Genetic variation in relation to adaptability of three mangrove species from the Indian Sundarbans assessed with RAPD and ISSR markers
Nirjhar Dasgupta•Paramita Nandy•Chandan Sengupta•Sauren Das
Introduction
Mangroves form a diverse plant community that thrive in tropical and subtropical estuaries.The main attribute of mangroves is in their ability to live under constant physiological stress(Chaudhuri 1996)through morphological,reproductive and physiological adaptations(Zimmermann 1983;Das 1999).The most important ability of mangroves is their tolerance of seawater levels of NaCl(~500 mM)(Takemura et al.2000;Yan and Guizhu 2007;Hogarth 2007).The coastal environment and inhabitants are largely dependent on mangrove forests for its enormously productive and protective ecosystem.Barbier(2007)opined that the economic annual value of mangrove forest is around US$12,392 ha-1a-1,when we include all production and protection by coastal and estuarine areas.
However,mangrove forest areas all over the world have confronted a major threat as the climate(precipitation and temperature),sea level and incidence of UV-β radiation are clearly changing(Spalding et al.1997;Spiers 1999).Mangrove ecosystems are the front-line defense against the adverse consequences of changes in sea level and also can absorb a considerable amount of UV radiation(Hiraishi and Harada 2003;Hogarth 2007).
In the Indian subcontinent,the Sundarbans delta is a mangrove swamp of approximately 2152 km2(FSI 2009)with the world’s richest species diversity.This mangrove forest is cradle of 36 true mangrove species,28 associates and 7 obligatory mangrove species belonging to 29 families and 49 genera(Naskar and GuhaBakshi 1983).Chanda and Datta(1986)commented that a disparity in the salinity level prevails between the western and eastern zones of the Sundarbans;freshwater in fl ux in the west(Indian Sundarbans)is much lower;thus,the salinity is higher than in the east(Bangladesh Sundarbans).The cumulative effects of the geomorphology,demographic interference,siltation in the riverbed and decrease in freshwater in fl ux(Hopson and Webster 2010;Chowdhury and Ward 2007)have been detrimental for some economically important plant species includingHeritiera fomes,Nypa fruticans,Xylocarpus mekongensisandX.granatumin the Indian Sundarbans(IUCN Red List 2014;Upadhyay et al.2002).
Genetic diversity has a major effect on biodiversity(Templeton 1993).Schaal et al.(1991)commented that genetic diversity is the most important criteria for any species to cope with changing environments or evolving competitors and parasites.Molecular markers can offer vital information to develop strategies for genetic conservation(Newbury and Ford-Lloyd 1993).The study of genetic diversity is a prerequisite to design the most suitable strategy for a conservation program.In the present study,the genetic polymorphisms of three true mangrove species have been studied by RAPD and ISSR markers.Excoecaria agallochaandPhoenix paludosagrow profusely in the Indian part of the Sundarbans forest,and the other,Xylocarpus granatum,is on the verge of extinction.In the background of the high salinity in Indian Sundarbans(Dasgupta et al.2010),this study aims to assess the sustainability of the three taxa in respect to their genetic diversity.Although genetic polymorphisms inAvicennia(Parani et al.1997;Schwarzbach and McDade 2002;Kader et al.2012)and Rhizophoraceae(Sheue et al.2009;Gurudeevan et al.2012)have been studied over the years,very limited work has been done on the three taxa presently studied in this paper.Parida et al.(1995)reported a high level of genetic polymorphism amongE.agallocha.Das and Jena(2008)reported eight ecotypes ofX.granatumin the Bhitarkanika mangrove forest,using RAPD markers.But so far there has been no report on the genetic polymorphism ofX.granatum,E.agallochaandP.paludosafrom the Indian Sundarbans.The present study on the comparative basis of genetic polymorphism of these three taxa might be helpful on understanding their current status in the Indian Sundarbans and inform strategies to conserve these taxa.
Materials and methods
Collection sites
LeafbudsofX.granatum,E.agallocha,andP.paludosawere collected from fi ve naturally occurring islands in the Sundarbans mangrove swamps.All sites are included in the core region,at the northeastern part of the western Sundarbans(Indian part)to the southern extremity toward the sea(Bay of Bengal),without any demographic interference,namely Sajnakhali (22°07′26′N, 88°49′51′E), Sudhanyakhali(22°06′05′N, 88°48′06′E), Sonargaon (22°06′29′N,88°46′23′E),Jharkhali(22°01′13′N,88°41′10′E)and Dobanki(21°59′04′N,88°44′38′E)(Fig.1).The substrate salinityof the collection sites occurred an increasing gradient(11.77±2.1,12.25±1.96,12.4±1.18,13.98±2.29 and 15.23±2.16 mg kg-1)(Dasguptaetal.2012).Fromeachof the sites mentioned,samples were collected from at least six plants10to30 mapart.Thecontrolledmesophyticreplicasof the studied species,well-matured to 15–17 years old,collected from the premises of the Indian Statistical Institute,Kolkata,where the soil salinity was 2 mg kg-1,(Dasgupta et al.2012)were also taken for a comparative study.
Isolation of genomic DNA
Young leaf buds were used to isolate total genomic DNA for all plant samples.The desired amount of leaf bud was weighed,washed with double-distilled water.Qiagen DNeasy Plant Mini Kits were used to extract DNA from these samples after they were aseptically ground in liquid nitrogen with a mortar and pestle.
RAPD and ISSR analysis
For RAPD and ISSR analysis,20 ng genomic DNA was used for ampli fi cation in a Primus 25 Advanced Thermocycler(Peqlab,Erlangen,Germany);26 RAPD primers and 20 ISSR primers were screened to assess the genetic diversity of the plant species based on better reproducibility(Tables 1,2).The 25 μL reaction mixture contained 1.5 μL 10×reaction buffer,25 mM MgCl2,20 pM RAPD primer,2 mM dNTPs,1 U Taq polymerase(Bioline,Taunton,MA,USA)and Millipore water to volume.The DNA in the reaction mixture was subjected to denaturation at 94°C for 5 min;45 cycles of denaturation for 45 s,primer annealing for 1 min and primer extension for 1.30 min at 72°C.The chemical concentrations and PCR condition were same for RAPD and ISSR analysis except for primer annealing.The temperature for primer annealing was adjusted based on the melting temperature(Tm)of the individual RAPD and ISSR primers and varied from 38 to 42°C for RAPDs and 50–54 °C for ISSRs.One last cycle of primer extension at 72°C for 7 min was followed by a temperature drop of 10°C.PCR products were separated by electrophoresis using 1.5% agarose gel containing ethidium bromide(0.5 μg/mL)in Tris–borate-EDTA(TBE)buffer.The sizes of the amplicons were determined using size standards(Hyper Ladder II,Bioline).DNA bands were visualized with a UV transilluminator and photographed for later analysis using various statistical programs.The PCR experiment was done thrice for each experiment.

Fig.1 Location of the study area
Statistical analyses
In RAPD and ISSR analysis,the presence or absence of the bands were scored as 1 or 0,respectively,to create a binary matrix using the multivariate analysis program NTSYS-PC(Rohlf 1993).Jaccard’s similarly matrix was constructed using the binary matrix and Jaccard’s coef ficient(Jaccard 1998).Following the unweighted pair group with arithmetic mean(UPGMA)method(Sneath and Sokal1973),a phylogenetic dendrogram was obtained from the similarity matrix by cluster analysis,using NTSYS version 2.1(Exeter Software,NY,NY,USA).POPGENE 32(Yeh et al.1997)statistical software was also used for calculating mean of observed number of alleles per loci(Na),mean effective number of allele per loci(Ne),Nei’s gene diversity value(h)(Nei 1973),and Shannon’s information index(I).

Table 1 List of RAPD primers and bands ampli fi ed using DNA from Excoecaria agallocha(Ea),Phoenix paludosa(Pp),and Xylocarpus granatum(Xg)

Table 2 Statistical analysis of DNA polymorphism by RAPD and ISSR molecular markers for the three mangrove species
Results
RAPD analysis
The 26 primers used for this study produced 223 bands inE.agallocha,of which 55 were polymorphic.Seven primers produced relatively more polymorphic bands:OPA3,OPA5,OPA 7,OPA12,OPA 15,OPA 19 and RAPD 22(Table 1).Of 223 bands generated forP.paludosa,55 were polymorphic(24.66%polymorphism)(Table 2).The total number of bands ranged between 7 and 11 per primer.P.paludosaproduced 52 polymorphic bands among 197 bands,with primers OPA3,OPA5,OPA7,OPA13 and OPA20 produced the most polymorphic bands(Table 1)for 26.4%polymorphism(Table 2).Total number of bands ranged from 6 to 10 bands using these primers.The endangeredX.granatumhad the least polymorphism(14.56%)(Table 2);of 206 bands generated,30 were polymorphic.Primers OPA3,OPA9,OPA14 and OPA 20 primers were noteworthy(Table 1)for this species for producing a relatively higher number of polymorphic bands.Figure 2a–c shows a gel with the DNA bands ampli fi ed by two RAPD primers for each of the three species.
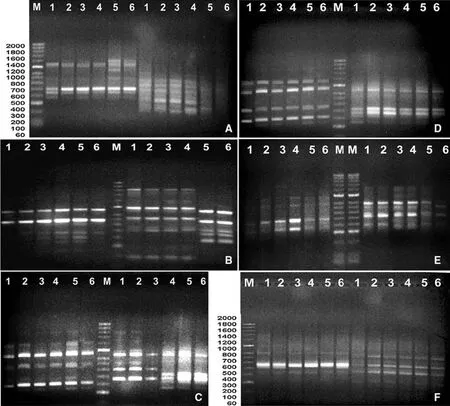
Fig.2 a–c RAPD gel,d–f ISSR gel of ampi fi ed DNA from three mangrove species from six sites:1,Mesophytic;2,Sajnekhali;3,Sudhanyakhali;4,Sonargaon;5,Jharkhali;6,Dobanki.a(OPA3,OPA 5)and d(ISSR1,ISSR4)Excoecaria agallocha;b(OPA3,OPA7)and e(ISSR7,ISSR 9)Phoenix paludosa;c(OPA9,OPA12)and f(ISSR4,ISSR12)Xylocarpus granatum
ISSR analysis
As in the RAPD analysis,polymorphism was relatively higher for the profusely growingE.agallochaandP.paludosa.The 20 primers were selected forE.agallochaproduced 24.87%polymorphism(Table 2)with 47 polymorphic among 189 ampli fi ed bands.Total number of bands per primer ranged from 7 to 12.ISSR1,ISSR4,ISSR7,ISSR12,ISSR 13,ISSR 15,ISSR 17,ISSR 18 AND ISSR 20 produced relatively more polymorphic bands(Table 3).P.paludosaproduced 187 bands,of which 38 were polymorphic,for 20.32%polymorphism(Table 2).Eight primers(ISSR2,ISSR3,ISSR7,ISSR9,ISSR13,ISSR15,ISSR16 and ISSR20)each produced three polymorphic bands(Table 3).Total number of bands per primer ranged between 7 and 12.X.granatumhad relatively less polymorphism with 12.92%,with 23 polymorphic bands of the 93 total bands(Table 2).ISSR4,ISSR7 and ISSR12 produced relatively more polymorphism among the samples(Table 3).Total number of bands expressed ranged between 8 and 12 per primer.Figure 2d–f shows a gel of the DNA bands ampli fi ed with the two ISSR primers for each of three species.
Statistical analyses
RAPD analysis ofE.agallochaproduced 55 polymorphic bands among 223 total bands.The mean observed number of alleles per locus(Na)was 1.1671,and the mean effective number of alleles per locis(Ne)was 1.1358.Nei’s gene diversity value(h)was 0.0794 and Shannon’s information index(I)was 0.1162.P.paludosaproduced 52 polymorphic bands among 197 bands expressed(Na=1.1753,Ne=1.1381,h=0.0818 andI=0.1187).ForX.granatum,Na=1.1135,Ne=1.0624,h=0.0637 and I=0.0857.The 30 of 206 bands were polymorphic.The UPGMA dendrogram based on the similarity matrix constructed for the RAPD analysis(Fig.3a–c)showed that forX.granatumthe non-saline site(site I)and the two most saline sites(site V and VI)clustered together,whereas in the other two taxa,the sites were in distant clusters.
ISSR analysis ofE.agallochaproduced 47 polymorphic bands among 189 bands;Na,Ne,h and I were 1.1679,1.1369,0.0799 and 0.1197,respectively.P.paludosaproduced 187 bands,38 of which were polymorphic;1.1469,1.1147,0.0688 and 0.1023 were the respective Na,Ne,h and I values.Na,Ne,h,I values forX.granatumwere 1.0847,1.0498,0.0583 and 0.0763,respectively.This species produced 23 polymorphic bands among a total of 193.For ISSR analysis,the UPGMA dendrogram based on the similarity matrix(Fig.3d–f)again showed that sites I,V and VI clustered together forX.granatum,whereas they were in distant clusters for the other two taxa.The same phenomenon was seen when a dendrogram was constructed using the combined RAPD and ISSR data(Fig.4a–c).

Table 3 List of ISSR primers and bands ampli fi ed using DNA from Excoecaria agallocha(Ea),Phoenix paludosa(Pp)and Xylocarpus granatum(Xg)
Discussion
The PCR-based RAPD technique is used very frequently by biologists because small amount of DNA is adequate and no prior knowledge about the DNA sequence of the species is needed(Williams et al.1990).It is a relatively fast,cheap method for estimating the genetic variation compared with morphological and allozyme methods.The molecular basis for RAPD was reported by Hadrys et al.(1992).This technique gives clear knowledge about the single base pair changes in the primer-binding sites or deletion in the areas between them.The only drawback of the RAPD technique is its lack of reproducibility for some ampli fi cation products,which can lead to faulty,controversial results.
Studies on mangroves have also been bene fi tted from these techniques. Lakshmi et al. (2000) reported intraspeci fi c genetic variation inE.agallocha.Both intraand interspeci fi c genetic variation inAvicenniawas revealed by Parani et al.(1997).In addition,the parentage of aRhizophorahybrid was analyzed using a RAPD marker.Teixeira et al.(2003)studied genetic polymorphism of 16 individuals ofAvicennia albafrom the Mekong Delta in Vietnam using 20 primer pairs for microsatellites.Six of the primers ampli fi ed polymorphic loci,seven of the loci ampli fi ed appeared to be monomorphic,and the other six pairs did not yield satisfactory products.Sheue et al.(2009)worked on the morphology and molecular basis of segregation of twoCeriopsspecies(C.zippelianaandC.decandra)from southeastern Asia using RAPD marker.Pawar et al.(2013)assessed the genetic diversity ofX.granatumandX.mekongensiscollected from different localities of the states of Tamil Nadu and Andhra Pradesh in India using the RAPD technique,where the authors reported that the two species ofXylocarpushad lower levels of similarity.
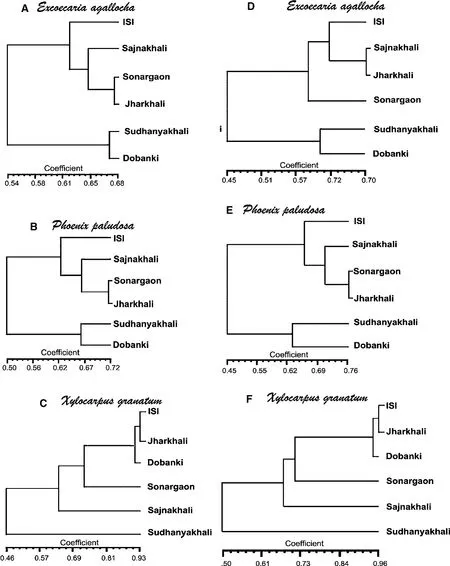
Fig.3 Dendrogram analysis.a–c RAPD analysis,d–f ISSR analysis,e–f RAPD and ISSR combined.a,d Excoecaria agallocha,b,e Phoenix paludosa,c,f Xylocarpus granatum
Dissimilarity found in the regions between microsatellites forms the basis of inter-simple sequence repeat(ISSR)analysis,which is frequently used to identify individuals,mostly plant species.Comparative studies of genetic variability of mangrove populations across large geographical ranges have often been done using ISSRs.Triest(2008)found signi fi cant differences in genetic diversity estimates between regions,despite the occasional low sample size of a population(10–20 individual mangrove trees).Genetic diversity within and among 10 mangrove and nonmangrove populations ofHeritiera littoraliscollected from three sites in China and one site in Australia was studied using ISSR by Jian et al.(2004),who reported a relatively high genetic diversity at the species level.Li and Chen(2004)estimated the genetic diversity ofSonneratia albafrom China using ISSR marker;103 polymorphic bands were obtained among 133 bands(77.44%polymorphism)using 11 ISSR primers,indicating a signi fi cant genetic variation at the species level.Tan et al.(2005)estimated the genetic diversity both within and among populations of a prevalent viviparous mangrove,Ceriops decandra,using ISSRs.
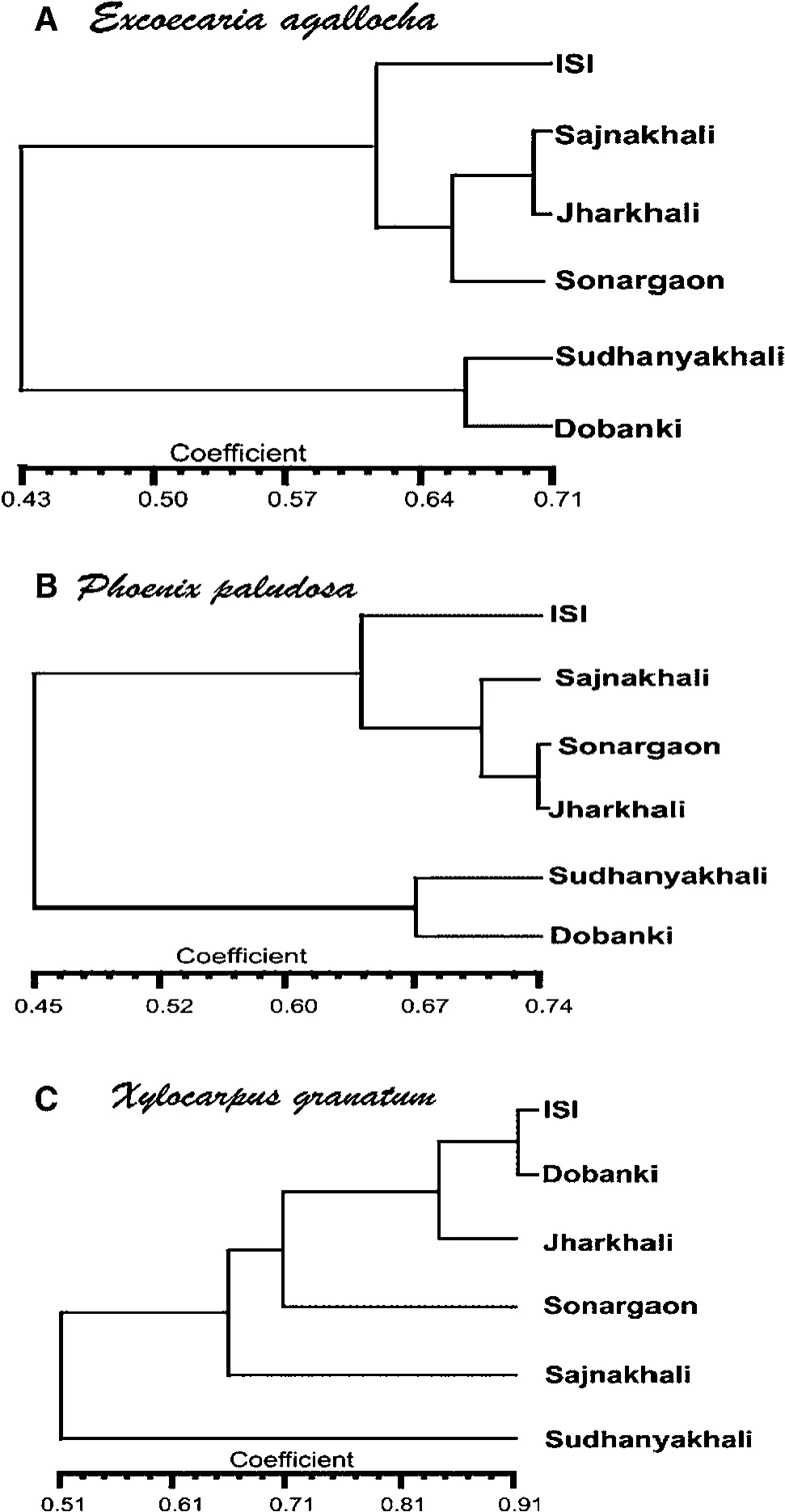
Fig.4 Dendrogram analysis.a–c RAPD and ISSR combined.a,d Excoecaria agallocha,b,e Phoenix paludosa,c,f Xylocarpus granatum
The present work on genetic polymorphism analysis using RAPD and ISSR revealed that relatively fewer polymorphicsms exist inX.granatumthan in the other two species.Kader et al.(2012)worked with a set of 10 RAPD and 10 ISSR markers to analyze the genetic diversity of the genusAvicenniafrom Sundarbans,India.In terms of polymorphism detection,polymorphic bands content per primer and total number of loci detected per primer,they found that ISSR markers were more ef fi cient than RAPD markers.In our study,although the level of polymorphism was in the same range in the ISSR and RAPD analyses of all three plant species,the total number of bands produced per primer was found to be high using ISSR.The result indicated that the percentage polymorphism was relatively low(14.56%for RAPD,12.92 for ISSR)inX.granatum,the stressed plant in the Indian Sundarbans compared with the other two taxa,which might be ascribed to the passive sustainability of this plant in the elevated saline regime.The profusely growing species,E.agallochaandP.paludosahad a relatively higher percentage polymorphism(24.66 and 26.4%for RAPD,24.87 and 20.32%for ISSR,respectively).From a genetic perspective,a higher level of genetic diversity results in a greater ability to adapt and evolve(Li and Chen 2004).Low genetic diversity can result in reduced adaptability and increased occurrence of less bene fi cial genes,leading to eventual extinction of the species;thus,the lower level of diversity within populations should receive special attention.The level of genetic diversity can provide very important information about the status of a species,an assessment of its conservation value,and ex situ conservation.
The salinity of Indian Sundarbans has risen over the years(Nandy(Dutta)et al.2009).Suf fi cient genetic polymorphism is a prerequisite for the plant to adapt to its environment through internal changes in its anatomy,physiology and biochemistry.The relatedness(proximity or distance)of genetic polymorphic expression of the studied taxa is evident from the prepared dendrogram(based on the similarity matrix).In the dendrogram prepared for the stressed species,X.granatum,from RAPD data,the extreme saline zone and mesophytic zone grouped into the same clade.The same phenomenon was observed in the dendrogram based on the ISSR data.On the contrary,in the two profusely growing species,the dendrograms prepared from RAPD and ISSR data showed that the most saline zones clustered in one clade.
The result depicted from this study agrees well with our previous study showing that the antioxidant activity ofX.granatumis much weaker thanP.paludosaandE.agallochabased on enzymatic and non-enzymatic variables(Dasgupta et al.2013)and the genetic polymorphism of another IUCN Red-listed speciesHeritiera fomeswas relatively lower than in the profusely growing speciesBruguieragymnorrhiza(Dasgupta et al.2015). The antioxidative response in plants is intricate,and scavenging of ROS takes place using enzymatic and non-enzymatic machinery.So the higher level of genetic polymorphism ofP.paludosaandE.agallochamight be attributed to superior protection against ROS;thus,growth of these two species is more sustainable in the increasing salinity of the present-day Sundarbans.
Conclusion
The relatively low polymorphism shown by the RAPD and ISSR analyses forX.granatummight be attributed to its relatively poorer ability to scavenge ROS in areas of higher salinity,which might be attributed to one of the major reasons for its gradual natural decline in the Indian Sundarbans.Currently,there are very limited resources in the natural habitats ofX.granatumpopulations in the Indian Sundarbans;thus,the most important aspect in protecting the resources,is maintaining higher genetic diversity in the species and thus its the ability to adapt and evolve,and it also provides very important information on the status of a species and conservation value.When selecting the best provenance for introduction and transplantation,quality groups,and individuals for genetic improvement,special attention should be paid to populations and individuals that come from special habitats or demonstrate irreplaceable uniqueness.
AcknowledgementsThe authors are sincerely indebted to the Director,Sundarbans Biosphere Reserve and Chief Principal Conservator of Forest and Wildlife,Government of West Bengal,for granting the required permission to conduct fi eldwork in the Sundarbans Reserved Forest.The authors thank the Indian Statistical Institute for funding this work.
Barbier EB(2007)Valuing ecosystem services as productive inputs.Econom Policy 22:177–229
Chanda S,Datta SC(1986)Prospects and problems of a mangrove ecosystem in western Sundarbans(India).Trans Bose Res Inst 49(3):47–57
Chaudhuri JK(1996)Mangrove forest management.Mangrove rehabilitation and management project in Sulawesi.p 297
Chowdhury MR,Ward MN(2007)Seasonal fl ooding in Bangladesh—variability and predictability.Hydrol Process 21:335–347
Das S(1999)An adaptive feature of some mangroves of Sundarbans,West Bengal.J Plant Biol 42(2):109–116
Das AB,Jena S(2008)Chromosome stability and interpopulation genetic variability in a tree mangroveXylocarpous granatumKoen.(Meliaceae)as revealed by RAPD markers.Cytologia 73:105–113
Dasgupta N,Nandy P,Tiwari C,Das S(2010)Salinity-imposed changes of some isozymes and total leaf protein expression in fi ve mangrovesfrom two differenthabitats.J Plant Int 5(3):211–221
Dasgupta N,Nandy P,Sengupta C,Das S(2012)Protein and enzymes regulations towards salt tolerance of some Indian mangroves in relation to adaptation.Trees 26(2):377–391
Dasgupta N,Nandy P,Das S(2013)Salt stress:a biochemical and physiological adaptation of some Indian halophytes of Sundarbans.In:Molecular stress physiology of plants.Springer India,pp 155–177
Dasgupta N,Nandy P,Sengupta C,Das S(2015)RAPD and ISSR marker mediated genetic polymorphism of two mangrovesBruguieragymnorrhizaandHeritierafomesfrom Indian Sundarbans in relation to their sustainability.Physiol Mol Biol Plants 21(3):375–384
Forest Survey of India(FSI)(2009)State forest report 2009.Ministry of Environment and Forests,Government of India,226p
Gurudeevan S,Satyavani K,Ramanathan T(2012)Genetic identiif cation ofCeriops decandra(ChiruKandal)using tRNA(Leu)molecular marker.Asian J Plant Sci 11:91–95
Hadrys H,Balick M,Schierwater B(1992)Applications of random ampli fi ed polymorphic DNA(RAPD)in molecular ecology.Genome 1(1):55–63
Hiraishi T,Harada K(2003)Greenbelt tsunami prevention in South-Paci fi c Region.http://eqtap.edm.bosai.go.jp/
Hogarth PJ(2007)The biology of Mangroves and sea grasses.Oxford University Press,New York,p 273
Hopson TM,Webster PJ(2010)A 1–10-Day ensemble forecasting scheme for the Major River Basins of Bangladesh:forecasting severe fl oods of 2003–07.J Hydrometeorol 11(3):618–641
Jaccard P(1998)Nouvelles researches sur la distribution forale.Bull Soc Sci Nat 44:223–270
Jian S,Tang T,Zhong Y,Shi S(2004)Variation in inter-simple sequence repeat(ISSR)in mangrove and non-mangrove populations ofHeritiera littoralis(Sterculiaceae)from China and Australia.Aquat Bot 79(1):75–86
Kader A,Sinha SN,Ghosh PD(2012)Evaluation of genetic diversity of Avicenniaceae family in Indian Sundarban by using RAPD and ISSR markers.Iranian J Genet Plant Breed 1:22–27
Lakshmi M,Parani M,Ram N,Parida A(2000)Molecular phylogeny of mangroves VI.Intraspeci fi c genetic variation in mangrove speciesExcoecaria agallochaL.(Euphorbiaceae).Genome 43(1):110–115
Li H-S,Chen G-Z(2004)Genetic diversity ofSonneratia albain China detected by Inter-simple Sequence Repeats(ISSR)analysis.Acta Bot Sinica 46:512–521
Nandy(Dutta)P,Dasgupta N,Das S(2009)Differential expression of physiologicalandbiochemicalcharactersofsomeIndianmangroves towards salt tolerance.Physiol Mol Biol Plants 15(2):151–160
Naskar KR,GuhaBakshi DN(1983)A brief review on some less familiar plants of the Sundarbans India.J Eco Taxon Bot 4(3):699–712
Nei M(1973)Analysis of gene diversity in subdivided populations.Proc Nat Acad Sci 70(12):3321–3323
Newbury HJ,Ford-Lloyd BV(1993)The use of RAPD for assessing variation in plants.Plant Growth Reg 12:43–51
Parani M,Lakshmi M,Elango S,Ram N,Anuratha CS,Parida A(1997)Molecular phylogeny of mangroves II.Intra and interspeci fi c variation inAvicenniarevealed by RAPD and RFLP markers.Genome 40(4):487–495
Parida A,Anuratha CS,Lakshmi M,Parani M,Kurjen J(1995)Application of molecular markers in assessing genetic diversity in Indian mangroves.In:Use of Induced Mutations and Molecular Techniques in Crop Improvement(FAO/IAEA,Vienna,Austria,595–600)
Pawar UR,Baskaran J,Ajithkumar IP,Panneerselvam R(2013)Genetic variation betweenXylocarpusspecies(Meliaceae)as revealed by Random Ampli fi ed Polymorphic DNA(RAPD)markers.Emirates J Food Agric 25(8):597–604
Rohlf FJ(1993)Ntsys-PC.Numerical taxonomy and multivariate analysis system Version I.80-Setauket,NY,Exeter Software
Schaal BA,Leverich WJ,Rogstad SH(1991)Comparison of methods for assessing genetic variation in plant conservation biology.Genetics and conservation of rare plants.Oxford University Press,New York,pp 123–134
Schwarzbach AE,McDade LA(2002)Phylogenetic relationships of the mangrove family Avicenniaceae based on chloroplast and nuclear ribosomal DNA sequences.Syst Bot 27:84–98
Sheue CR,Liu HY,Tasi CC,Rashid SMA,Yong JWH,Yang YP(2009)On the morphology and molecular basis of segregation of two speciesCeriops zippelianaBlum.andC.decandra(Griff.)Ding Hou(Rhizophoraceae)from Southeastern Asia.Blumea 54:220–227
Sneath PH,Sokal RR(1973)Numerical taxonomy.The principles and practice of numerical classi fi cation
Spalding M,Blasco F,Field C(1997)World Mangrove Atlas.The International Society for Mangrove Ecosystems.Okinawa,Japan,p 178
Spiers AG(1999)Review of International continental wetland resources.Global review of wetland resources and priorities for wetland inventory.Finlayson,CM and Spiers,AG(eds.),supervising Scientist Report 144.Canberra,Australia:63–104
Takemura T,Hanagata N,Sugihara K,Baba S,Karube I,Dubinsky Z(2000)Physiological and Biochemical Responses to Salt Stress in the Mangrove,Bruguiera gymnorrhiza.Aquat Bot 68:15–28
Tan F,Huang Y,Ge X,Su G,Ni X,Shi S(2005)Population genetic structure and conservation implications ofCeriops decandrain Malay Peninsula and North Australia.Aqua Bot 81(2):175–188
Teixeira S,Arnaud-Haond S,Duarte CM,Serrao E(2003)Polymorphic microsatellite DNA markers in the mangrove treeAvicennia alba.Mol Ecol Notes 3:544–546
Templeton AR (1993)Translocation as conservation tool.In:Biodiversity in mangrove landscapes,theory and practice(ed.Szaro R.I.N)(Oxford University Press)
The IUCN Red List of Threatened Species.Version(2014)<www.iucnredlist.org>.Downloaded on 22 March 2015
Triest L(2008)Molecular ecology and biogeography of mangrove trees towards conceptual insights on gene fl ow and barriers:a review.Aqua Bot 89(2):138–154
Upadhyay VP,Ranjan R,Singh JS(2002)The Human Mangrove Con fl icts-The way out.CurrSci 83:1328–1336
Williams JGK,Kubelik AR,Livak KJ,Rafalski JA,Tingey SV(1990)DNA polymorphisms ampli fi ed by arbitrary primers are useful as genetic markers.Nucleic Acids Res 18:6531–6535
Yan L,Guizhu C(2007)Physiological adaptability of three mangrove species to salt stress.Acta Ecol Sinica 27(6):2208–2214
Yeh FC,Yang RC,Boyle TB,Ye ZH,Mao JX(1997)POPGENE,the user-friendly shareware for population genetic analysis.Molecular biology and biotechnology center.University of Alberta,Canada,p 10
Zimmermann MH(1983)Xylem structure and the ascent of sap.Springer,Berlin
 Journal of Forestry Research2018年2期
Journal of Forestry Research2018年2期
- Journal of Forestry Research的其它文章
- Effect of species composition on ecosystem services in European boreal forest
- Analysis of SSR loci and development of SSR primers in Eucalyptus
- Optimal and synchronized germination of Robinia pseudoacacia,Acacia dealbata and other woody Fabaceae using a handheld rotary tool:concomitant reduction of physical and physiological seed dormancy
- Genetic effects of historical anthropogenic disturbance on a longlived endangered tropical tree Vatica mangachapoi
- Cloning and characterization of geranylgeranyl diphosphate synthetase from Pinus massoniana and its correlation with resin productivity
- In vitro anther culture and Agrobacterium-mediated transformation of the AP1 gene from Salix integra Linn.in haploid poplar(Populus simonii×P.nigra)
