High-throughput sequencing analysis of differentially expressed miRNAs and target genes in ischemia/reperfusion injury and apelin-13 neuroprotection
Chun-mei Wang, Xue-lu Yang, Ming-hui Liu, Bao-hua Cheng, Jing Chen, Bo Bai
Neurobiology Institute, Jining Medical University, Jining, Shandong Province, China
Introduction
Apelin separated and purified from bovine stomach is composed of apelin-13, -17, and -36, including 13, 17, and 36 amino acids at the C-terminal, respectively (Tatemoto et al.,1998; Kawamata et al., 2001). In the cardiovascular system,apelin has been found to protect myocardiocytes in a myocardial injury model (Jia et al., 2006). Within the central nervous system, apelin can alter blood pressure, feeding behavior, and hormone release, suggesting that it plays a vital role in neurons (Tatemoto et al., 2001; Bao et al., 2016). Furthermore, apelin-13 was shown to display many more biological functions than apelin-36 (Simpkin et al., 2007). Khaksari et al. (2012) found that apelin-13 markedly decreased brain infarct volume and postischemic cerebral edema by inhibiting apoptosis in a rat focal stroke model. Our previous results also showed that apelin-13 had a neuroprotective effect against cerebral ischemia/reperfusion injury (IRI) through inhibiting neuronal apoptosis (Yan et al., 2015). However,the precise mechanisms by which apelin-13 exerts its neuroprotective effects have not yet been revealed.
MicroRNAs (miRNAs) could mediate the expression of genes mainlyviaimperfect pairing with untranslated regions at the 3′ end of target mRNAs (Carrington and Ambros,2003; Bartel, 2004; Matinez and Peplow, 2016), thus regu-lating a variety of biological processes (Hwang and Mendell,2006; Zhao et al., 2007). miRNAs have been reported to be involved in the development and function of the brain (Sempere et al., 2004). They have also been found to participate in the regulation of some drugs, which have been proven to exert neuroprotective effects in cerebral ischemia (Cao et al.,2012; Shi et al., 2013). For example, miR-29b overexpression has been shown to reduce blood-brain barrier disruption after ischemic stroke (Wang et al., 2015). Solexa high-throughput sequencing (HiSeq) has recently been developed for the identification of miRNAs based on sequencing (Zhao et al., 2010;Xie et al., 2011). To date, HiSeq has been widely used for global miRNA profiling in a large number of organisms (Liu et al.,2010; Farazi et al., 2011; Chen et al., 2012; Ji et al., 2012). However, it has not been reported whether apelin-13 reperfusion could cause changes of miRNA expression, as determined using HiSeq. Therefore, in this study, we test the hypothesis that apelin-13 reperfusion would cause the up- or downregulation of miRNAs, which subsequently play important roles through their targets in IRI and the neuroprotective effect of apelin-13.To evaluate the miRNA prolife after IRI and apelin-13 reperfusion, we performed HiSeq after apelin-13 reperfusion in a rat middle cerebral artery occlusion (MCAO) model. The obtained results should help to reveal the neuroprotective and therapeutic effects of apelin-13 on ischemic stroke by clarifying the upstream regulatory mechanism of miRNAs.
Materials and Methods
Animals
Thirty specific-pathogen-free male Wistar rats aged 6–8 weeks and weighing about 300 g were purchased from Lukang Pharmaceutical Co., Ltd., China [animal certification SCXK (Lu)20130001] as a model of MCAO. These rats were raised in standard cages where they were allowed free access to food and water. The environment was controlled at 22–26°C and 50–60% humidity under a 12-hour light/dark cycle. The study protocol was approved by the Ethics Committee of Jining Medical University of China (approval number: 2017-KY-021).
Establishment of transient focal MCAO and apelin-13 intervention
Eighteen rats were randomly and equally divided into three experimental groups: (1) sham group: filament was only inserted into the common carotid artery, but not into the middle cerebral artery; (2) IRI group: rats were exposed to 2-hour occlusion followed by 24-hour reperfusion with 0.9% NaCl saline (10 μL per rat); and (3) apelin-13 group: rats were treated with intracerebroventricular injection of apelin-13 (50 μg/kg; Phoenix Pharmaceuticals, Inc., Burlingame, CA, USA) after ischemia at the beginning of reperfusion. Rats were kept in separate cages for 24 hours with free feeding.
Transient focal MCAO was made by the intraluminal filament method (Longa et al., 1989; Xu et al., 2006; Vakili and Zahedi Khorasani, 2007; Wyman et al., 2009; Xin et al., 2015).All rats were anesthetized by the intraperitoneal injection of chloral hydrate (10%; 350 mg/kg). The rats were fixed and disinfected. The right common carotid, external carotid, and internal carotid arteries were isolated. Subsequently, the carotid arteries and external carotid arteries were ligated. A carotid artery was cut and filament was inserted at a depth of 18 mm. The skin incision was then sutured. Subsequently, the filament was removed after 2 hours of ischemia and 24 hours of reperfusion.
RNA sequencing for miRNA
Total RNA from the hippocampus was obtained from the apelin-13, IRI, and sham groups using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The quality and concentration of RNA were evaluated using BioSpec-nano (Shimadzu Corporation, Kyoto, Japan). Total RNA was then separated by denaturing polyacrylamide gel electrophoresis (15%)and sRNAs in the size range of 18–30 nucleotides (nt) were excised from the denaturing gel and recovered. Subsequently, proprietary (Solexa) adapters were ligated to the 3′- and 5′-terminals of these sRNAs using T4 ligase. The ligated products were purified and subsequently reverse-transcribed into cDNA. The cDNA fragments were amplified by quantitative real-time polymerase chain reaction (qRT-PCR) to construct sRNA libraries, which were sequenced with a Solexa sequencer (Illumina, San Diego, CA, USA).
Sequence analysis
The original data after sequencing were converted into raw data. The raw data were purged of contaminant reads and named clean reads, which were then analyzed for the length distribution using software developed by BGI (Beijing, China). Subsequently, clean reads were screened in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) and Rfam databases (http://rfam.sanger.ac.uk/) to determine the type of small RNA. The tag2repeat software (BGI) was used to annotate the repeat overlapping sequences as repeat-associated small RNA. The unique sRNA sequences were analyzed to confirm conserved miRNA in the rat database of miRBase (http://www.mirbase.org/index.shtml). Those that could not be confirmed were predicted as novel miRNAs according to the hairpin structure of miRNA precursor using Mireap software (BGI).
qRT-PCR for differential miRNA
To evaluate the accuracy of data from HiSeq, qRT-PCR(Wang et al., 2011) was used to quantitatively detect several miRNAs differentially expressed among the sham group,IRI group, and apelin-13 group. In brief, RNAs were isolated and cDNA was reverse-transcribed using an NCode™VILO™ miRNA cDNA Synthesis Kit (Invitrogen). The PCR amplification was performed in LightCycle 480II (Roche,Basel, Switzerland) with the NCode™ EXPRESS SYBR GreenER™ Kit (Invitrogen). The reaction procedure was performed as follows: 2 minutes at 50°C, 2 minutes at 95°C,and then 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 60 seconds. The nuclear RNA U6 was used as a loading control. All reactions were carried out in triplicate. The relative expression level of miRNA was calculated and normalized using the 2−ΔΔCTmethod.
Prediction of target genes
The putative targets of miRNAs were predicted using RNA-hybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/).Two criteria were applied for the prediction. One was the absence of mismatch between 2 and 8 nt at the 5′ end. The other was that G-U was allowed, but no more than three of them. The required minimum free energy of the duplex formed by miRNA and the target was ≥ 75%.
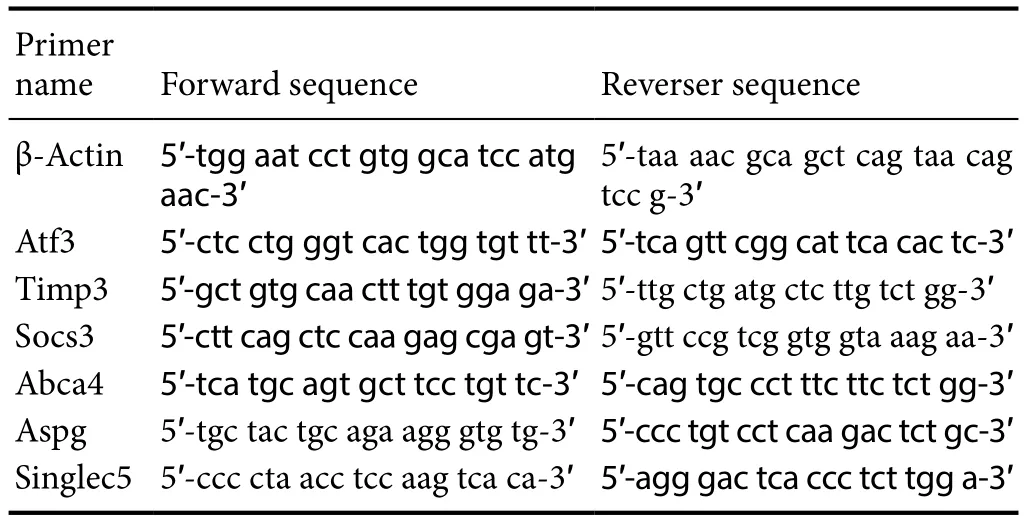
Table 1 Primer sequences used for gene amplification in the rat model
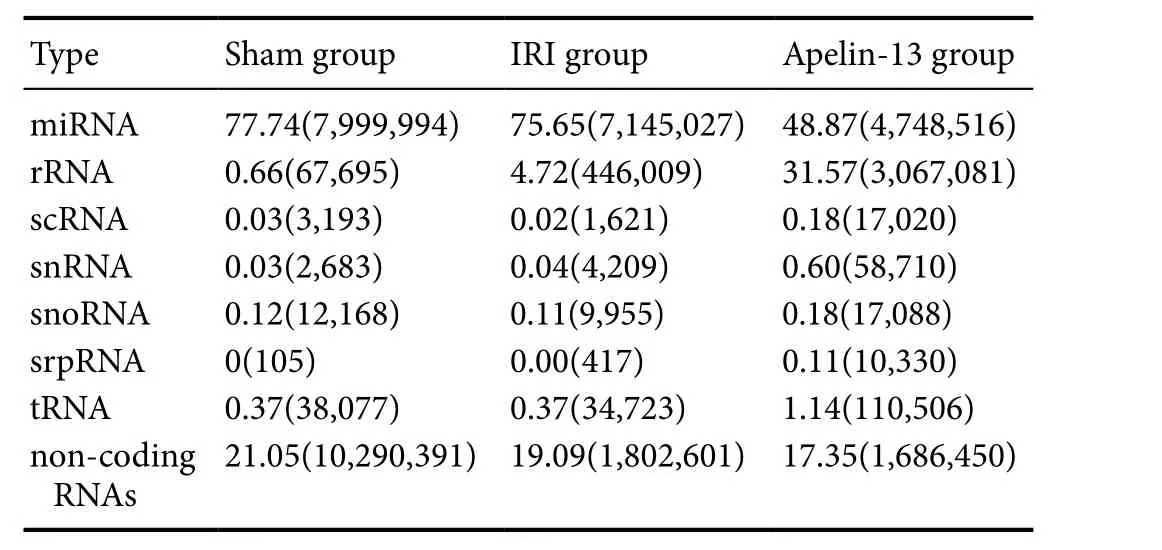
Table 2 Type and frequency of sRNAs

Table 3 Some of the differentially expressed miRNAs (miR)
qRT-PCR for predicted target genes
qRT-PCR was used to detect the predicted target genes that were differentially expressed. The quantitative PCR reaction was carried out with SuperReal PreMix Plus kit (Beijing Tiangen Biology Co., Ltd., Beijing, China). The process was performed on the LightCycle 480II (Roche) as follows: 95°C for 15 minutes; and then 40 cycles of 95°C for 10 seconds,60°C for 20 seconds, and 72°C for 20 seconds. The relative expression of target genes compared with β-actin was determined using the 2−ΔΔCTmethod. The primers used in the current study are listed inTable 1.
Gene Ontology (GO) enrichment analysis
GO enrichment analysis was used to predict miRNA targets.In this method, all of the target candidates were first assigned terms in the GO database (http://www.geneontology.org/), and then the numbers of genes for each term were determined, after which a hypergeometric test was used to select significantly enriched GO terms.
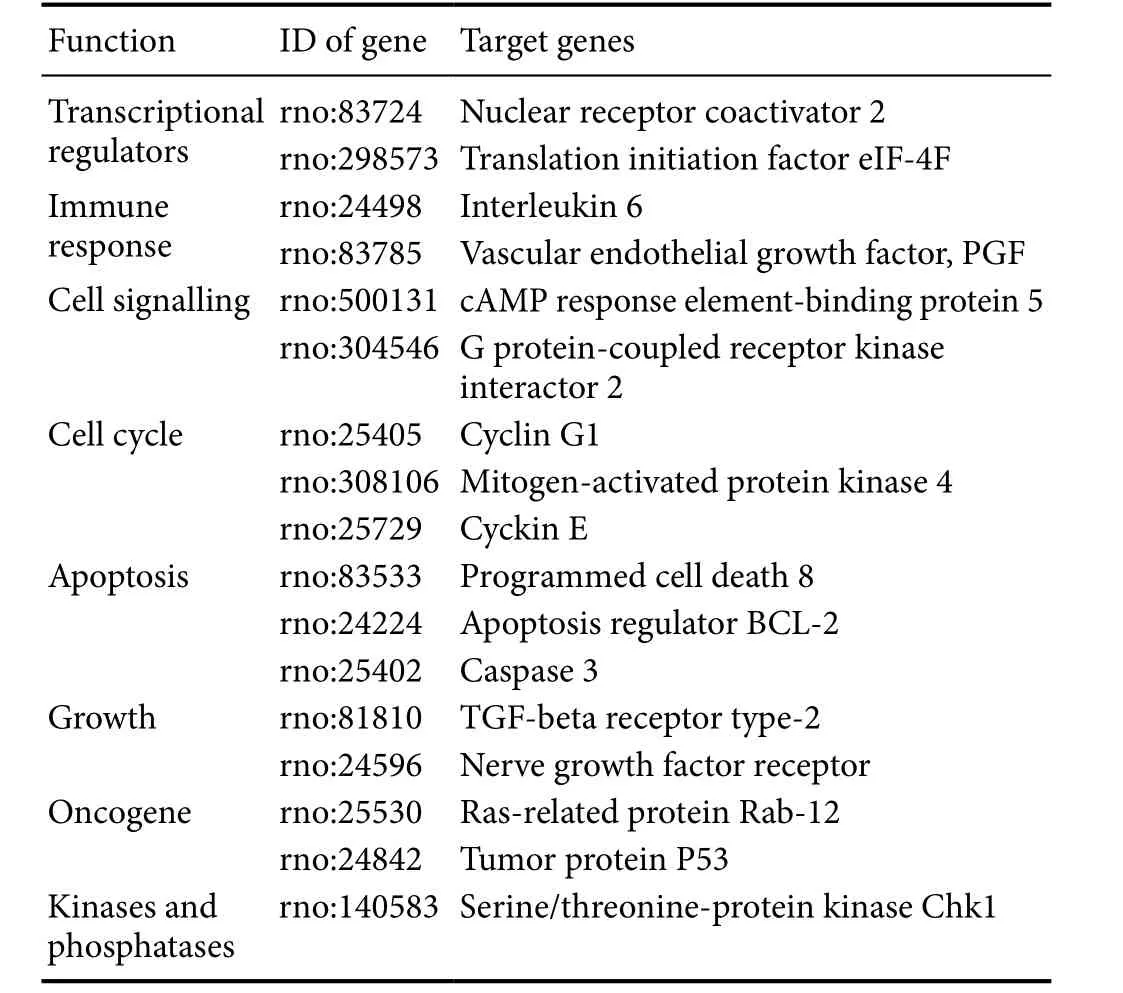
Table 4 Some of the predicted targets of miRNAs
KEGG pathway analysis
We also utilized the Kyoto Encyclopedia of Gene and Genome (KEGG) pathway database to evaluate the main pathways associated with the miRNA targets. KEGG analysis might select significant metabolic pathways and signal pathways of target candidates based on all reference genes.A false discovery rate ≤ 0.05 was regarded as representing differential enrichment of the target candidates.
Statistical analysis
Data, expressed as the mean ± SD, were analyzed using Graph-Pad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA,USA). One-way analysis of variance followed by Tukey’spost hoctest was used for comparisons among multiple groups. A value ofP< 0.05 was considered statistically significant.
Results
Deep sequencing
After sequencing, we obtained numerous sequence reads.The length distribution of the sequence reads was evaluated.The mean read size was in the range of 18–28 nt (Figure 1).Those 22 nt in size were the most abundant group of small RNAs, followed by those sequences 23 and 21 nt in length.These results are consistent with those previously reported(Agarwal et al., 2016; Hong et al., 2016). miRNAs with a sequence length of 22 bp also formed the largest class.
The type and number of sRNAs were searched according to sequence similarity using several databases (miRNAs,rRNAs, scRNAs, sn/snoRNAs, srpRNAs, tRNAs, and other noncoding RNAs). miRNAs constituted the majority of sRNAs in the sham group (7,999,994/10,290,391 = 77.74%),IRI group (7,145,027/9,444,562 = 75.65%), and apelin-13 group (4,748, 516/9,715,707 = 48.87%) (Table 2), suggesting that these miRNAs could be functional and represent another level of regulation in IRI and neuroprotection by apelin-13.
Identification of conserved miRNAs
miRNAs are generally known to be conserved across species.In the current study, 296 miRNAs were downregulated and 61 miRNAs were upregulated in IRI model compared withtheir levels in sham model (Figure 2). Overall, 197 downregulated and 155 upregulated miRNAs were detected in apelin-13 model compared with IRI model. Some of the miRNAs that differed in their expression levels among sham, IRI, and apelin-13 groups are listed inTable 3. Compared with the sham group, Rno-miR-27-5p, Rno-miR-34c-5p, Rno-miR-143-3p,Rno-miR-223-3p, Rno-miR-448-3p, and Rno-miR-1298 were most markedly downregulated in IRI model, exceeding twofold change. However, these miRNAs were clearly upregulated in apelin-13 model. Rno-miR-328a-5p, Rno-miR-503-3p,Rno-miR-875-3p, and Rno-879-5p were most markedly upregulated in IRI model, but were downregulated in apelin-13 model. These results indicate that these miRNAs might play vital roles in the neuroprotection by apelin-13.
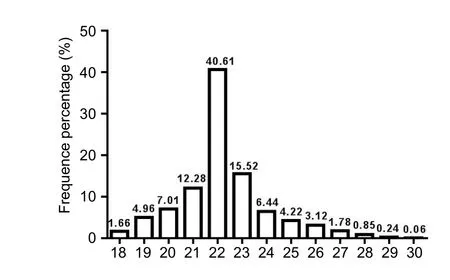
Figure 1 Sequence length of short sRNAs from apelin-13 model detected by Solexa sequencer.

Figure 2 Known miRNAs are differentially expressed among the models.
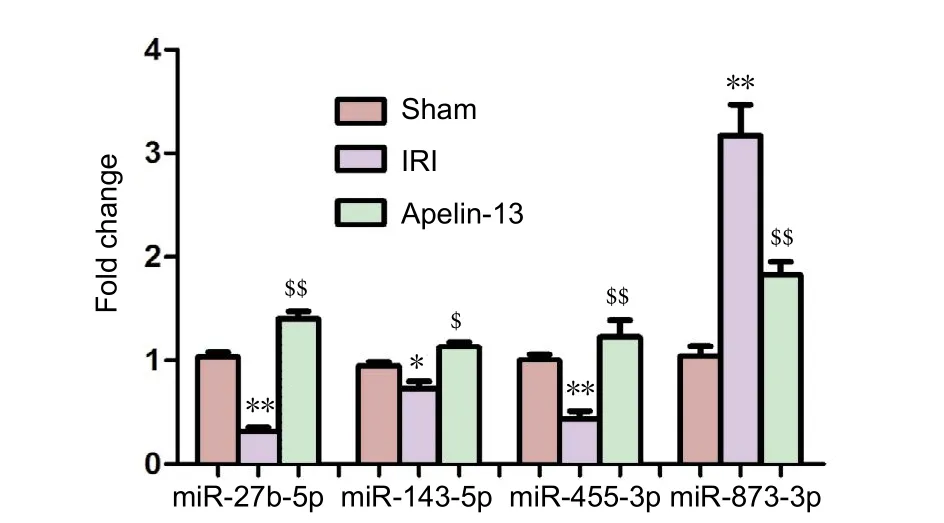
Figure 3 Four differentially expressed miRNAs quantified using quantitative real-time polymerase chain reaction.

Figure 4 Expression of target genes quantified using quantitativereal-time polymerase chain reaction.
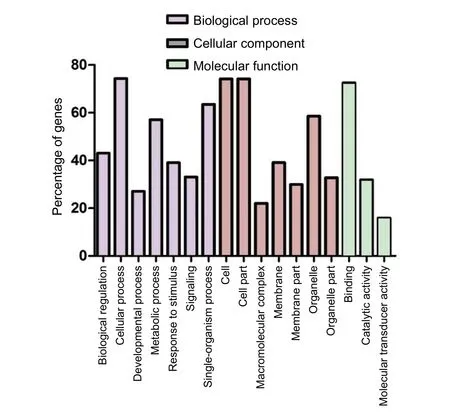
Figure 5 Categories of the GO terms of miRNA targets.
qRT-PCR for differential expression of miRNAs
To validate the sequencing data, several differentially expressed miRNAs were selected for amplification using qRTPCR. The expression of three miRNAs, Rno-miR-27b-5p,Rno-miR-143-5p, and Rno-miR-455-3p, was downregulated in IRI model, but upregulated after apelin-13 reperfusion.The reverse pattern was noted for the expression of rnomiR-873-3p (Figure 3). Generally, there was good consistency between the results of qRT-PCR and HiSeq.
Prediction of targets of miRNAs
Analysis of the predicted targets of miRNAs may aid our understanding of the regulatory role of abnormal miRNAs.In the present study, for known miRNAs that were differentially expressed, a total of 34,252 putative targets that were mapped to 259,856 sites were predicted. A number of predicted targets of miRNA are listed inTable 4, such as eIF-4F, interleukin 6, PGF, cyclin G1, cyclin E, nerve growth factor receptor, TGF-β receptor, BCL-2, caspase 3, Rab-12,and P53. It was shown that miRNAs probably regulate the role of apelin-13 by changing inflammatory factors, apop-
tosis factors, and transcription factors. However, bioinformatic predictions for targets must be subjected to further verificationvia in vitroandvivoexperiments. Therefore, the in-depth analysis of targets was performed to reveal the regulatory mechanisms of differently expressed miRNAs.
To determine whether there were changes in the expression of targets, qRT-PCR was used to quantify the target expression level of four miRNAs (Rno-miR-27b-5p, RnomiR-143-5p, Rno-miR-455-3p, and Rno-miR-873-3p). The sequences of the primers used are listed inTable 1. Four predicted targets (Af3, Timps, Socs3, and Aspg) were upregulated in IRI model compared with those in sham model,but were downregulated after apelin-13 reperfusion. The expression of two targets, Abca4 and Siglec5, was decreased in IRI model and increased to nearly normal in apelin-13 model (Figure 4). It was indicated that the predicted targets are involved in IRI and neuroprotection by apelin-13.
GO analysis
To obtain further insight into the functions of different miRNAs, we evaluated the targets of the miRNAs in GO.miRNA targets were assigned to 51 categories as follows: 23 were associated with biological processes, 15 were associated with cellular components, and 13 were involved in molecular functions. The top 17 GO terms are shown inFigure 5. The majority of targets were enriched in the categories of metabolic process, cellular process, single-organism process,cell, organelle, cell part, and binding, indicating that apelin-13 probably exerts its neuroprotective effect by altering an extensive range of physiological processes.
KEGG pathway analysis

Figure 6 KEGG pathway analysis of predicted target genes.
The KEGG pathway analysis indicated that the predicted targets of the miRNAs are involved in 304 biological processes,such as metabolic pathway, signal transduction, and disease.The top pathway was metabolic pathways, containing 2,190 targets accounting for 10.41% of the total. The second was olfactory transduction, in which there were 1,479 targets accounting for 7.03%, followed by biosynthesis of secondary metabolites (786 accounting for 3.74%), pathways in cancer (768 accounting for 3.65%), and regulation of actin cytoskeleton (740 accounting for 3.52%). The predicted targets of miRNAs were annotated as being involved in the MAPK signaling pathway or JAK-ATAT signaling pathway (Figure 6). These results indicate that some underlying biological processes participate in apelin-13 reperfusion and provide meaningful clues to research the targets of miRNAs in apelin-13 neuroprotection.
Discussion
As HiSeq has been shown to have pronounced advantages compared with the use of a microarray, an increasing number of studies have used this method to evaluate the regulation of miRNA under different conditions (Wyman et al.,2009; Vaz et al., 2010). In the present study, Rno-miR-1298,Rno-miR-223-3p, Rno-miR-27-5p, and Rno-miR-381-5p were markedly downregulated in IRI model, but upregulated after apelin-13 reperfusion. Rno-miR-328a-5p, Rno-miR-875-3p, and Rno-miR-879-5p were clearly upregulated in IRI model, but downregulated after apelin-13 reperfusion.This indicates that these abnormal miRNAs should be involved in IRI and the neuroprotective effect of apelin-13.Furthermore, several different miRNAs in the present study have also been reported to play important roles in other diseases. For example, Mmu-miR-27a-5p by upregulation of MCPIP1, which decreased the secretion of IL-6, IL-1β, and IL-10, inhibited the inflammatory response in macrophage cells induced by lipopolysaccharide (Cheng et al., 2015).It has also been shown that the overexpression of miR-223 may protect the brain from neuronal death induced by global ischemia and excitotoxic injury by regulating the glutamate receptor subunits GluR2 and NR2B (Harraz et al.,2012). miR-381 had a major role in glioma progression by targeting LRRC4 (Tang et al., 2011). Several other miRNAs have not been reported to be associated with the central nervous system, but were found to be abnormally expressed in the current study, suggesting that they specifically participate in the neuroprotection mediated by apelin-13.
miRNAs were reported to play critical roles by controlling a number of targets of mRNAs (Zhao et al., 2007; Liu et al.,2010; Li and Tang, 2016). Thus, the identification of potential targets should be significant in shedding light on miRNA regulation. In the present study, 34,252 targets of conserved miRNAs were predicted and these genes were mapped on 259,856 loci. We found many predicted target genes associated with inflammation, apoptosis, anti-apoptosis, proliferation, and immune response. For example, the inflammatory factors encoded by genes that were predicted to be target genes here included tumor necrosis factor-α, interleukin-6,prostaglandin F, and intercellular adhesion molecule-1.Some apoptosis-related genes and anti-apoptotic genes were also found, including caspase 3, Bcl-2, p53, and HSP.Furthermore, our lab has focused on the neuroprotective effects of apelin-13 in rat cerebral ischemic models and found many differentially expressed genes (Xin et al., 2015; Yan et al., 2015). In this study, we also verified that the expression of Af3, Timps, Socs3, and Aspg was downregulated in apelin-13 model compared with their levels in IRI model. The expression of Abca4 and Siglec5 was upregulated after apelin-13 reperfusion. We are currently selecting more targets to validate whether they are targets of miRNAs.
GO analysis indicated that the predicted targets of different miRNAs participate in a number of biological processes. Specifically, this analysis revealed that approximately 34.8% of the predicted targets were associated with catalytic activity in the molecular function group, approximately 80% of the predicted targets were associated with a cellular component, and 38% of the predicted targets could respond to stimuli in the biological process group. In the KEGG pathway analysis, the most represented pathways included regulation of actin cytoskeleton,mitogen-activated protein kinase signaling pathway, calcium signaling pathway, and especially nuclear factor-κB signaling pathway and Wnt signaling pathway. Xu et al. (2012) found that matrine has neuroprotective effects against focal cerebral ischemiaviainhibition of the nuclear factor-κB signaling pathway. The Wnt signaling pathway also plays important roles in neuronal migration, synaptic differentiation, synaptic plasticity, and cerebral ischemia (Cerpa et al., 2009).
Although numerous different miRNAs were detected, and their putative targets and signaling pathways were predictedviaa bioinformatic approach, there is still a need for further experimental validation of whether they do indeed participate in the regulation of IRI and apelin-13 reperfusion. This will thus be the focus of our future research. Therefore, we plan to perform further experiments to verify the biological functions of these miRNAsin vitroandin vivo, as well as to determine the relationship between miRNA and their targets.
In summary, our results illustrate for the first time the miRNA profile after apelin-13 reperfusion of a rat MCAO model, as determined using HiSeq. We identified 352 differentially expressed miRNAs that could be used as potential regulators of IRI and the neuroprotection by apelin-13,among which 78 showed a change of expression greater than twofold change. Moreover, the predicted targets of different miRNAs were further evaluated using GO and KEGG analyses. These analyses indicated that the targets are mainly involved in biological processes and metabolic pathways. Our results revealed the abnormally expressed miRNAs and shed light on the regulatory mechanism behind apelin-13 neuroprotection, thereby providing an experimental foundation for the study of nerve regeneration in ischemic stroke.
Author contributions:CMW wrote this paper. XLY prepared the MACO models. MHL assisted the preparation of animal models. BHC carried out qRT-PCR for target genes. JC was responsible for the revision of this paper.BB was responsible for study design. All authors approved the final version of the paper.
Conflicts of interest:None declared.
Financial support:This study was supported by the National Natural Science Foundation of China, No. 81501018 and 816712276; the Natural Science Foundation of Shandong Province of China, No. ZR2013CQ031 and ZR2014HL040. Funders had no involvement in the study design; data collection, management, analysis, and interpretation; paper writing; or decision to submit the paper for publication.
Research ethics:The study protocol was approved by the Ethics Committee of Jining Medical University of China (approval number: 2017-KY-021).The experimental procedure followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No.8023, revised 1985).
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under identical terms.
Agarwal S, Nagpure NS, Srivastava P, Kushwaha B, Kumar R, Pandey M,Srivastava S (2016) In silico genome wide mining of conserved and novel miRNAs in the brain and pineal gland of Danio rerio using small RNA sequencing data. Genom Data 7:46-53.
Bao HJ, Qiu HY, Kuai JX, Song CJ, Wang SX, Wang CQ, Peng HB, Han WC,Wu YP (2016) Apelin-13 as a novel target for intervention in secondary injury after traumatic brain injury. Neural Regen Res 11:1128-1133.
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297.
Cao L, Feng C, Li L, Zuo Z (2012) Contribution of microRNA-203 to the isoflurane preconditioning-induced neuroprotection. Brain Res Bull 88:525-528.
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336-338.
Cerpa W, Toledo EM, Varela-Nallar L, Inestrosa NC (2009) The role of Wnt signaling in neuroprotection. Drug News Perspect 22:579-591.
Chen C, Deng B, Qiao M, Zheng R, Chai J, Ding Y, Peng J, Jiang S (2012)Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs. PLoS One 7:e31426.
Cheng Y, Du L, Jiao H, Zhu H, Xu K, Guo S, Shi Q, Zhao T, Pang F, Jia X,Wang F (2015) Mmu-miR-27a-5p-dependent upregulation of MCPIP1 inhibits the inflammatory response in LPS-induced RAW264.7 macrophage cells. Biomed Res Int 2015:607692.
Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, Kreike B, Sie D, Hovestadt V, Wessels LF, van de Vijver MJ, Tuschl T (2011)MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res 71:4443-4453.
Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL (2012) MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A 109:18962-18967.
Hong X, Qin J, Chen R, Yuan L, Zha J, Wang Z (2016) Identification and characterization of novel and conserved microRNAs in several tissues of the Chinese rare minnow (Gobiocypris rarus) based on illumina deep sequencing technology. BMC Genomics 17:283.
Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776-780.
Ji Z, Wang G, Xie Z, Wang J, Zhang C, Dong F, Chen C (2012) Identification of novel and differentially expressed MicroRNAs of dairy goat mammary gland tissues using solexa sequencing and bioinformatics.PLoS One 7:e49463.
Jia YX, Pan CS, Zhang J, Geng B, Zhao J, Gerns H, Yang J, Chang JK,Tang CS, Qi YF (2006) Apelin protects myocardial injury induced by isoproterenol in rats. Regul Pept 133:147-154.
Kawamata Y, Habata Y, Fukusumi S, Hosoya M, Fujii R, Hinuma S,Nishizawa N, Kitada C, Onda H, Nishimura O, Fujino M (2001) Molecular properties of apelin: tissue distribution and receptor binding.Biochim Biophys Acta 1538:162-171.
Khaksari M, Aboutaleb N, Nasirinezhad F, Vakili A, Madjd Z (2012)Apelin-13 protects the brain against ischemic reperfusion injury and cerebral edema in a transient model of focal cerebral ischemia. J Mol Neurosci 48:201-208.
Li Y, Tang PF (2016) Can exosomal micro-RNAs be as biomarkers of diseases? Zhongguo Zuzhi Gongcheng Yanjiu 20:7738-7745.
Liu S, Li D, Li Q, Zhao P, Xiang Z, Xia Q (2010) MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genomics 11:148.
Longa EZ, Weinstein PR, Carlson S, Cummins R (1989) Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 20:84-91.
Martinez B, Peplow PV (2016) Blood microRNAs as potential diagnostic and prognostic markers in cerebral ischemic injury. Neural Regen Res 11:1375-1378.
Reaux A, De Mota N, Skultetyova I, Lenkei Z, El Messari S, Gallatz K, Corvol P, Palkovits M, Llorens-Cortès C (2001) Physiological role of a novel neuropeptide, apelin, and its receptor in the rat brain. J Neurochem 77:1085-1096.
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5:R13.
Shi H, Sun BL, Zhang J, Lu S, Zhang P, Wang H, Yu Q, Stetler RA, Vosler PS, Chen J, Gao Y (2013) miR-15b suppression of Bcl-2 contributes to cerebral ischemic injury and is reversed by sevoflurane preconditioning.CNS Neurol Disord Drug Targets 12:381-391.
Simpkin JC, Yellon DM, Davidson SM, Lim SY, Wynne AM, Smith CC(2007) Apelin-13 and apelin-36 exhibit direct cardioprotective activity against ischemia-reperfusion injury. Basic Res Cardiol 102:518-528.
Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, Liao Q, Guo X, Wang R,Li X, Zeng F, Wu M, Li G (2011) Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res 1390:21-32.
Tatemoto K, Takayama K, Zou MX, Kumaki I, Zhang W, Kumano K, Fujimiya M (2001) The novel peptide apelin lowers blood pressure via a nitric oxide-dependent mechanism. Regul Pept 99:87-92.
Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, Zou MX, Kawamata Y, Fukusumi S, Hinuma S, Kitada C, Kurokawa T, Onda H, Fujino M (1998) Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun 251:471-476.
Vakili A, Zahedi khorasani M (2007) Post-ischemic treatment of pentoxifylline reduces cortical not striatal infarct volume in transient model of focal cerebral ischemia in rat. Brain Res 1144:186-191.
Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, Bhattacharya A (2010) Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 11:288.
Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH (2011) miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 411:586-592.
Wang Y, Huang J, Ma Y, Tang G, Liu Y, Chen X, Zhang Z, Zeng L, Wang Y, Ouyang YB, Yang GY (2015) MicroRNA-29b is a therapeutic target in cerebral ischemia associated with aquaporin 4. J Cereb Blood Flow Metab 35:1977-1984.
Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK,Urban N, Drescher CW, Knudsen BS, Tewari M (2009) Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One 4:e5311.
Xie SS, Li XY, Liu T, Cao JH, Zhong Q, Zhao SH (2011) Discovery of porcine microRNAs in multiple tissues by a Solexa deep sequencing approach. PLoS One 6:e16235.
Xin Q, Cheng B, Pan Y, Liu H, Yang C, Chen J, Bai B (2015) Neuroprotective effects of apelin-13 on experimental ischemic stroke through suppression of inflammation. Peptides 63:55-62.
Xu M, Yang L, Hong LZ, Zhao XY, Zhang HL (2012) Direct protection of neurons and astrocytes by matrine via inhibition of the NF-kappaB signaling pathway contributes to neuroprotection against focal cerebral ischemia. Brain Res 1454:48-64.
Xu X, Chua CC, Gao J, Hamdy RC, Chua BH (2006) Humanin is a novel neuroprotective agent against stroke. Stroke 37:2613-2619.
Yan XG, Cheng BH, Wang X, Ding LC, Liu HQ, Chen J, Bai B (2015) Lateral intracerebroventricular injection of Apelin-13 inhibits apoptosis after cerebral ischemia/reperfusion injury. Neural Regen Res 10:766-771.
Zhao CZ, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ, Li MJ, Li CS, Zhang BH, Wang XJ (2010) Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol 10:3.
Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129:303-317.
- 中国神经再生研究(英文版)的其它文章
- Neuroprotective effects of statins against amyloid βinduced neurotoxicity
- Detection of thinned corticospinal tract and corticoreticular pathway in a patient with a calf circumference discrepancy
- Dyslipidemia modulates Müller glial sensing and transduction of ambient information
- Mitochondrial transplantation strategies as potential therapeutics for central nervous system trauma
- A new direction for Alzheimer’s research
- DNA plasticity and damage in amyotrophic lateral sclerosis

