Production, Purification and Characterization of Alkaline β-Glucosidase Isolated from Agrobacterium sp. PJD-1-1
YAO Jian, CHEN Qing-long, ZHONG Guo-xiang, GUI Lun, BI Wen-hui
(1. Inst. of Agric. Appl. Microbiol., Jiangxi Acad. of Agric. Sci., Nanchang 330200; 2. Dept. of Food Sci. & Engin., Shangdong Agric. & Engin. Uni., Jinan 250100)
Cellulose,amajorcomponentofplantcellwalls,isthemostabundantpolysaccharideinnatureandavirtuallyinexhaustiblesourceofrenewablebioenergy[1].Thehydrolysisofcellulosedependsonatleastthreeenzymes,includingendoglucanase(EC3.2.1.4),cellobiohydrolase(EC3.2.1.91)andβ-glucosidase(EC3.2.1.21)[2].Theendoglucanaserandomlyattackcelluloseinamorphouszonesandreleaseoligomerswhilethecellobiohydrolaseliberatecellobiosefromreducingandnon-reducingend[3],andtheβ-glucosidasehydrolyzesthedellobioseandcellooligosaccharidestoglucose,whichisessentialforthefinalstepofcellulosesaccharificationbecauseitreducestheinhibitionofendoglucanaseandcellobiohydrolase[4].Furthermore,β-glucosidaseplaysimportantrolesinmanybiologicalprocesses,suchasbiogenesisofvariousfunctionalmoleculesfromglycosideprecursors,andcyanide-basedbiologicaldefensemechanismsinplants[5].Inaddition,β-glucosidasehaspotentialapplicationsinthepharmaceutical,cosmeticanddetergentindustries.Todate,manyβ-glucosidasesfromdifferentmicroorganismshavebeenisolatedandinvestigatedfortheirphysicochemicalproperties,suchasβ-glucosidasefromPenicilliumitalicum[6],AspergillusfumigatusZ5[2],Saccharomycescerevisiae[7],Chaetomellaraphigera[8],Myceliophthorathermophila[9],Colletotrichumgraminicola[10],Fusariumproliferatum[3],Thermoanaerobacteriumthermosaccharolyticum[11],OenococcusoeniATCC BAA-1163[12]andBifdobacteriumbreveUCC2003[13]. However, the properties of β-glucosidases from these microorganisms did not always meet the requirements for a given application. In this study, anAgrobacteriumsp. strain that produces β-glucosidase with novel properties was isolated from putrefied sugarcane leaves and its production of β-glucosidase was optimized. The enzyme was purified and its biochemical properties, including molecular mass, optimum pH and temperature, metal ions were investigated.
1 Materials and methods
1.1 Materials
1.1.1 Sample sources Putrefied sugarcane leaves were used for isolating β-glucosidase producing strains.
1.1.2 Medium(g/L) ①Screening medium: lactose 5.0, NaNO32.0, K2PO31.0; KCl 0.5, MgSO40.5, FeSO40.01, esculin hydrate 1.0, ammonium iron (III) citrate 2.5, 115 ℃ sterilization for 20 min;②Fermentation medium: lactose 5.0, NaNO32.0, K2PO31.0, KCl 0.5, MgSO40.5, FeSO40.01,115 ℃ sterilization for 20 min.
1.1.3 Reagent 4-Nitrophenyl-β-D-Galactopyranoside (pNPG) was purchased from Sigma-Aldrich.
1.2 Methods
1.2.1 Isolation of β-glucosidase producingAgrobacteriumsp. PJD-1-1 strain Bacteria strains from putrefied sugarcane leaves were plated on screening medium plates, and incubated at 30 ℃ for 4 days. Isolates were maintained purely at 30 ℃ after 4 times of subsequent transfers on the same plates. For the β-glucosidase production, the isolate was cultivated in the fermentation medium. Then the culture was incubated at 20 ℃ (pH 7.0, 120 r/min) for 5 days and the culture supernatant was used for enzyme activity.
1.2.2 DNA extraction and 16S rDNA sequence analysis Genomic DNA extraction was carried out according to a method described previously[14]. Amplification of the 16S rDNA was performed by using 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-TACCTTGTTACGACTT-3′). PCR products were ligated to pGEM-T easy vector and transformed intoE.coliDH5α. The insert DNA sequencing was performed using a BigDye sequencing kit and ABI 377 DNA sequencer (Applied Biosystems, Inc.). The sequence was analyzed using the gapped BLAST (http://www. ncbi. nlm.nih.gov) search algorithms and aligned to their nearest neighbors. Phylogenetic tree was constructed by neighbor-joining method using Molecular Evolutionary Genetics Analysis 5.0 software (MAGE ver. 5.0).
1.2.3 Enzyme assay β-Glucosidase activity was determined according to the method described previously with minor modification. The reaction mixture containing 100 μL of purified β-glucosidase and 100 μL of 10 mmol/LpNPG in 50 mmol/L phosphate buffer (pH 8.0) was incubated at 50 ℃ for 15 min. Then the reaction was terminated by adding 200 μL of 200 mmol/L Na2CO3. The absorbance ofp-nitrophenol released was measured at 410 nm using a spectrophotometer. One unit of β-glucosidase activity was defined as the amount of enzyme required to release 1 μmol/L of p-nitrophenol in 1 min under specific conditions.
1.2.4 Optimization of β-glucosidase production β-Glucosidase production byAgrobacteriumsp. PJD-1-1 under different incubation time (0-8 days) in shake (120 r/min) culture conditions was investigated. Various carbon sources (0.5%, w/w) such as pectin, lactose, cellulose, avicel, xylose, carboxy methylated cellulose (CMC) as well as various nitrogen sources (0.2%, w/w) such as NH4Cl, urea, NaNO3, beef extract, yeast extract, peptone, (NH4)2SO4and different concentrations of lactose ranging from 0.25% to 2.5% (w/w) were tested for β-glucosidase production. Based on the conditions described above, a set of experiments was conducted at different temperatures (15-35 ℃) and pHs (pH 5.0-9.0) to obtain maximum β-glucosidase production.
1.2.5 Purification of β-glucosidase ①Ammonium sulfate precipitation and ultrafiltration: Culture filtrate containing β-glucosidase was saturated with gradient ammonium sulfate at 4 ℃ for 12 h. The precipitate was collected by centrifugation at 8 000 r/min for 20 min and suspended in 50 mmol/L phosphate buffer (pH 8.0). ②Gel-filtration chromatography: Five milliliter of the concentrated sample was loaded on to Sephadex G-100 column with a bed size of 1.5 cm×60 cm pre-equilibrated with 50 mmol/L phosphate buffer (pH 8.0). The column was eluted with the same buffer at a flow rate of 1 mL/min. Fractions of one milliliter size were collected and estimated at 280 nm for protein content and at 410 nm for β-glucosidase activity. The fractions containing β-glucosidase activity were pooled. ③Ion exchange chromatography: Further purification was carried out on DEAE Sepharose Fast Flow column (GE Healthcare) pre-equilibrated with 10 mmol/L phosphate buffer (pH 8.0). The column was eluted using step gradients of 0.2, 0.4, 0.6, 0.8, 1.0 mol/L NaCl in 10 mmol/L phosphate buffer (pH 8.0) at a flow rate of 0.75 mL/min. All factions were analyzed for β-glucosidase activity and fractions corresponding to the activity peaks were pooled and concentrated.
1.2.6 Enzyme characterization The molecular mass and effect of temperature, pH, and additives on β-glucosidase activity were studied and the activity was assayed as described above unless otherwise stated. All the experiments were done in triplicate for each tube at each sample condition.①Protein determination and SDS-PAGE analysis: Protein concentrations were determined according to Bradford′s method[15]with bovine serum albumin (BSA) used as standard. The molecular mass of purified β-glucosidase was analyzed by SDS-PAGE according to the method of Laemmli[16]with a 12% separating gel and 5% concentrating gel used in this study. After electrophoresis, the SDS-PAGE gel was stained with Coomassie brilliant blue G-250 for molecular mass analysis. The molecular weights of the protein were determined by the standard protein mixture of 205, 97, 66, 45 and 25 kDa. ②Optimum temperature and pH for the enzyme activity: the optimum temperature of β-glucosidase was determined at a temperature range of 25-60 ℃. The optimum pH was determined by measured β-glucosidase activity in a pH values ranging from 5.5 to 9.5 at 50 ℃. The following buffers were used: 50 mmol/L sodium acetate buffer (pH 4.0-6.0); 50 mmol/L phosphate buffer (pH 5.5-8.0); 50 mmol/L Tris-HCl buffer (pH 7.5-9.0); 50 mmol/L glycine-NaOH buffer (pH 8.5-10.0). Then the residual activity was measured as described above. ③ Effect of temperature on the stability of β-glucosidase: The thermostability of β-glucosidase was determined by incubating the purified enzyme in phosphate buffer (50 mmol/L, pH 8.0) at diferent temperatures (30, 35, 40, 45, 50, 55, 60, 65, and 70 ℃) for 1 h. Then, the residual activity was measured as described above. ④Effects of various chemical additives on β-glucosidase activity: The effects of various metal ions (Na+, K+, Mg2+, Ca2+, Mn2+, Ba2+, Fe2+, Co2+, Ni2+, Pb2+, Cu2+, and Fe3+), ethylene diamine tetraacetic acid (EDTA), urea, and sodium dodecyl sulfate (SDS) on β-glucosidase activity were investigated. The enzyme was incubated with 1 mmol/L of the respective chemicals at room temperature for 0.5 h. Then, the residual activity was assayed as described above.
2 Results
2.1 Isolation and identification of β-glucosidase producing bacteria
A β-glucosidase producing bacteria strain named PJD-1-1 was isolated from putrefied sugarcane leaves. Comparison of almost complete 16S rRNA sequence of PJD -1-1 against the GenBank database indicates that PJD-1-1 was included in the genusAgrobacterium. Strain PJD-1-1 shares highest percentage nucleotide sequence similarities withAgrobacteriumsp. 2382 (identity of 100%),Agrobacteriumtumefaciensstrain 12b3 (identity of 100%),Agrobacteriumtumefaciensstrain D19(identity of 100%) andAgrobacteriumsp. APW21 (identity of 100%). A phylogenetic tree illustrating the relationship of strain PJD-1-1 to related bacterial species was shown in Fig.1, and the selected strain was indentified asAgrobacteriumsp. PJD-1-1 on the basis of 16S rRNA sequence.

Fig.1 Phylogenetic tree derived from the analysis of 16S rDNA sequence using neighbor-joiningmethod, showing the relationship among Agrobacterium sp. PJD-1-1 and representatives of some related texa
2.2 Optimization of parameters for the enzyme production
Various carbon sources such as pectin, lactose, cellulose, avicel, xylose, CMC and nitrogen sources such as NH4Cl, urea, NaNO3, beef extract, yeast extract, peptone (NH4)2SO4were tested for β-glucosidase production byAgrobacteriumsp.PJD-1-1. All substances induced β-glucosidase production at different levels. β-Glucosidase activity reached the highest level with lactose as carbon source, followed by avicel, cellulose, pectin, CMC and xylose (Fig.2A). Lactose at the concentration of 0.5% showed the maximum enzyme production, and further increase caused a decrease in enzyme production (Fig.2B). With lactose as carbon source, NaNO3as nitrogen source showed maximum enzyme production, while other inorganic nitrogen sources result in least production of the enzyme. Among the organic nitrogen sources, peptone was found to be the best for enzyme production, while urea resulted in least enzyme production (Fig.2C). Besides carbon and nitrogen sources, initial pH, incubation temperature, and incubation time also influenced enzyme production. Initial pH of 7.0 was found to be the optimum pH for enzyme production, an increase and decrease in pH would cause the decrease in the enzyme production (Fig.2D). Temperature of 20 ℃ was found to be the optimum temperature for enzyme production and higher temperature was found to reduce the enzyme production (Fig.2E). TheAgrobacteriumsp.PJD-1-1 was found to have maximum enzyme production when it was incubated at pH 7.0 and 20 ℃ for 5 days, longer incubation time would decrease enzyme activity (Fig.2F).

Fig.2 Effect of carbon sources (0.5%) (A), lactose concentration (B), nitrogen sources (C), pH (D), temperature (E) and incubation time (F) on enzyme production
2.3 Partial purification of β-glucosidase
The strainAgrobacteriumsp.PJD-1-1 secreted β-glucosidase into the culture filtrate when grown in liquid medium and the extracellular extract was harvested after 5 days incubation. The partial purification process was summarized in Table 1. In first step of the purification with ammonium sulfate fractionation, about 75.5% of total β-glucosidase activities were recovered in the fraction of 20%-60% ammonium sulfate. This step removed the greater part of the contaminants and decreased total protein amount from 12.09 mg to 5.072 mg. The precipitate with β-glucosidase was dissolved in 10 mmol/L phosphate buffer (pH 8.0) and further purified with gel-filtration chromatography and ion exchange chromatography, retaining 10.6% of the activity from the previously step. SDS-PAGE analysis of the purified enzyme showed the presence of a single band with an apparent molecular mass of ca.40 kDa when stained with Coomassie brilliant blue (Fig.3), so further purification steps were not required.
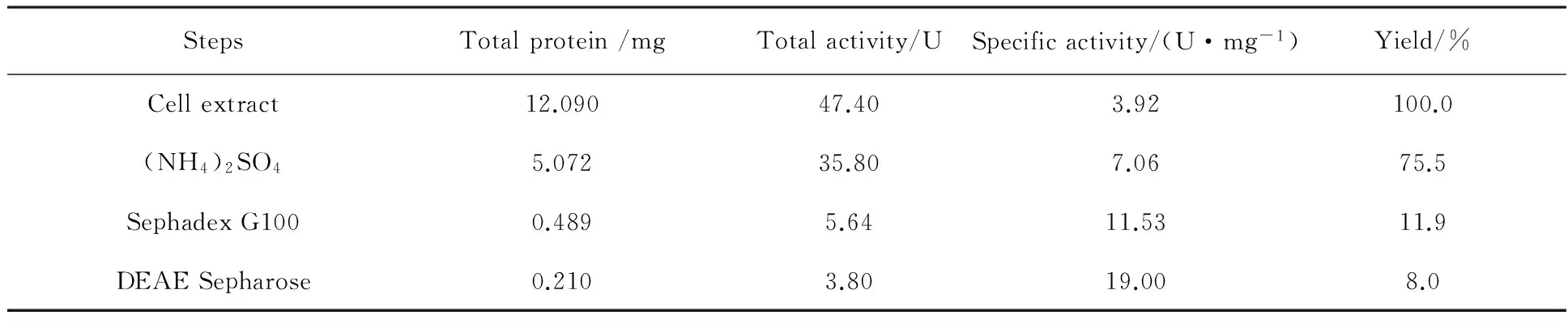
Table 1 Purification of β-glucosidase from Agrobacterium sp. PJD -1-1

Fig.3 SDS-PAGE analysis of the purified enzyme
2.4 Effect of pH and temperature on β-glucosidase activity
The effect of pH and temperature on β-glucosidase activity was measured at a pH rang of 5.5 to 9.5 and a temperature rang of 25 to 60 ℃. The enzyme showed its highest activity at pH of 8.0 (Fig.4A) and temperature of 50 ℃ (Fig.4B). Thermal stability analysis revealed that the enzyme was stable below 50 ℃, no obvious loss of activity was found when incubated below 50 ℃, and it kept more than 70% of its activity after incubation at 60 ℃ for 30 min (Fig.4C).
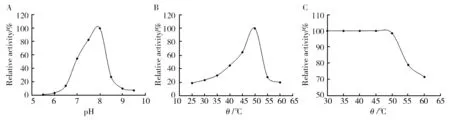
Fig.4 Effect of pH (A), temperature (B) on enzyme activity and temperature on the stability (C) of enzyme
2.5 Effects of different chemicals on β-glucosidase activity
Effects of different chemicals on activity of β-glucosidase was tested by pre-incubating the enzyme with 1 mmol/L of each chemical, respectively, at room temperature for 30 min. Ba2+, Ca2+, Mn2+and urea only slightly affected the enzyme activity compared with that in the absence of any added chemicals (Table 2). The relative activity of the enzyme was decreased to 64.1% and 21.2% by the presence of Ni2+, Cu2+, respectively. Hg2+and Ag+strongly affected the enzyme activity to 11.2% and 9.4%, respectively.

Table 2 Effect of chemicals on β-glucosidase activity from Agrobacterium sp. PJD -1-1
3 Discussion
β-Glucosidase plays a major role in the conversion of cellulosic biomass to ethanol, being the rate-limiting enzyme that determines the action of all cellulose components. A large number of β-glucosidases from microorganism have being produced, purified and studied, but most of these enzymes show maximal activity in the acidic pH range (pH 4.0-6.5)[17]. In this study, a new strainAgrobacteriumsp. PJD-1-1 producing β-glucosidase with its optimum activity at pH 8.0 was isolated. As carbon and nitrogen are energy sources which are essential for the growth of microorganisms. The effects of supplementation of different carbon and nitrogen sources viz. lactose, pectin, cellulose, avicel, xylose and CMC for carbon sources, and NH4Cl, urea, NaNO3, beef extract, yeast extract, peptone (NH4)2SO4for nitrogen sources on production of β-glucosidase were evaluated. The resulted showed that lactose and NaNO3at the concentration of 0.5% and 0.2%, separately, obtained maximum enzyme production. Different to our results, Saha et al.[18]reported that β-glucosidase fromCandidapeltataobtained its highest activity with xylose as carbon sources. Besides carbon and nitrogen sources, initial pH, incubation temperature and time were also tested for enzyme production. TheAgrobacteriumsp. PJD-1-1 in this study was found to have maximum enzyme production when it was incubated at pH 7.0 and 20 ℃ for 5 days. However, the enzyme fromCandidapeltataincreased up to 4 days when it was incubated at 28 ℃, after which it decreased gradually[18].
Upon fractionation of the β-glucosidase active fractions with ammonium sulfate, 75.5% of total β-glucosidase activities were recovered in the fraction of 20%-60% ammonium sulfate. After gel filtration chromatography on Sephadex G-100 column, the enzyme was found in fractions 20-60 tubes and pooled. Anion-exchange chromatography of the combined active fraction on a DEAE Sepharose column removed most of the contaminants, and the effluent containing β-glucosidase activity was concentrated by ultrafiltration. The enzyme was purified 4.85- fold to homogeneity with overall enzyme yield of 8.0% and a specific activity of 19.0 U/mg proteins. SDS-PAGE analysis of the purified enzyme showed a single band with molecular mass of ca.40 kDa which was lower than most of analogous β-glucosidases[2,18-21]. The optimum temperature of purified enzyme was 50 ℃, which was comparable to that of β-glucosidases fromSporidioboluspararoseus(55 ℃)[21],Rhynchophoruspalmarum(55 ℃)[1],Pichiaetchellsii(50 ℃)[22],Candidasake(52 ℃)[23], andAspergillusoryzae(50 ℃)[24], higher than that of β-glucosidases fromWickerhamomycesanomalus(35 ℃)[25],Pichiapastoris(40 ℃)[26], andGalleriamellonella(42 ℃)[27], but lower that of β-glucosidases fromThermoanaerobacteriumthermosaccharolyticum(70 ℃)[11],Penicilliumitalicum(65 ℃)[6],Aspergillusunguis(60 ℃)[17]. Most β-glucosidases characterized so far show maximal activity in the acidic pH range (pH 4.0-6.5), while the β-glucosidase screened in this study showed its highest activity at alkaline conditions with optimum pH of 8.0.
The effects of various mental ions and agents on the enzyme activity were tested. Ba2+, Ca2+, Pb2+, Co2+, Zn2+, Mn2+, Na+, K+, urea and EDTA were slightly effected the enzyme activity which was consistent with the properties of β-glucosidase fromAspergillusniger[27]. However, the activity of β-glucosidases fromPenicilliumfuniculosumandSporidioboluspararoseuswas increased by Mn2+, K+and Na+[20-21],Zn2+and SDS strongly inhibited the activity of β-glucosidase fromSporidioboluspararoseus[21]while K+, Ca2+, EDTA and Ba2+enhanced the activity of β-glucosidase from a marineStreptomycete[19]. The activity of β-glucosidase in this study was heavily inhibited by Cu2+, Hg2+and Ag+. Cu2+and Hg2+also inhibit HGT-BG[24]and BglA[29], suggesting that the active catalytic sites of these enzymes might possess thiol groups that cause sensitivity to inhibition by Hg2+. However, the activity of β-glucosidase fromMyceliophthorathermophilawas activated by Cu2+[9].
In conclusion, a novelAgrobacteriumsp. PJD-1-1 producing β-glucosidase was isolated. The high level of enzyme production on lactose and NaNO3as carbon and nitrogen sources, respectively, and it showed its optimum activity in the alkaline pH range and also showed considerable thermal stabilities, which make it an interesting candidate for biological applications.
Reference
[1] Yapi DYA, Gankri D, Niamke SL, et al. Purification and biochemical characterization of a specific β-glucosidase from the digestive fluid of larvae of the palm weevil,Rhynchophoruspalmarum[J]. J Insect Sci, 2009, 9(4): 1-13.
[2] Liu DY, Zhang RF, Yang XM, et al. Characterization of a thermostable β-glucosidase fromAspergillusfumigatusZ5, and its functional expression inPichiapastorisX33[J]. Microb Cell Fact, 2012, 11(1):25.
[3] Gao ZQ, Hop DV, Yen LTH, et al. The production of β-glucosidases byFusariumproliferatumNBRC109045 isolated from Vietnamese forest [J]. AMB Express, 2012, 2(1): 1-13.
[4] Uchima CA, Tokudag, Watanabe H, et al. Heterologous expression inPichiapastorisand characterization of an endogenous thermostable and high-glucose-tolerant β-glucosidasefrom the termiteNasutitermestakasagoensis[J]. Appl Environ Microb, 2012, 78(12): 4288-4293.
[5] Uchiyama T, Miyazake K, Yaoi K. Characterization of a novel β-glucosidase from a compost microbial metagenome with strong transglycosylation activity[J]. J Biol Chem, 2013, 288(25): 18325-18334.
[6] Park A, Hong JH, Kim J-J, et al. Biochemical characterization of an extracellular β-glucosidase from the fungus,Penicilliumitalicum, isolated from rotten citrus peel[J]. Mycobiology, 2012, 40(3): 173-180.
[7] Schmidt S, RAinieri S, Witte S, et al. Identification of aSaccharomycescerevisiaeglucosidase that hydrolyzes flavonoid glucosides [J]. Appl Environ Microb, 2011, 77(5): 1751-1757.
[8] Yoneda A, Kuo H-W D, Ishihara M, et al. Glycosylation variants of a β-glucosidase secreted by a taiwanese fungus,Chaetomellaraphigera, exhibit variant-specific catalytic and biochemical properties[J]. PLoS ONE, 2014, 9(9): e106306.
[9] Karnaouri A, Topakas E, Paschos T, et al. Cloning, expression and characterization of an ethanol tolerant GH3 β-glucosidase fromMyceliophthorathermophila[J]. Peer J, 2013, 1:e46.
[10] Zimbardi ALRL, Sehn C, Meleiro LP, et al. Optimization of β-glucosidase, β-xylosidase and xylanase production byColletotrichumgraminicolaunder solid-state fermentation and application in raw sugarcane trash saccharification[J]. Int J Mol Sci, 2013, 14(2): 2875-2902.
[11] Pei JJ, Pang Q, Zhao LG, et al.Thermoanaerobacteriumthermosaccharolyticumβ-glucosidase: a glucose-tolerant enzyme with high specific activity for cellobiose[J]. Biotechnol Biofuels, 2012, 5(1): 31.
[12] Michlmayr H, Schümann C, Wurbs P, et al. A β-glucosidase fromOenococcusoeniATCC BAA-1163 with potential for aroma release in wine: cloning and expression inE.coli[J]. World J Microb Biot, 2010, 26(7): 1281-1289.
[13] Pokusaeva K, Connell-motherway MO, Zomer A, et al. Characterization of two novel β-glucosidases fromBifidobacteriumbreveUCC2003[J]. Appl Environ Microb, 2009, 75(4): 1135-1143.
[14] Yao J, Fan X-J, Lu Y, et al. Isolation and characterization of a novel tannase from a metagenomic library[J]. J Agr Food Chem, 2011, 59(8): 3812-3818.
[15] Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding[J]. Anal Biochem, 1976, 72:248-254.
[16] Laemmli UK,King J. Polypeptides of the tail fibers of bacteriophage T4[J]. J MolCell Biol, 1971, 62(3): 465-477.
[17] Rajasree KP, Mathew GM, Pandey A, et al. Highly glucose tolerant β-glucosidase fromAspergillusunguis: NII 08123 for enhanced hydrolysis of biomass[J]. World J Microb Biot, 2013, 40(9):967-975.
[18] Saha BC, Bothast R J. Production, purification, and characterization of a highly glucose-tolerant novel β-glucosidase fromCandidapeltata[J]. Appl Environ Microb,1996, 62(9): 3165-3170.
[19] Mai ZM, Yang J, Tian XP, et al. Gene cloning and characterization of a novel salt-tolerant and glucose-enhanced β-glucosidase from a marineStreptomycete[J]. Appl Biochem Biot, 2013, 169(5):1512-1522.
[20] Ramani G, Meera B, Vanitha C, et al. Molecular cloning and expression of thermostable glucose tolerant β-glucosidase ofPenicilliumfuniculosumNCL1 inPichiapastorisand its characterization[J]. J Ind Microbiol Biot, 2015, 42(4):553-565.
[21] Baffi M A, Martin N, Tobal TM, et al. Purification and characterization of an ethanol-tolerant β-glucosidase fromSporidioboluspararoseusand its potential for hydrolysis of wine aroma precursors [J]. Appl Biochem Biot, 2013, 171(7):1681-1691.
[22] Wallecha A, Mishra S. Purification and characterization of two β-glucosidases from a thermo-tolerant yeastPichiaetchellsii[J]. BBA-Proteins Proteom, 2003, 1649 (1):74- 84.
[23] Gueguen Y, Chemardin P, Arnaud A. Purification and Characterization of an intracellular β-glucosidase from aCandidasakestrain isolated from fruit juices[J]. Appl Biochem Biot, 2001, 95(3): 151-162.
[24] Riou C, Salmon J-M, Vallier M-J, et al. Purification, characterization, and substrate specifi city of a novel highly glucose-tolerant β-glucosidase fromAspergillusoryzae[J]. Appl Environ Microb, 1998, 64(10): 3607-3614.
[25] Restuccia C, Muccilli S, Palmeri R, et al. Analkaline β-glucosidase isolated froman olive brine strain ofWickerhamomycesanomalus[J]. FEMS Yeast Res, 2011, 11(6): 487-493.
[26] Turan Y, Zheng M. Purification and characterization of an intracellular β-glucosidase from theMethylotrophicYeast Pichia pastoris[J]. Biochemistry (Moscow), 2005, 70(12):1363-1368.
[27] Turan Y, Er A, Acar M, et al. Purification and characterization of β-glucosidase from greater wax mothGalleriamellonellal. (lepidoptera: pyralidae)[J]. Arch Insect Biochem, 2014, 86(4): 209-219.
[28] Yan T-R, Lin C-L. Purification and characterization of a glucose-tolerant β-glucosidase formAspergillusnigerccrc 31494[J]. Biosci Biotech Bioch, 1997, 61(6): 965-970.
[29] Eric P, SalvadoR V, Julio P, et al. Purification and characterization of aBacilluspolymyxabeta-glucosidase expressed inEscherichiacoli[J]. J Bacteriol, 1992, 174(9):3087-3091.