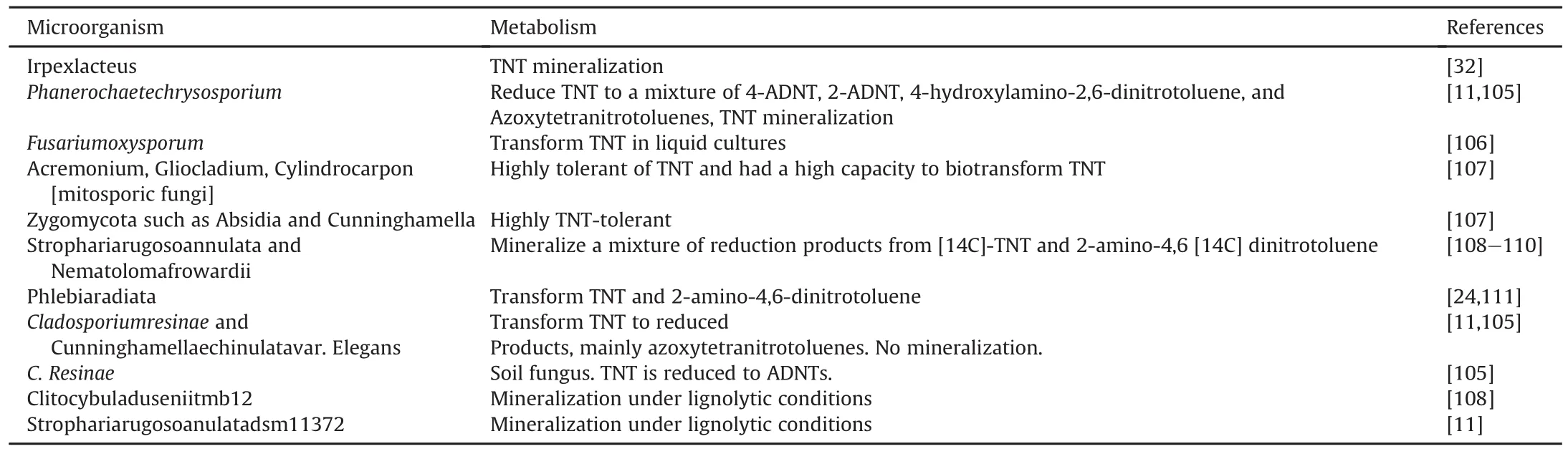
Table 3Microorganisms involve in TNT degradation by fungi under aerobic condition.
4.1.3.1.Enzymatic degradation of TNT.TNT degradation can be done,through enzymatic process by nitroreductases involved in the yeast's NADPH dehydrogenase[16-19].Nitroreductases are present in bacteria and in some higher organisms such as plants,fungi,and animals.Nitroreductases are either Oxygen-insensitive Type I or Type II Hydride Transferases.
4.1.3.1.1.Oxygen-insensitive type I nitroreductases.Monomeric nitroreductases or homodimeric flavinmononucleotide(FMN)can be found in cell cytoplasm.Nitroreductases contain proteins with a subunit size of 25 kDa and take an electron from the NAD(P)H coenzyme[5].They are able to catalyze,by two electron increments,the reduction of aromatic nitro-groups to amino-groups.The process produces two intermediates(nitroso groups and hydroxylamine)that can interact easily with themselves or other organic molecules,such as proteins and humic acids,and finally produce polymeric compounds[59].A small group of these enzymes act over the aromatic ring by a hydride-ion addition to TNT and other nitroaromatics[60].Type I nitroreductases are present in Gram-negative bacteria:Enterobacter cloacae[61],Escherichia coli[62],Helicobacter pylori[63],Klebsiella sp.[64],Rhodobacter capsulatus[65],Salmonella enterica[66],Pseudomonas fluorescens[67],Pseudomonas pseudoalcaligenes[68],Pseudomonas putida[58,69],Selenomonas ruminatium[70],Vibrio fisherii[71],and Vibrio harveyi[72].Gram-negative bacteria are the best candidates for the microbial incorporation in selfcleaning explosive formulations since they are more tolerant to TNT than Gram-positive bacteria.The efficiency of microbial remediation of TNT is constrained bytwomajor factors:(1)lowrate of degradation hampering time-efficient clean-up of contaminants(particularly at high loading rates);and(2)the relatively low tolerance of microbes tochemical toxicity.Studies on the E.coli TNT tolerance have reported various multiple thresholds ranging from 66 to 200 mg.
4.1.3.1.2.Type II Hydride Transferases.The nitroreductase of Type II is a flavoprotein of the family of the(β/α)8barrel NAD(P)H dehydrogenase[5].The enzymatic catalysis can be identified by a double-displacement reaction (a temporary non-sequential mechanism),where an enzyme is changed into an intermediate form when the first substrate-to-product reaction occurs.It works in two-half reactions.First,the enzyme produces bound FMNH2by NAD(P)H reduction.Second,an oxidative half-reaction of the TNT reoxidizes the FMNH2to yield the formation of mono-and dihydride-Meisenheimer-complexes,by two possible pathways in mutual competition:1)nitro-reduction present on TNT and 2)the specific nucleophilic addition of hydride-ions to TNT.The nitrogroup reaction occurs faster because of the different redoxpotential,and in most the cases this reaction starts at the paranitro group[19,67,71].However,some reports suggested that ortho-derivate(2-ADNT)can be produced by some bacterial strains due to the gradients in substrate specificities of the degrading enzymes[9,73].TNT reduction by Clostridium acetobutylicum reduces TNT to its dihydroxylamino derivate[23];meanwhile enzymes from Raoultella terrigena strain HB use the nitro-group reduction pathway[74,75].R.Terrigena,HB transforms dinitrotoluene and nitrobenzenes.
4.1.3.1.3.Laccases.When whole cells are compared with cellfree extract for microbial transformation,it seems plausible that nitrophenolic compounds are substrates for the reducing enzymes.Also it can be inferred that these compounds do not pass the bacterial cell membrane,acting as metabolic inhibitors.On the other hand,nitrobenzenes could be good substrates either for whole cells or for cell extracts.Laccases(benzenediol:oxygen oxidoreductases EC 1.10.3.2)are multi-copper proteins that oxidizes various aromatic and non-aromatic compounds using oxygen for a radicalcatalyzed reaction mechanism.These enzymes are found in many bacteria and eukaryotes(fungi,insect and higher plants)[38],and catalyze the secondary transformations of TNT metabolites.Laccases use oxygen as the terminal electron acceptor apart from copper,and don't need co-factors or peroxide.TNT did not act as substrate for these oxidative enzymes,but after conversion by nitroreductases the reduced metabolites such as aminodinitrotoluenes (ADNT),azoxy-compounds,and diaminonitrotoluenes can be oxidized by laccase to polymeric products[75].The existence of phenolic compounds during enzymatic activity of laccases was very effective in immobilizing the typical TNT metabolites (ADNTs and 4,4′-dimethyl-3,3′,5,5′-tetranitro-azoxybenzene).The addition of phenolic compounds during the reductive conversion of TNT by the fungus Trametes modesta inhibited the accumulation of stable TNT metabolites by at least 92%[76].When laccase from Trametes villosa was supplemented to a solution containing 4-ADNTand TNT,only 30%of the 4-ADNTand none of the TNT was transformed.The same experiment was done in the presence of catechol,4-ADNT,and up to 80%of TNT was removed from the solution.This was attained at close to a neutral pH,which is helpful for treatment in natural environments[77].
To reduce TNT contamination,an advanced microbiological method was developed and involved a TNT-transforming Bacillus sp.Strain SF,whose spores were amalgamated into an explosive formulation containing TNTand ammonium nitrate[78].By adding water,this mixture and Bacillus cells propagated out instantaneously,and initiated TNTconversion even after a 5-year storage of the bio-explosive at room temperature[78].These self-cleaning explosive preparations open new opportunities for the use of specific TNT-transforming microorganisms,such as spores of Clostridium bifermentans KMR-1,which can be used as a relativelystable inoculant for TNT biodegradation[79].The opportunity to lyophilize a P.Putida strain in the presence of cryoprotectants was also investigated for the application of non-sporulating microorganisms into TNT-based explosive formulations[78].However,the survival of P.Putida cells was limited in the bioexplosive formulation,underlining the need to optimize the cryoprotective media and the lyophilization conditions[78].Summarizing,the development of self-cleaning explosive preparations using microbial catabolic capabilities has recently appeared as an attractive strategy to prevent further TNT contamination[5].
4.2.Other process
Saupe reported lab-scale efforts on the alkaline hydrolysis of TNTand mixtures of TNT derivatives[80].At a temperature of 80°C and initial pH 14 for 4 h,a complete deactivation of TNT and removal of 90%of DOC(dissolve organic carbon)was obtained.The authors suggest that alkaline hydrolysis before thermal and biological treatments works effectively as remediation for TNT[80].More recently,Mills et al.Found evidence that a Meisenheimer complex is formed during the TNT removal under alkaline conditions[81].In conclusion,different physical methods,such as treatment with activated carbon,incineration techniques,and alkaline hydrolysis present some disadvantages:(1)these treatments are very expensive;(2)require additional ex-situ treatment;(3)possible production of new chemical by-products or waste gas emissions.
Chemical processes,such as advanced oxidation,for the treatment of soils and waters contaminated with TNT have been extensively applied.Some of these processes are associated with the generation of different radical species,principally hydroxyl radicals.Other methods such as the use of H2O2,O3,and UV,Fenton's reagent,and ionizing radiation have been applied.Fentonbased reactions are effective to achieve TNT degradation in different aqueous matrices;however,they need to adjust the pH consistently during the Fenton oxidation process.Chemical treatment of TNT also present other disadvantages that limit its applicability and efficiency:scavenging of hydroxyl radicals by natural organic matter or halides and resistance to oxidation by some compounds such as perchlorinated compounds[1].
4.3.Future prospect of TNT removal through bioelectrochemical treatment
Microbial fuel cells(MFCs)are well studied platform[112-115]and they and more recently extended into variousBio-Electrochemical treatment(BET).BETs have evolved as a promising technology for waste remediation.In a bioelectrochemical system the electrochemically active microorganisms degrade a complex organic compound and transfer electrons from a reduced electron donor to an electrode,and finally to an oxidized electron acceptor generating power.Therefore,it can be presumed that coupling of bio-anode to a counter electrode(cathode)will have a positive in fluence on the overall wastewater treatment efficiency,which has to be tapped[82].During a BEToperation,the possibility ofintegratingdiversecomponents(biological,physical,and chemical)in anodic chambers could trigger multiple reactions,namely biochemical,physical,physicochemical,electrochemical,or oxidation.These can be classed as bioelectrochemical reactions resulting from the metabolic activity of a substrate and subsequent secondary reactions,[84].The anode chamber of a microbial fuel cell(MFC)resembles a conventional anaerobic bioreactor and mimics the function of a conventional electrochemical cell used for wastewater treatment in the degradation of organic matter and toxic/xenobiotic pollutants[83-85].Potential differences between anodic oxidation and cathodic reduction have positive in fluence on the pollutant removal in MFC.The in situ generated biopotential enhances the degradation of different pollutants in both the anode and cathode chambers.Due to the anodic oxidation potential,reactive species like ·OH and ·O2 are generated at the anode surface,which helps to break the complex chemical structures present in wastewater.Sometimes,pollutants themselves act as mediators in electron transfer;for example,elemental sulfur present in wastewater converts itself to sulfate in the MFC,which is easier for degradation.Biohazardous toxic compounds such as endocrinedisrupting estrogens can also be considered as mediator molecules in MFC[86].BET systems can be easily integrated with the TNT degradation.Further these BETs can be easily integrated with laccases through a laccases-based electrode system,[85].The advantage of BETs over chemical oxidation,physical,and biological processes,is their “environmentally-friendly”capacity as they transform pollutants from waste to energy.
5.Conclusions
The presence of TNT molecules in the environment creates the need to clean contaminated sites.A combined aerobic-anaerobic system can degrade and mineralize TNT to a larger extent than an individual process.The resultant molecules of this degradation can be further degraded by fungal and enzymatic systems.Some advanced microbial strategies are currently under development to minimize the danger of TNT contamination.Further BET can be easily developed and integrated with laccases through laccasesbased electrode systems to develop an effective biological treatment processes.
Con flicts of interest
The author declares no con flict of interests.The work reported in the manuscript is the original work of the authors and this manuscript has not been previously submitted to any journal.
Acknowledgement
The authors want to thank the Director of the School of Engineering and Science,Tecnolo′gico de Monterrey,Campus Monterrey for his encouragement and financial support.We acknowledge the financial support from CONACYT(Mexico)PhD scholarship No.309171 for CCZ,SNI-C fellowship to RC and MR,FR.Author also thanks Aavesh green sustainability solutions S.De R.L.De.C.V.for their support.
[1]Ayoub K,van Hullebusch ED,Cassir M,Bermond A.Application of advanced oxidation processes for TNT removal:a review.,.J Hazard Mater 2010;178:10-28.https://doi.org/10.1016/j.jhazmat.2010.02.042.
[2]Meagher RB.Plants tackle explosive contamination.Nat Biotechnol 2006;24:161-3.
[3]Gómez-Garde~nes J,Lotero L,Taraskin SN,P′erez-Reche FJ.Explosive contagion in networks.Sci Rep 2016;6,19767.
[4]Miura G.Plant toxicology:defusing the explosive.Nat Chem Biol 2015;11:829.
[5]Stenuit BA,Agathos SN.Microbial 2,4,6-trinitrotoluene degradation:could we learn from(bio)chemistry for bioremediation and vice versa?Appl MicrobiolBiotechnol2010;88:1043-64.https://doi.org/10.1007/s00253-010-2830-x.
[6]Anasonye F,Winquist E,R¨as¨anen M,Kontro J,Bj¨orkl¨of K,Vasilyeva G,et al.Bioremediation of TNT contaminated soil with fungi under laboratory and pilot scale conditions.Int Biodeterior Biodegrad 2015;105:7-12.https://doi.org/10.1016/j.ibiod.2015.08.003.
[7]Claus H.Biological remediation of explosive residues.Cham:Springer International Publishing;2014.https://doi.org/10.1007/978-3-319-01083-0.
[8]Rodgers JD,Bunce NJ.Treatment methods for the remediation of nitroaromatic explosives.Water Res 2001;35:2101-11.http://www.ncbi.nlm.nih.gov/pubmed/11358288.[Accessed 15 February 2016].
[9]Oh B-T,Shea PJ,Drijber RA,Vasilyeva GK,Sarath G.TNT biotransformation and detoxification by a Pseudomonas Aeruginosa strain.Biodegradation 2003;14:309-19.https://doi.org/10.1023/A:1025656325834.
[10]Erkelens M,Adetutu EM,Taha M,Tudararo-Aherobo L,Antiabong J,Provatas A,et al.Sustainable remediation-the application of bioremediated soil for use in the degradation of TNT chips.J Environ Manag 2012;110:69-76.https://doi.org/10.1016/j.jenvman.2012.05.022.
[11]Esteve-Nú~nez A,Caballero A,Ramos JL.Biological degradation of 2,4,6-trinitrotoluene.Microbiol Mol Biol Rev 2001;65:335-52.https://doi.org/10.1128/MMBR.65.3.335-352.2001.table of contents.
[12]McFarlan G,Sara,Yao.2,4,6-Trinitrotoluene degradation pathway.http://eawag-bbd.ethz.ch/tnt/tnt_map.html.[Accessed 9 March 2016].
[13]Hawari J.Biodegradation of RDX and HMX:from basic research to field application In:biodegradation of nitroaromatic by compounds and explosives.Biodegrad Nitroaromatic Compd Explos 2000:277-310.https://www.researchgate.net/publication/44068770_Biodegradation_of_RDX_and_HMX_from_basic_research_to_ field_application_In_biodegradation_of_nitroaromatic_by_compounds_and_explosives.[Accessed 20 April 2016].
[14]Koch C,Müller S,Harms H,Harnisch F.Microbiomes in bioenergy production:from analysis to management.Curr Opin Biotechnol 2014;27:65-72.https://doi.org/10.1016/j.copbio.2013.11.006.
[15]Bumpus JA,Tatarko M.Biodegradation of 2,4,6-trinitrotoluene by Phanerochaete chrysosporium:identification of initial degradation products and the discovery of a TNT metabolite that inhibits lignin peroxidases.Curr Microbiol 1994;28:185-90.https://doi.org/10.1007/BF01571063.
[16]French CE,Nicklin S,Bruce NC.Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase.Appl Environ Microbiol 1998;64:2864-8.http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=106784&tool=pmcentrez&rendertype=abstract.[Accessed 5April 2016].
[17]Klausmeier RE,Appleton JA,DuPre ES,Tenbarge K.The enzymology of trinitrotoluene reduction.Int Biodeterior Biodegrad 2001;48:67-73.https://doi.org/10.1016/S0964-8305(01)00067-1.
[18]Williams RE,Rathbone DA,Scrutton NS,Bruce NC.Biotransformation of explosives by the old yellow enzyme family of flavoproteins.Appl Environ Microbiol 2004;70:3566-74. https://doi.org/10.1128/AEM.70.6.3566-3574.2004.
[19]Kim H-Y,Song H-G.Purification and characterization of NAD(P)H-dependent nitroreductase I from Klebsiella sp.C1 and enzymatic transformation of 2,4,6-trinitrotoluene.Appl Microbiol Biotechnol 2005;68:766-73.https://doi.org/10.1007/s00253-005-1950-1.
[20]Whitacre DM,editor.Reviews of environmental contamination and toxicology|David M.Whitacre.Springer;2012.http://www.springer.com/gp/book/9781461414629.
[21]Montgomery MT,Cof fin RB,Boyd TJ,Smith JP,Walker SE,Osburn CL.2,4,6-Trinitrotoluene mineralization and bacterial production rates of natural microbial assemblages from coastal sediments.Environ Pollut 2011;159:3673-80.https://doi.org/10.1016/j.envpol.2011.07.018.
[22]U.S Government Accountability Of fice(GAO).U.S.GAO-military munitions:DOD needs to develop a comprehensive approach for cleaning up contaminated sites,December 19.2003.http://www.gao.gov/products/GAO-04-147.[Accessed 20 April 2016].
[23]Symons ZC,Bruce NC.Bacterial pathways for degradation of nitroaromatics.Nat Prod Rep 2006;23:845-50.https://doi.org/10.1039/b502796a.
[24]Van Aken B,Skubisz K,Naveau H,et al.Biodegradation of 2,4,6-trinitrotoluene(TNT)by the white-rot basidiomycete Phlebia radiata.Biotechnol Lett 1997;19:813.https://doi.org/10.1023/A:1018360814703.
[25]George I,Eyers L,Stenuit B,Agathos SN.Effect of 2,4,6-trinitrotoluene on soil bacterial communities.J Ind Microbiol Biotechnol 2008;35:225-36.https://doi.org/10.1007/s10295-007-0289-2.
[26]Lewis TA,Newcombe DA,Crawford RL.Bioremediation of soils contaminated with explosives.JEnviron Manag 2004;70:291-307.https://doi.org/10.1016/j.jenvman.2003.12.005.
[27]Travis ER,Bruce NC,Rosser SJ.Short term exposure to elevated trinitrotoluene concentrations induced structural and functional changes in the soil bacterialcommunity.Environ Pollut2008;153:432-9.https://doi.org/10.1016/j.envpol.2007.08.006.
[28]Travis ER,Bruce NC,Rosser SJ.Microbial and plant ecology of a long-term TNT-contaminated site.Environ Pollut 2008;153:119-26.https://doi.org/10.1016/j.envpol.2007.07.015.
[29]Pennington JC,Brannon JM.Environmental fate of explosives.Thermochim Acta 2002;384:163-72.https://doi.org/10.1016/S0040-6031(01)00801-2.
[30]Lee B,Sheng-Yih,Brodman.Biodegradation of 1,3,5-Trinitro-1,3,5-triazine(RDX).J Environ Sci Heal 2004;A39(No.1):61-75.http://www.tandfonline.com/doi/pdf/10.1081/ESE-120027368.[Accessed 20 April 2016].
[31]Snellinx Z,Nepovím A,Taghavi S,Vangronsveld J,Vanek T,van der Lelie D.Biological remediation of explosives and related nitroaromatic compounds.Environ Sci Pollut Res 2002;9:48-61.https://doi.org/10.1007/BF02987316.
[32]Kim H-Y,Song H-G.Transformation and mineralization of2,4,6-trinitrotoluene by the white rot fungus Irpex lacteus.Appl Microbiol Biotechnol 2003;61:150-6.https://doi.org/10.1007/s00253-002-1211-5.
[33]Martin JL,Comfort SD,Shea PJ,Drijber RA,Kokjohn TA.Denitration of 2,4,6-trinitrotoluene by Pseudomonas savastanoi.Can J Microbiol 1997;43:447-55.https://doi.org/10.1139/m97-063.
[34]Vorbeck C,Lenke H,Fischer P,Knackmuss HJ.Identification of a hydride-Meisenheimer complex as a metabolite of 2,4,6-trinitrotoluene by a Mycobacterium strain.J Bacteriol 1994;176:932-4.http://jb.asm.org/content/176/3/932.abstract.[Accessed 29 March 2016].
[35]Boopathy R,Kulpa CF.Biotransformation of 2,4,6-trinitrotoluene(TNT)by a Methanococcus sp.(strain B)isolated from a lake sediment.Can J Microbiol 1994;40:273-8.http://www.ncbi.nlm.nih.gov/pubmed/8039051.[Accessed 5 April 2016].
[36]Crawford R.The microbiology and treatment of nitroaromatic compounds.Curr Opin Biotechnol 1995;6:329-36. https://doi.org/10.1016/0958-1669(95)80055-7.
[37]Ederer MM,Lewis TA,Crawford RL.2,4,6-Trinitrotoluene(TNT)transformation by clostridia isolated from a munition-fed bioreactor:comparison with non-adapted bacteria.J Ind Microbiol Biotechnol 1997;18:82-8.https://doi.org/10.1038/sj.jim.2900257.
[38]Claus H.Microbial degradation of 2,4,6-trinitrotoluene in vitro and in natural environments.Biol Remediat Explos Residues 2014.http://www.gao.gov/new.items/d04147.pdf.[Accessed 20 April 2016].
[39]Ederer MM,Lewis TA,Crawford RL.2,4,6-Trinitrotoluene(TNT)transformation by clostridia isolated from a munition-fed bioreactor:comparison with non-adapted bacteria.J Ind Microbiol Biotechnol 18:82-88.http://www.ncbi.nlm.nih.gov/pubmed/9134759.[Accessed 1 April 2016].
[40]PreussA,FimpelJ,DiekertG.Anaerobictransformation of2,4,6-trinitrotoluene(TNT).Arch Microbiol 1993;159:345-53.https://doi.org/10.1007/BF00290917.
[41]Lewis TA,Ederer MM,Crawford RL,Crawford DL.Microbial transformation of 2,4,6-trinitrotoluene.J Ind Microbiol Biotechnol 1997;18:89-96.https://doi.org/10.1038/sj.jim.2900258.
[42]Kalderis D,Juhasz AL,Boopathy R,Comfort S.Soils contaminated with explosives:environmental fate and evaluation of state-of-the-art remediation processes(IUPAC Technical Report).Pure Appl Chem 2011;83:1407-84.https://doi.org/10.1351/PAC-REP-10-01-05.
[43]M.W,Ramaraj Boopathy CFK.Anaerobic removal of 2,4,6-trinitrotoluene(TNT)under different electron accepting conditions:laboratory study on JSTOR.In:Anaerob.Remov.2,4,6-Trinitrotoluene under differ.Electron accept.Cond.Lab.Study,water environment federation;1993.p.271-5.http://www.jstor.org/stable/25044298?seq=1#page_scan_tab_contents.[Accessed 24 April 2016].
[44]Ha¨Idour A,Ramos JL.Identification of products resulting from the biological reduction of 2,4,6-trinitrotoluene,2,4-dinitrotoluene,and 2,6-dinitrotoluene by Pseudomonas sp.Environ Sci Technol 1996;30:2365-70.https://doi.org/10.1021/es950824u.
[45]Advances in applied microbiology.Adv Appl Microbiol 2016;94.https://doi.org/10.1016/S0065-2164(16)30004-1.
[46]Arora D.Fungal biotechnology in agricultural,food,and environmental applications.CRC Press Book;2003.https://www.crcpress.com/Fungal-Biotechnology-in-Agricultural-Food-and-Environmental-Applications/Arora/9780824747701.[Accessed 20 April 2016].
[47]Singh SN,editor.Biological remediation of explosive residues.Cham:Springer International Publishing;2014.https://doi.org/10.1007/978-3-319-01083-0.
[48]Spain JC.Biodegradation of nitroaromatic compounds.Annu Rev Microbiol 1995;49:523-55.https://doi.org/10.1146/annurev.mi.49.100195.002515.
[49]Hodgson J,Rho D,Guiot SR,Ampleman G,Thiboutot S,Hawari J.Tween 80 enhanced TNT mineralization by Phanerochaete chrysosporium.Can J Microbiol 2000;46:110-8.http://www.ncbi.nlm.nih.gov/pubmed/10721478.[Accessed 24 April 2016].
[50]Kim HY,Song HG.Comparison of 2,4,6-trinitrotoluene degradation by seven strains of white rot fungi.Curr Microbiol 2000;41:317-20.http://www.ncbi.nlm.nih.gov/pubmed/11014867.[Accessed 24 April 2016].
[51]Ziganshin AM,Ziganshina EE,Byrne J,Gerlach R,Struve E,Biktagirov T,et al.Fe(III)mineral reduction followed by partial dissolution and reactive oxygen species generation during 2,4,6-trinitrotoluene transformation by the aerobicyeastYarrowialipolytica.AMB Express2015;5:8.https://doi.org/10.1186/s13568-014-0094-z.
[52]Khilyas IV,Ziganshin AM,Pannier AJ,Gerlach R.Effect of ferrihydrite on 2,4,6-trinitrotoluene biotransformation by an aerobic yeast.Biodegradation 2013;24:631-44.https://doi.org/10.1007/s10532-012-9611-4.
[53]Ziganshin AM,Naumova RP,Pannier AJ,Gerlach R.In fluence of pH on 2,4,6-trinitrotoluene degradation by Yarrowia lipolytica.Chemosphere 2010;79:426-33.https://doi.org/10.1016/j.chemosphere.2010.01.051.
[54]Ziganshin AM,Gerlach R,Naumenko EA,Naumova RP.Aerobic degradation of 2,4,6-trinitrotoluene by the yeast strain Geotrichum candidum AN-Z4.Microbiology 2010;79:178-83. https://doi.org/10.1134/S0026261710020086.
[55]Solyanikova IP,Baskunov BP,Baboshin MA,Saralov AI,Golovleva LA.Detoxification of high concentrations of trinitrotoluene by bacteria.Appl Biochem Microbiol 2011;48:21-7. https://doi.org/10.1134/S0003683812010152.
[56]Duque E,Haidour A,Godoy F,Ramos JL.Construction of a Pseudomonas hybrid strain that mineralizes 2,4,6-trinitrotoluene.J Bacteriol 1993;175:2278-83. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=204515&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[57]Wittich R-M,Ramos JL,van Dillewijn P.Microorganisms and explosives:mechanisms of nitrogen release from TNT for use as an N-Source for growth.Environ Sci Technol 2009;43:2773-6.https://doi.org/10.1021/es803372n.
[58]Caballero A,Lázaro JJ,Ramos JL,Esteve-Nú~nez A.PnrA,a new nitroreductasefamily enzyme in the TNT-degrading strain Pseudomonas putida JLR11.Environ Microbiol 2005;7:1211-9. https://doi.org/10.1111/j.1462-2920.2005.00801.x.
[59]Sarlauskas J,Nemeikaite-Ceniene A,Anusevicius Z,Miseviciene L,Julvez MM,Medina M,et al.Flavoenzyme-catalyzed redox cycling of hydroxylaminoand amino metabolites of 2,4,6-trinitrotoluene:implications for their cytotoxicity.Arch Biochem Biophys 2004;425:184-92.https://doi.org/10.1016/j.abb.2004.02.043.
[60]Ramos JL,González-P′erez MM,Caballero A,van Dillewijn P.Bioremediation of polynitrated aromatic compounds:plants and microbes put up a fight.Curr Opin Biotechnol 2005;16:275-81. https://doi.org/10.1016/j.copbio.2005.03.010.
[61]Haynes CA,Koder RL,Miller A-F,Rodgers DW.Structures of nitroreductase in three states:effects of inhibitor binding and reduction.J Biol Chem 2002;277:11513-20.https://doi.org/10.1074/jbc.M111334200.
[62]Whiteway J,Koziarz P,Veall J,Sandhu N,Kumar P,Hoecher B,et al.Oxygeninsensitive nitroreductases:analysis of the roles of nfsA and nfsB in developmentofresistance to 5-nitrofuran derivativesin Escherichiacoli.J Bacteriol 1998;180:5529-39.http://jb.asm.org/content/180/21/5529.full.[Accessed 24 April 2016].
[63]Goodwin A,Kersulyte D,Sisson G,Veldhuyzen van Zanten SJ,Berg DE,Hoffman PS.Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene(rdxA)that encodes an oxygen-insensitive NADPH nitroreductase.Mol Microbiol 1998;28:383-93.http://www.ncbi.nlm.nih.gov/pubmed/9622362.[Accessed 24 April 2016].
[64]Shin J-H,Song H-G.Nitroreductase II involved in 2,4,6-trinitrotoluene degradation:purification and characterization from Klebsiellasp.Cl.J Microbiol 2009;47:536-41.https://doi.org/10.1007/s12275-008-0171-6.
[65]Blasco R,Castillo F.Characterization of a nitrophenol reductase from the phototrophic bacterium Rhodobacter capsulatus E1F1.Appl Environ Microbiol 1993;59:1774-8. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=182160&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[66]Nokhbeh MR,Boroumandi S,Pokorny N,Koziarz P,Paterson ES,Lambert IB.Identification and characterization of SnrA,an inducible oxygen-insensitive nitroreductase in Salmonella enterica serovar Typhimurium TA1535.Mutat Res 2002;508:59-70. http://www.ncbi.nlm.nih.gov/pubmed/12379462.[Accessed 24 April 2016].
[67]Pak JW,Knoke KL,Noguera DR,Fox BG,Chambliss GH.Transformation of 2,4,6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C.Appl Environ Microbiol 2000;66:4742-50.http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=92374&tool=pmcentrez&rendertype=abstract.[Accessed 24April 2016].
[68]Somerville CC,Nishino SF,Spain JC.Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45.J Bacteriol 1995;177:3837-42. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=177104&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[69]Caballero A,Esteve-Nú~nez A,Zylstra GJ,Ramos JL.Assimilation of nitrogen from nitrite and trinitrotoluene in Pseudomonas putida JLR11.J Bacteriol 2005;187:396-9.https://doi.org/10.1128/JB.187.1.396-399.2005.
[70]Anderson PJ,Cole LJ,McKay DB,Entsch B.A flavoprotein encoded in Selenomonas ruminantium is characterized after expression in Escherichia coli.Protein Expr Purif 2002;24:429-38.https://doi.org/10.1006/prep.2001.1581.
[71]Rie fler RG,Smets BF.NAD(P)H: flavin mononucleotide oxidoreductase inactivation during 2,4,6-trinitrotoluene reduction.Appl Environ Microbiol 2002;68:1690-6.https://doi.org/10.1128/AEM.68.4.1690-1696.2002.
[72]Lei B,Liu M,Huang S,Tu SC.Vibrio harveyi NADPH- flavin oxidoreductase:cloning,sequencing and overexpression of the gene and purification and characterization of the cloned enzyme.J Bacteriol 1994;176:3552-8.http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=205543&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[73]K.K,Toshinari HO,Maed.Characterization of 2,4,6-trinitrotoluene(TNT)-metabolizing bacteria isolated from TNT-polluted soils in the Yamada Green Zone,Kitakyushu,Japan.J Environ Biotechnol 2006;6:33-9.http://www.jseb.jp/jeb/06-01/06-01-033.pdf.[Accessed 24 April 2016].
[74]Claus H,Bausinger T,Lehmler I,Perret N,Fels G,Dehner U,et al.Transformation of 2,4,6-trinitrotoluene(TNT)by Raoultella terrigena.Biodegradation 2007;18:27-35.https://doi.org/10.1007/s10532-005-9033-7.
[75]Claus H,Perret N,Bausinger T,Fels G,Preuss J,K¨onig H.TNT transformation products are affected by the growth conditions of Raoultella terrigena.Biotechnol Lett 2007;29:411-9.https://doi.org/10.1007/s10529-006-9244-y.
[76]Nyanhongo GS,Couto SR,Guebitz GM.Coupling of 2,4,6-trinitrotoluene(TNT)metabolites onto humic monomers by a new laccase from Trametes modesta. Chemosphere 2006;64:359-70. https://doi.org/10.1016/j.chemosphere.2005.12.034.
[77]Wang C-J,Thiele S,Bollag J-M.Interaction of 2,4,6-trinitrotoluene(TNT)and 4-amino-2,6-dinitrotoluene with humic monomers in the presence of oxidative enzymes.Arch Environ Contam Toxicol 2002;42:1-8.https://doi.org/10.1007/s002440010284.
[78]Nyanhongo GS,Aichernig N,Ortner M,Steiner W,Guebitz GM.Incorporation of 2,4,6-trinitrotoluene(TNT)transforming bacteria into explosive formulations. J Hazard Mater 2009;165:285-90. https://doi.org/10.1016/j.jhazmat.2008.09.107.
[79]Sembries S,Crawford RL.Production of Clostridium bifermentans spores as inoculum for bioremediation of nitroaromatic contaminants.Appl Environ Microbiol 1997;63:2100-4. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1389175&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[80]Saupe A,Garvens HJ,Heinze L.Alkaline hydrolysis of TNT and TNT in soil followed by thermal treatment of the hydrolysates.Chemosphere 1998;36:1725-44.https://doi.org/10.1016/S0045-6535(97)10063-7.
[81]Mills A,Seth A,Peters G.Alkaline hydrolysis of trinitrotoluene,TNT.Phys Chem Chem Phys 2003;5:3921-7.https://doi.org/10.1039/b304616h.
[82]Mohan SV,Chandrasekhar K.Self-induced bio-potential and graphite electron accepting conditions enhances petroleum sludge degradation in bioelectrochemical system with simultaneous power generation.Bioresour Technol 2011;102:9532-41.https://doi.org/10.1016/j.biortech.2011.07.038.
[83]Katuri KP,Venkata Mohan S,Sridhar S,Pati BR,Sarma PN.Laccase-membrane reactors for decolorization of an acid azo dye in aqueous phase:process optimization.Water Res 2009;43:3647-58.https://doi.org/10.1016/j.watres.2009.05.028.
[84]Mohan SV,Raghavulu SV,Peri D,Sarma PN.Integrated function of microbial fuel cell(MFC)as bio-electrochemical treatment system associated with bioelectricity generation under higher substrate load.Biosens Bioelectron 2009;24:2021-7.https://doi.org/10.1016/j.bios.2008.10.011.
[85]Venkata Mohan S,Falkentoft C,Venkata Nancharaiah Y,Sturm BSM,Wattiau P,Wilderer PA,et al.Bioaugmentation of microbial communities in laboratory and pilot scale sequencing batch bio film reactors using the TOL plasmid.Bioresour Technol 2009;100:1746-53.https://doi.org/10.1016/j.biortech.2008.09.048.
[86]Kumar AK,Reddy MV,Chandrasekhar K,Srikanth S,Mohan SV.Endocrine disruptive estrogens role in electron transfer:bio-electrochemical remediation with microbial mediated electrogenesis.Bioresour Technol 2012;104:547-56.https://doi.org/10.1016/j.biortech.2011.10.037.
[87]Kalafut T,Wales ME,Rastogi VK,Naumova RP,Zaripova SK,Wild JR.Biotransformation patterns of 2,4,6-trinitrotoluene by aerobic bacteria.Curr Microbiol 1998;36:45-54.http://www.ncbi.nlm.nih.gov/pubmed/9405746.[Accessed 24 April 2016].
[88]Vanderberg LA,Perry JJ,Unkefer PJ.Catabolism of 2,4,6-trinitrotoluene by Mycobacterium vaccae.Appl Microbiol Biotechnol 1995;43:937-45.http://www.ncbi.nlm.nih.gov/pubmed/7576561.[Accessed 24 April 2016].
[89]Vorbeck C,Lenke H,Fischer P,Spain JC,Knackmuss HJ.Initial reductive reactions in aerobic microbial metabolism of 2,4,6-trinitrotoluene.Appl Environ Microbiol 1998;64:246-52. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=124701&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[90]Van Aken B,Yoon JM,Schnoor JL.Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene,hexahydro-1,3,5-trinitro-1,3,5-triazine,and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp.associated with poplar tissues(Populus deltoides x).Appl Environ Microbiol 2004;70:508-17.http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=321275&tool=pmcentrez&rendertype=abstract.[Accessed 5 April 2016].
[91]Alvarez MA,Kitts CL,Botsford JL,Unkefer PJ.Pseudomonas aeruginosa strain MA01 aerobically metabolizes the aminodinitrotoluenes produced by 2,4,6-trinitrotoluene nitro group reduction.Can J Microbiol 1995;41:984-91.http://www.ncbi.nlm.nih.gov/pubmed/7497356.[Accessed 24 April 2016].
[92]Gilcrease PC,Murphy VG.Bioconversion of 2,4-diamino-6-nitrotoluene to a novel metabolite under anoxic and aerobic conditions.Appl Environ Microbiol 1995;61:4209-14. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=167732&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[93]Fiorella PD, Spain JC. Transformation of 2,4,6-trinitrotoluene by Pseudomonas pseudoalcaligenes JS52.Appl Environ Microbiol 1997;63:2007-15. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1389165&tool=pmcentrez&rendertype=abstract.[Accessed 24 April 2016].
[94]Schackmann A,Müller R.Reduction of nitroaromatic compounds by different Pseudomonas species under aerobic conditions.Appl Microbiol Biotechnol 1991;34.https://doi.org/10.1007/BF00169355.
[95]McCormick NG,Feeherry FE,Levinson HS.Microbial transformation of 2,4,6-trinitrotoluene and other nitroaromatic compounds.Appl Environ Microbiol 1976;31:949-58. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=169861&tool=pmcentrez&rendertype=abstract.[Accessed 5 April 2016].
[96]Jones AM,Greer CW,Hawari J,Ampleman G,Lavigne J.Biodegradability of selected highly energetic pollutants under aerobic conditions.1995.http://www.osti.gov/scitech/biblio/490985-biodegradability-selected-highly-energetic-pollutants-under-aerobic-conditions.[Accessed 24 April 2016].
[97]Montpas S,Samson J,Langlois′E,Lei J,Pich′e Y,Ch^enevert R.Degradation of 2,4,6-trinitrotoluene by Serratia marcescens.Biotechnol Lett 1997;19:291-4.https://doi.org/10.1023/A:1018326228448.
[98]Cai X,Servinsky M,Kiel J,Sund C,Bennett GN.Analysis of redox responses during TNT transformation by Clostridium acetobutylicum ATCC 824 and mutants exhibiting altered metabolism.Appl Microbiol Biotechnol 2013;97:4651-63.https://doi.org/10.1007/s00253-012-4253-3.
[99]Khan TA,Bhadra R,Hughes J.Anaerobic transformation of 2,4,6-TNT and related nitroaromatic compounds by Clostridium acetobutylicum.J Ind Microbiol Biotechnol 1997;18:198-203. https://doi.org/10.1038/sj.jim.2900328.
[100]Shin CY,Crawford DL.Biodegradation of trinitrotoluene(TNT)by a strain of Clostridium bifermentans. http://www.osti.gov/scitech/biblio/474237.[Accessed 24 April 2016].
[101]Hughes JB,Wang CY,Bhadra R,Richardson A,Bennett GN,Rudolph FB.Reduction of 2,4,6-trinitrotoluene by Clostridium acetobutylicum through hydroxylamino-nitrotoluene intermediates.Environ Toxicol Chem 1998;17:343-8.https://doi.org/10.1002/etc.5620170301.
[102]Boopathy R,Kulpa CF.Trinitrotoluene(TNT)as a sole nitrogen source for a sulfate-reducing bacterium Desulfovibrio sp.(B strain)isolated from an anaerobic digester.Curr Microbiol 1992;25:235-41.http://www.ncbi.nlm.nih.gov/pubmed/1368976.[Accessed 24 April 2016].
[103]PreussA,FimpelJ,DiekertG.Anaerobictransformation of2,4,6-trinitrotoluene(TNT).Arch Microbiol 1993;159:345-53.https://doi.org/10.1007/BF00290917.
[104]Drzyzga O,Bruns-Nagel D,Gorontzy T,Blotevogel KH,Gemsa D,von L¨ow E.Mass balance studies with 14C-labeled 2,4,6-trinitrotoluene(TNT)mediated by an Anaerobic desulfovibrio species and an Aerobic serratia species.Curr Microbiol 1998;37:380-6.http://www.ncbi.nlm.nih.gov/pubmed/9806975.[Accessed 24 April 2016].
[105]Bayman P,Radkar GV.Transformation and tolerance of TNT(2,4,6-trinitrotoluene)by fungi.IntBiodeterior Biodegrad 1997;39:45-53.https://doi.org/10.1016/S0964-8305(96)00066-2.
[106]HoehamerCF,Wolfe NL,Eriksson KEL.Biotransformation of2,4,6-trinitrotoluene(TNT)by the fungus Fusarium oxysporum.Int J Phytoremediation 2006;8:95-105.https://doi.org/10.1080/15226510600678423.
[107]Weber RWS,Ridderbusch DC,Anke H.2,4,6-Trinitrotoluene(TNT)tolerance and biotransformation potentialofmicrofungiisolated from TNT-contaminated soil.Mycol Res 2002;106:336-44.https://doi.org/10.1017/S0953756202005609.
[108]Scheibner K,Hofrichter M,Herre A,Michels J,Fritsche W.Screening for fungi intensively mineralizing 2,4,6-trinitrotoluene.Appl Microbiol Biotechnol 1997;47:452-7.http://www.ncbi.nlm.nih.gov/pubmed/9163958.[Accessed 24 April 2016].
[109]Scheibner K,Hofrichter M,Fritsche W.Mineralization of 2-amino-4,6-dinitrotoluene by manganese peroxidase of the white-rot fungus Nematoloma frowardii.Biotechnol Lett 1997;19:835.https://doi.org/10.1023/A:1018369116521.
[110]Scheibner K,Hofrichter M.Conversion of aminonitrotoluenes by fungal manganese peroxidase.J Basic Microbiol 1998;38:51-9.http://www.ncbi.nlm.nih.gov/pubmed/9575043.[Accessed 24 April 2016].
[111]Van Aken B,Hofrichter M,Scheibner K,Hatakka AI,Naveau H,Agathos SN.Transformation and mineralization of 2,4,6-trinitrotoluene(TNT)by manganese peroxidase from the white-rot basidiomycete Phlebia radiata.Biodegradation 1999;10:83-91. http://www.ncbi.nlm.nih.gov/pubmed/10466197.[Accessed 24 April 2016].
[112]Chandra R,Annie Modestra J,Venkata Mohan S.Biophotovoltaic cell to harness bioelectricity from acidogenic wastewater associated with Microbial Community Pro filing.Fuel 2015;160:502-12.
[113]Chandra R,Sravan JS,Hemalatha M,Kishore Butti S,Venkata Mohan S.Photosynthetic synergism for sustained power production with microalgae and photobacteria in a biophotovoltaic cell.Energy Fuels 2017.https://doi.org/10.1021/acs.energyfuels.7b00486.
[114]Chandra R,Venkata Subhash G,Venkata Mohan S.Mixotrophic operation of photo-bioelectrocatalytic fuel cell under anoxygenic microenvironment enhances the light dependent bioelectrogenic activity.Bioresour.Technol.2012;109:46-56.
[115]Venkata Mohan S,Mohanakrishna G,Velvizhi G,Babu VL,Sarma PN.Biocatalyzed electrochemical treatment of real field dairy wastewater with simultaneous power generation.Biochem.Eng.J.2010;51:32-9.
- Defence Technology的其它文章
- Evaluating location specific strain rates,temperatures,and accumulated strains in friction welds through microstructure modeling
- Crystal lattice free volume in a study of initiation reactivity of nitramines:Impact sensitivity
- The effects of compressibility and strength on penetration of long rod and jet
- A DFT study on TNGU isomers and aluminized cis-TNGU composites
- Numerical simulation and optimized design of cased telescoped ammunition interior ballistic
- The kinetic of mass loss of grades A and B of melted TNT by isothermal and non-isothermal gravimetric methods

