仔猪肠道微生物研究进展
夏耀耀,任文凯*,黄瑞林,曾本华,魏泓,印遇龙*
(1.中国科学院亚热带农业生态研究所畜禽健康养殖研究中心,中国科学院亚热带农业生态过程重点实验室,畜禽养殖污染控制与资源化技术国家工程实验室,湖南省畜禽健康养殖工程技术研究中心,农业部中南动物营养与饲料科学观测实验站,长沙 410125; 2.中国科学院大学,北京 100049; 3.第三军医大学基础部实验动物学教研室,重庆 400038)
研究进展
仔猪肠道微生物研究进展
夏耀耀1,2,任文凯1,2*,黄瑞林1,曾本华3,魏泓3,印遇龙1*
(1.中国科学院亚热带农业生态研究所畜禽健康养殖研究中心,中国科学院亚热带农业生态过程重点实验室,畜禽养殖污染控制与资源化技术国家工程实验室,湖南省畜禽健康养殖工程技术研究中心,农业部中南动物营养与饲料科学观测实验站,长沙 410125; 2.中国科学院大学,北京 100049; 3.第三军医大学基础部实验动物学教研室,重庆 400038)
肠道菌群在哺乳动物及人类健康的作用正日益受到重视。仔猪的健康生长需要一个动态平衡的肠道微生态环境。然而,在猪的生命周期中,从食管到直肠的微生物分布与组成存在时间和空间的变化。健康的肠道菌群具有促进猪的营养代谢,维持肠黏膜屏障,调节免疫应答,抑制病原菌感染等功能。多种因素对猪肠道菌群的形成与稳定具有重要的作用,包括分娩方式(经过产道或剖宫产)、幼龄时期饮食(母乳或配方饲料)、抗生素或抗生素样分子的使用等。本文主要从仔猪肠道微生物组成与定植、功能、影响肠道微生物的因素等方面论述了仔猪肠道微生物与仔猪肠道健康的关系,从而加深肠道微生物对维护仔猪肠道健康作用的认识。
肠道微生物菌群;肠道健康;仔猪
哺乳动物的肠道中存在大量的微生物[1-4]。肠道微生物间以及宿主和肠道微生物之间存在着复杂的动态平衡关系。多年来,研究发现单胃动物的肠道生理功能受其肠道菌群的影响[5,6]。仔猪在出生后,通过接触源自外界以及母体的各种微生物,其胃肠道逐渐形成一个复杂的微生态环境。这种复杂的微生态环境对仔猪的健康生长极其重要。保持动态平衡的肠道微生态环境不仅有助于仔猪对营养物质的消化、吸收,还能形成微生物屏障以阻止病原菌的入侵并促进其肠道发挥免疫功能[7]。
营养因素与肠道菌群之间存在复杂的关系[8]。肠道微生物会影响宿主对营养物质的消化和吸收,同时,微生物的发酵产物可为宿主提供营养成分或者干预宿主的健康。此外,某些微生物代谢产物(丁酸)如具有特定的生物活性,可以影响肠道微生物的数量与组成[9],或者对宿主有潜在的不利影响[10]。仔猪出生后,首先通过吮食母乳在肠道内建立一个相对稳定的微生态环境,而在断奶阶段,由于产生断奶应激以及日粮成分发生改变,其肠道内的微生态环境会发生巨大的变化[11]。本文主要从仔猪不同生长阶段和肠道不同部位的微生物组成、定植情况以及影响肠道微生物的因素等方面,阐述仔猪肠道微生物与仔猪肠道健康的关系,旨在为健康养殖和仔猪肠道健康生态营养调控等方面研究提供帮助。
1 仔猪肠道微生物的定植与组成
新生仔猪肠道处于无菌状态,仔猪通过与外界环境和母亲的产道、粪便接触[12],造成需氧、兼性厌氧和专性厌氧菌在其肠道内按照一定次序定植[13]。这是因为随着需氧菌和肠杆菌、肠球菌等兼性厌氧菌的定植,胃肠道的氧气被消耗完毕,继而逐渐形成厌氧环境,导致专性厌氧菌开始定植和生长。仔猪出生2 d后,乳酸杆菌等便逐渐替代需氧菌成为其肠道内的优势菌群。整个哺乳期阶段,每克食糜所含乳酸杆菌的数量保持在107~109范围内[14]。在仔猪的整个生命过程中,由于断奶应激、仔猪日粮营养物质的变化、胃肠道环境在不同生理阶段的改变以及肠道内特定微生物的定植位点发生转变等,其肠道微生物区系的种类和数量也发生变化[13]。据报道,哺乳仔猪肠道中乳酸杆菌的数量较断奶仔猪更高,而大肠杆菌数量则相反[15],这是由于断奶仔猪胃内pH值升高,乳酸杆菌减少,使大肠杆菌开始大量增殖,进而抑制乳酸杆菌的生长。哺乳仔猪胃内pH值较低,会导致经口病原菌无法像断奶仔猪中的病原菌那样通过胃部而定植于肠道内[16]。Konstantinov等[17-19]研究发现,仔猪断奶后回肠中乳酸杆菌数量较断奶前相比显着下降,大肠杆菌数量却显着增加。在仔猪断奶后的2周,其肠道的生理及微生态环境会发生巨大的变化[20,21]。
有研究表明,仔猪不同胃肠道部分的微生物分布和组成有很大差异。胃和小肠内的优势菌群主要是梭菌IX群、链球菌和乳酸杆菌等,而后肠部位分布着多样性更高的菌群,其中盲肠、结肠和直肠内优势菌群以厚壁菌门中的梭菌IV群、XIV群和拟杆菌门细菌为主[22]。
2 肠道微生物功能
宿主的首要任务是抵御肠道微生物引起的持续性感染。尽管肠道微生物具有多种有益于宿主健康的功能,如碳水化合物的消化和发酵、合成维生素、维持肠绒毛的正常功能、调节免疫反应以及保护肠道免受病原菌感染[23,24],然而,肠道微生物也会通过其代谢产物、基因产物或潜在致病性给宿主带来不利影响[25]。因此,肠道微生物的有益功能和对宿主产生的危害之间的平衡取决于其分布、多样性以及代谢产物的总体状态(如图1)。
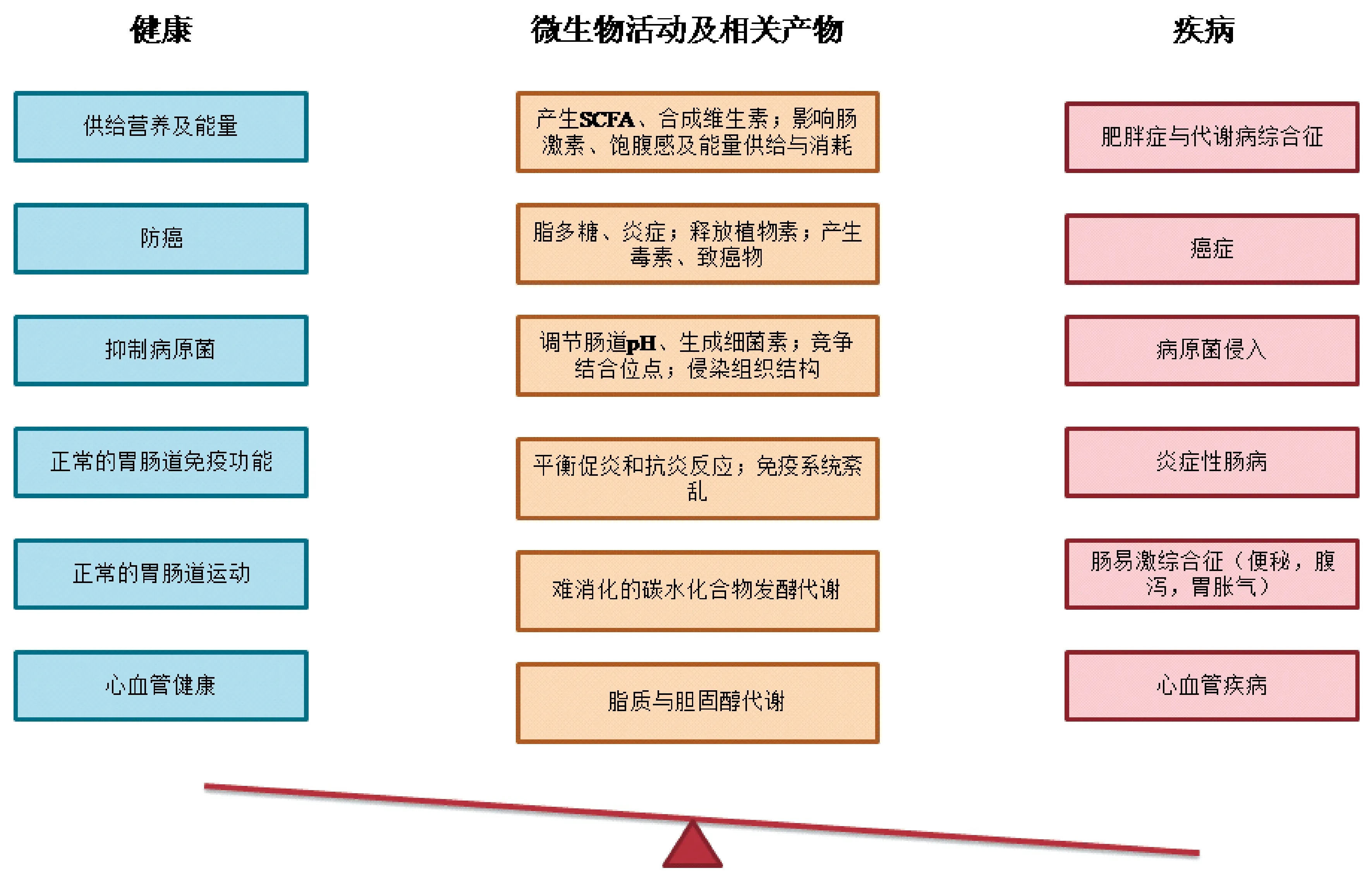
图1 肠道菌群对肠道及宿主健康的影响[26]Fig.1 Effects of intestinal microbiota on intestinal and host health[26]
2.1 营养物质代谢
日粮碳水化合物是肠道菌群最主要的营养来源。未经近端小肠消化的碳水化合物及结肠内微生物(如拟杆菌属、双歧杆菌属和肠杆菌等)难以消化的寡糖经过发酵生成乙酸、丙酸、丁酸等短链脂肪酸[27,28]。拟杆菌属是参与碳水化合物代谢的主要微生物,它通过表达糖基转移酶、糖苷水解酶和多糖酶等酶来实现这一功能。肠道菌群也通过抑制脂肪细胞的脂肪酶活性对脂质代谢产生积极影响。此外,多形拟杆菌可以通过上调胰脂肪酶消化脂质时所需的辅脂酶的表达来增加脂质的水解效率[29]。
肠道菌群具有有效的蛋白质代谢机制,即通过微生物蛋白酶和肽酶与宿主蛋白酶产生协同作用。一些基因产物可将氨基酸转化成小信号分子和抗微生物肽,如通过细菌hdcA基因编码的组胺脱羧酶可将L-组氨酸转化成组胺[30],通过细菌gadB基因编码的谷氨酸脱羧酶可将谷氨酸转化为γ-氨基丁酸[31]。
肠道菌群还能合成维生素K和部分维生素B。研究表明,拟杆菌能合成具有抗糖尿病、抗动脉粥样硬化、降血脂、免疫调节等特性的共轭亚油酸[32-34]。正常的肠道菌群还能增加血清中丙酮酸、柠檬酸、延胡索酸和苹果酸的浓度来促进代谢[35]。
2.2 抗菌保护作用
肠道菌群能够通过提高对病原菌的定植抑制和促进对病原菌的清除这两个作用来减弱肠道病原菌感染[36,37]。如在长期使用抗生素后,通常伴随着肠道固有菌群的破坏,导致肠道微生物对艰难梭菌的定植抑制作用丧失[37-39]。成功治疗艰难梭菌感染的策略是来自健康个体的粪便微生物移植,在粪便移植后,其肠道菌群结构得以恢复,从而抑制艰难梭菌[40,41]。肠道菌群还可通过调控宿主先天和获得性免疫应答来影响肠道感染[42,43]。如无菌小鼠由于其吞噬细胞活化功能及积聚到感染部位的功能受损,导致其对李斯特菌感染非常敏感[43]。本课题组前期研究也表明,肠产毒素性大肠杆菌感染时,肠道L.lactissubsp.lactis增加,通过产生过量γ-氨基丁酸而激活mTORC1-S6K1-EGR-2-GFI-1信号通路从而促进IL-17表达[44]。然而,有一些肠道病原菌感染,肠道菌群在感染期间几乎不会减弱病原菌的感染性或者反而使其增强[45,46]。因此,肠道菌群在肠道感染中所起的作用可能不仅取决于感染模式还取决于入侵的病原体。
2.3 免疫调节
肠道菌群与宿主免疫系统的发育有关[47]。与正常小鼠相比,无菌小鼠体内T 细胞数量更少,sIgA和抗体等水平也较低,而先天免疫反应细胞更活跃且体内溶菌酶活性更强[48,49]。分节丝状菌可诱导调节性T细胞反应[50,51],而Foxp3+T细胞对肠道炎症反应具有重要的调节作用[52]。肠道微生物对于Foxp3+T细胞的正常发育和功能也是必不可少的,然而,这种介导的机制尚不清楚[53,54]。研究发现,调节性T细胞和梭状芽孢杆菌均呈高密度状态分布在近端结肠,而无菌小鼠体内调节性T细胞的水平可因梭状芽胞杆菌的定植得以恢复[56]。有报道称细菌中的LPS可与其结合蛋白LBP、CD14结合形成三联复合物(LPS-LBP-sCD14),作用于TLR4,激活MyD-88依赖性信号通路机制,从而刺激T 细胞增生[55]。
肠道黏膜相关淋巴组织主要由派伊尔结、孤立淋巴滤泡和肠系膜淋巴结等部分构成[57]。肠道菌群可作为抗原刺激派伊尔结和肠系膜淋巴结等发育成熟,并且其代谢物及组成也会影响相关淋巴组织产生免疫反应[58-60]。脆弱类拟杆菌等能激活肠道树突状细胞诱导肠道黏膜中的浆细胞产生sIgA[61]。无菌仔猪体内分泌IgA 的浆细胞发育不成熟,因此血清中IgA水平极低,但在ASF (altered Schaedler flora)定植其肠道后,血清IgA水平得到极大提升[62,63]。本试验室前期研究表明添加谷氨酰胺可促进含有正常肠道微生物的小鼠分泌sIgA,然而在抗生素清除肠道微生物的小鼠模型下,则无影响,进一步表明肠道微生物在肠道sIgA分泌中的作用[64]。
先天淋巴细胞可快速对上皮细胞产生的信号做出响应[65],具有类似Th1、Th2及Th17细胞的表达模式,但其分化更依赖于微生物组成而非体细胞重组[66]。肠道微生物可直接或间接地调控先天淋巴细胞,如细菌代谢物吲哚-3-甲醛可通过芳烃受体直接刺激先天淋巴细胞合成IL-22[67],而间接调控则通过招募其他免疫细胞(如肠巨噬细胞CX3CR1+)来实现[68]。固有层巨噬细胞的免疫调节作用是在稳定的状态下表达IL-1β前体,这有助于在病原体入侵过程中肠道免疫系统迅速产生成熟的IL-1β。由共生菌群诱导激活的MyD-88依赖机制对上述作用至关重要,而微生物菌群调节巨噬细胞产生的IL-10则需要MyD-88的非依赖机制[69,70]。
2.4 胃肠道结构和肠道屏障的完整性
目前已有足够的证据证明肠道菌群能够维持胃肠功能和结构的完整性。据报道,多形拟杆菌可诱导小富脯氨酸蛋白2 A表达,维持上皮绒毛中细胞桥粒正常生长[71]。微生物细胞壁肽聚糖可刺激TLR2和维持紧密连接[72]。此外,鼠李糖乳杆菌GG可产生p40和p75这两种可溶性蛋白,其可以避免依赖于上皮细胞生长因子受体EGFR、蛋白激酶C这两条途径的肠上皮细胞发生程序性凋亡[73]。内源性大麻素系统同样能调节肠道菌群以维持肠道屏障功能。例如,革兰氏阴性菌Akkermansiamuciniphilla可通过减少血氧毒素的代谢来增加内源性大麻素的水平,从而控制肠道屏障的功能[74]。
肠道微生物通过诱导与肠道微血管系统发育相关的转录因子血管生成素-3的生成来促进肠黏膜结构的发育,该结论也已在多个无菌小鼠试验中得到验证[75-79]。肠道菌群也可在细胞表面和亚细胞水平上调节黏膜上微生物附着位点的糖基化模式,如由多形拟杆菌分泌的一种信号分子能刺激位于细胞表面糖缀合物的海藻糖基团的表达[80]。
3 影响仔猪肠道菌群的因素
在仔猪整个动态持续的生命过程中,多种因素会对其肠道菌群的形成与组成造成影响。
3.1 仔猪不同生长阶段
猪肠道菌群是一个非常复杂的生态系统,其组成成分动态且多样,并会随时间及肠段的变化而发生转变[81]。肠道菌群从仔猪出生开始就产生定植,仔猪通过消耗母乳为乳酸菌群体提供营养优势,从而构建乳汁依赖型的菌群[82]。大肠杆菌和链球菌属细菌能够在肠道内建立一个有利于拟杆菌属、乳杆菌属和梭菌属等专性厌氧细菌定植的厌氧环境[83]。依据哺乳动物进行的一项研究表明,母乳喂养和宿主遗传学的转变对肠道菌群的发展形成有很大的影响[84]。因此,哺乳期为肠道菌群的变化提供了一个特殊的渠道窗口。
3.2 日粮营养成分
在断奶阶段,仔猪开始采食谷物和粗蛋白浓度相对较高的饲料。许多研究表明,仔猪断奶时,其肠道中的乳酸菌属细菌含量减少且微生物多样性降低,然而梭菌属、普氏菌属、大肠杆菌和包括变形杆菌科在内的一些兼性厌氧菌含量升高。不同来源和水平的蛋白或纤维会影响断奶仔猪肠道微生物的相对丰度[85,86]。如断奶仔猪采食富含果胶和豆粕的日粮,会导致结肠中乳酸杆菌的相对丰度降低,普氏菌属相对丰度增加[87],而采食富含鱼粉的日粮,则会使得埃希氏菌和志贺氏杆菌的含量增加[88]。许多试验表明,哺乳动物肠道微生物的数量同样受到日粮中不同脂肪来源及成分的影响。刘忠臣[89]发现在日粮中添加椰子油、鱼油和猪油均可扰乱肠道菌群平衡,具体表现为降低盲肠内容物中大肠杆菌的数量,增加乳酸杆菌、双歧杆菌的数量以及乳酸杆菌/大肠杆菌、双歧杆菌/大肠杆菌的比值,但三种不同的脂肪造成的影响具有差异性。本组研究也发现,日粮中单一氨基酸的比例变化(如精氨酸或谷氨酰胺)也显着影响肠道微生物的数量[90,91]。在断奶仔猪日粮中添加高铜能够抑制仔猪肠道有害菌的正常生长,添加氧化锌却可以维持仔猪肠道正常的微生态环境;而在断奶仔猪日粮中添加高锌,则会降低仔猪胃肠道前段及回肠中乳酸杆菌的数量,提高大肠杆菌和肠球菌的数量[92]。
3.3 抗生素、微生态制剂及其他物质
饲用抗生素具有广谱抗菌性以及潜在的杀死或阻止病原菌和有益微生物生长的能力,可以引起断奶仔猪肠道菌群发生改变[82,93,94],微生物群落的多样性可能会进一步下降[93,95]。长期使用亚治疗剂量的抗生素会导致病原菌在肠道中定植并引发疾病[96]。
胡远亮[97]研究表明,益生菌能增加断奶仔猪肠道微生物的多样性。Mair等[98]通过给断奶仔猪饲喂肠球菌、乳酸杆菌、双歧杆菌和菊粉得出上述有益菌和菊粉二者组成的合生元制剂可在一定程度上调节仔猪的胃肠道功能,如降低胃、空肠和回肠中的pH值,影响其肠道菌群的生长和组成。Ren等[99]发现褪黑素可提高肠道乳酸杆菌的比例及代谢,并能减少肠产毒素大肠杆菌在肠道的定植。除上述影响因素,其他物质,如酶、植物提取物、酸化剂等通常在仔猪肠道菌群的形成和功能中也发挥着重要的作用[100-102]。
4 展望
驻留在人和动物肠道内的微生物与宿主之间存在着一种复杂的互惠共生关系。目前,虽然有许多研究报道了肠道微生物与宿主健康相关的功能,但是我们只对其中的一些基本功能有一个大致的了解。许多问题都值得我们深入研究,如与宿主营养物质代谢相关的特定微生物的作用机制;微生物组成及其代谢产物与机体免疫细胞种类、数量和分化的关系;机体表面(除肠道外)存在的微生物是否影响宿主健康以及与肠道微生物间是否存在互作关系等。
微生物平衡的破坏可影响肠道的健康进而导致宿主的疾病,通过维持肠道微生物的平衡可以保障肠道的健康,减少仔猪腹泻,从而促进养猪业的健康稳定发展。因此,深入研究肠道微生物与肠道健康相关的调控机制,不仅对维护仔猪健康具有重要的意义,还为我们预防人类相关疾病提供了更宽广的思路。
[1] Moore WE,Cato EP,Holdeman LV.Some current concepts in intestinal bacteriology [J].Am J Clin Nutr,1978,31(10 Suppl): S33-S42.
[2] Backhed F,Ley RE,Sonnenburg JL,et al.Host-bacterial mutualism in the human intestine [J].Science,2005,307(5717): 1915-1920.
[3] Louis P,Flint HJ.Diversity,metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine [J].FEMS Microbiol Lett,2009,294(1): 1-8.
[4] Savage DC.Microbial ecology of the gastrointestinal tract [J].Annu Rev Microbiol.1977,31: 107-133.
[5] Cummings JH,Macfarlane GT.Colonic microflora: nutrition and health [J].Nutrition,1997,13(5): 476-478.
[6] Roberfroid MB,Bornet F,Bouley C,et al.Colonic microflora: nutrition and health.Summary and conclusions of an International Life Sciences Institute (ILSI) [Europe] workshop held in Barcelona,Spain[J].Nutr Rev,1995,53(5): 127-130.
[7] Blaut M,Clavel T.Metabolic diversity of the intestinal microbiota: implications for health and disease [J].J Nutr,2007,137(3 Suppl 2): 751S-755S.
[8] Edwards C.Interactions between nutrition and the intestinal microflora [J].Proc Nutr Soc,1993,52(2): 375-382.
[9] Augeron C,Laboisse CL.Emergence of permanently differentiated cell clones in a human colonic cancer cell line in culture after treatment with sodium butyrate [J].Cancer Res,1984,44(9): 3961-3969.
[10] Owen RW,Thompson MH,Hill MJ,et al.The importance of the ratio of lithocholic to deoxycholic acid in large bowel carcinogenesis [J].Nutr Cancer,1987,9(2-3): 67-71.
[11] Wellock IJ,Fortomaris PD,Houdijk JG,et al.Effects of dietary protein supply,weaning age and experimental enterotoxigenic Escherichia coli infection on newly weaned pigs: health [J].Animal,2008,2(6): 834-842.
[12] Collins SM,Bercik P.The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease [J].Gastroenterology,2009,136(6): 2003-2014.
[13] 张柏林,秦贵信,孙泽威,等.仔猪胃肠道微生物菌群定植规律及其功能的研究进展 [J].中国畜牧杂志,2009(19): 66-69.
[14] Swords WE,Wu CC,Champlin FR,et al.Postnatal changes in selected bacterial groups of the pig colonic microflora [J].Biol Neonate,1993,63(3): 191-200.
[15] Mathew AG,Robbins CM,Chattin SE,et al.Influence of galactosyl lactose on energy and protein digestibility,enteric microflora,and performance of weanling pigs [J].J Anim Sci,1997,75(4): 1009-1016.
[16] 赵桂英,杨亮宇,段纲,等.断奶仔猪胃肠道正常菌群的数量和分区 [J].黑龙江畜牧兽医,2003(09): 26-27.
[17] Konstantinov SR,Zhu WY,Williams BA,et al.Effect of fermentable carbohydrates on piglet faecal bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA [J].FEMS Microbiol Ecol,2003,43(2): 225-235.
[18] Konstantinov SR,Poznanski E,Fuentes S,et al.Lactobacillus sobrius sp.nov.,abundant in the intestine of weaning piglets [J].Int J Syst Evol Microbiol,2006,56(Pt 1): 29-32.
[19] Luyer MD,Buurman WA,Hadfoune M,et al.Pretreatment with high-fat enteral nutrition reduces endotoxin and tumor necrosis factor-alpha and preserves gut barrier function early after hemorrhagic shock [J].Shock,2004,21(1): 65-71.
[20] Boudry G,Peron V,Le Huerou-Luron I,et al.Weaning induces both transient and long-lasting modifications of absorptive,secretory,and barrier properties of piglet intestine [J].J Nutr,2004,134(9): 2256-2262.
[21] Inoue R,Tsukahara T,Nakanishi N,et al.Development of the intestinal microbiota in the piglet [J].J Gen Appl Microbiol,2005,51(4): 257-265.
[22] Ley RE,Peterson DA,Gordon JI.Ecological and evolutionary forces shaping microbial diversity in the human intestine [J].Cell,2006,124(4): 837-848.
[23] Buffie CG,Pamer EG.Microbiota-mediated colonization resistance against intestinal pathogens [J].Nat Rev Immunol,2013,13(11): 790-801.
[24] Kamada N,Seo SU,Chen GY,et al.Role of the gut microbiota in immunity and inflammatory disease [J].Nat Rev Immunol,2013,13(5): 321-335.
[25] Blaser MJ,Kirschner D.The equilibria that allow bacterial persistence in human hosts [J].Nature,2007,449(7164): 843-849.
[26] Flint HJ,Scott KP,Louis P,et al.The role of the gut microbiota in nutrition and health [J].Nat Rev Gastroenterol Hepatol,2012,9(10): 577-589.
[27] Macfarlane S,Macfarlane GT.Regulation of short-chain fatty acid production [J].Proc Nutr Soc,2003,62(1): 67-72.
[28] Sartor RB.Microbial-host interactions in inflammatory bowel diseases and experimental colitis [J].Nestle Nutr Workshop Ser Pediatr Program,2009,64: 121-132,132-137,251-257.
[29] Hooper LV,Wong MH,Thelin A,et al.Molecular analysis of commensal host-microbial relationships in the intestine [J].Science,2001,291(5505): 881-884.
[30] Thomas CM,Hong T,van Pijkeren JP,et al.Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling [J].PLoS One,2012,7(2): e31951.
[31] De Biase D,Pennacchietti E.Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function,distribution and biomedical implications of the gadBC operon [J].Mol Microbiol,2012,86(4): 770-786.
[32] Feitoza BA,Pereira FA,da Costa FN,et al.Conjugated linoleic acid (CLA): effect modulation of body composition and lipid profile [J].Nutr Hosp,2009,24(4): 422-428.
[33] Devillard E,Mcintosh FM,Duncan SH,et al.Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid[J].J Bacteriol,2007,189(6): 2566-2570.
[34] Devillard E,Mcintosh FM,Paillard D,et al.Differences between human subjects in the composition of the faecal bacterial community and faecal metabolism of linoleic acid[J].Microbiology,2009,155(Pt 2): 513-520.
[35] Velagapudi VR,Hezaveh R,Reigstad CS,et al.The gut microbiota modulates host energy and lipid metabolism in mice [J].J Lipid Res,2010,51(5): 1101-1112.
[36] Endt K,Stecher B,Chaffron S,et al.The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea [J].PLoS Pathog,2010,6(9): e1001097.
[37] Seekatz AM,Young VB.Clostridium difficile and the microbiota [J].J Clin Invest,2014,124(10): 4182-4189.
[38] Britton RA,Young VB.Role of the intestinal microbiota in resistance to colonization by Clostridium difficile [J].Gastroenterology,2014,146(6): 1547-1553.
[39] Theriot CM,Koenigsknecht MJ,Carlson PJ,et al.Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection [J].Nat Commun,2014,5: 3114.
[40] Fuentes S,van Nood E,Tims S,et al.Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection [J].ISME J,2014,8(8): 1621-1633.
[41] Seekatz AM,Aas J,Gessert CE,et al.Recovery of the gut microbiome following fecal microbiota transplantation[J].MBio,2014,5(3): e814-e893.
[42] Khosravi A,Yanez A,Price JG,et al.Gut microbiota promote hematopoiesis to control bacterial infection [J].Cell Host Microbe,2014,15(3): 374-381.
[43] Mittrucker HW,Seidel D,Bland PW,et al.Lack of microbiota reduces innate responses and enhances adaptive immunity against Listeria monocytogenes infection [J].Eur J Immunol,2014,44(6): 1710-1715.
[44] Ren W,Yin J,Xiao H,et al.Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection [J].Front Immunol,2016,7: 685.
[45] Cantacessi C,Giacomin P,Croese J,et al.Impact of experimental hookworm infection on the human gut microbiota [J].J Infect Dis,2014,210(9): 1431-1434.
[46] Uchiyama R,Chassaing B,Zhang B,et al.Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity [J].J Infect Dis,2014,210(2): 171-182.
[47] Littman DR,Pamer EG.Role of the commensal microbiota in normal and pathogenic host immune responses [J].Cell Host Microbe,2011,10(4): 311-323.
[48] Xu X,Xu P,Ma C,et al.Gut microbiota,host health,and polysaccharides [J].Biotechnol Adv,2013,31(2): 318-337.
[49] Morland B,Midtvedt T.Phagocytosis,peritoneal influx,and enzyme activities in peritoneal macrophages from germfree,conventional,and ex-germfree mice [J].Infect Immun,1984,44(3): 750-752.
[50] Gaboriau-Routhiau V,Rakotobe S,Lecuyer E,et al.The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses [J].Immunity,2009,31(4): 677-689.
[51] Ivanov II,Atarashi K,Manel N,et al.Induction of intestinal Th17 cells by segmented filamentous bacteria [J].Cell,2009,139(3): 485-498.
[52] Izcue A,Coombes JL,Powrie F.Regulatory lymphocytes and intestinal inflammation [J].Annu Rev Immunol,2009,27: 313-338.
[53] Geuking MB,Cahenzli J,Lawson MA,et al.Intestinal bacterial colonization induces mutualistic regulatory T cell responses [J].Immunity,2011,34(5): 794-806.
[54] Round JL,Lee SM,Li J,et al.The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota [J].Science,2011,332(6032): 974-977.
[55] Feng T,Wang L,Schoeb TR,et al.Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis [J].J Exp Med,2010,207(6): 1321-1332.
[56] Atarashi K,Tanoue T,Shima T,et al.Induction of colonic regulatory T cells by indigenous Clostridium species [J].Science,2011,331(6015): 337-341.
[57] Koboziev I,Karlsson F,Grisham MB.Gut-associated lymphoid tissue,T cell trafficking,and chronic intestinal inflammation [J].Ann N Y Acad Sci,2010,1207 Suppl 1: E86-E93.
[58] Round JL,Mazmanian SK.The gut microbiota shapes intestinal immune responses during health and disease[J].Nat Rev Immunol,2009,9(5): 313-323.
[59] Renz H,Brandtzaeg P,Hornef M.The impact of perinatal immune development on mucosal homeostasis and chronic inflammation [J].Nat Rev Immunol,2011,12(1): 9-23.
[60] Maynard CL,Elson CO,Hatton RD,et al.Reciprocal interactions of the intestinal microbiota and immune system [J].Nature,2012,489(7415): 231-241.
[61] He B,Xu W,Santini PA,et al.Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL [J].Immunity,2007,26(6): 812-826.
[62] Butler JE,Lager KM,Splichal I,et al.The piglet as a model for B cell and immune system development [J].Vet Immunol Immunopathol,2009,128(1-3): 147-170.
[63] Laycock G,Sait L,Inman C,et al.A defined intestinal colonization microbiota for gnotobiotic pigs[J].Vet Immunol Immunopathol,2012,149(3-4): 216-224.
[64] Wu M,Xiao H,Liu G,et al.Glutamine promotes intestinal SIgA secretion through intestinal microbiota and IL-13 [J].Mol Nutr Food Res,2016,60(7): 1637-1648.
[65] Spits H,Di Santo JP.The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling [J].Nat Immunol,2011,12(1): 21-27.
[66] Spits H,Cupedo T.Innate lymphoid cells: emerging insights in development,lineage relationships,and function [J].Annu Rev Immunol,2012,30: 647-675.
[67] Zelante T,Iannitti RG,Cunha C,et al.Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22 [J].Immunity,2013,39(2): 372-385.
[68] Manta C,Heupel E,Radulovic K,et al.CX3CR1+macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium [J].Mucosal Immunol,2013,6(1): 177-188.
[69] Franchi L,Kamada N,Nakamura Y,et al.NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense [J].Nat Immunol,2012,13(5): 449-456.
[70] Rivollier A,He J,Kole A,et al.Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon [J].J Exp Med,2012,209(1): 139-155.
[71] Lutgendorff F,Akkermans LM,Soderholm JD.The role of microbiota and probiotics in stress-induced gastro-intestinal damage [J].Curr Mol Med,2008,8(4): 282-298.
[72] Cario E,Gerken G,Podolsky DK.Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function [J].Gastroenterology,2007,132(4): 1359-1374.
[73] Yan F,Cao H,Cover TL,et al.Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism [J].J Clin Invest,2011,121(6): 2242-2253.
[74] Cani PD,Possemiers S,Van de Wiele T,et al.Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability [J].Gut,2009,58(8): 1091-1103.
[75] Stappenbeck TS,Hooper LV,Gordon JI.Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells [J].Proc Natl Acad Sci U S A,2002,99(24): 15451-15455.
[76] Gordon HA,Bruckner-Kardoss E.Effect of normal microbial flora on intestinal surface area [J].Am J Physiol,1961,201: 175-178.
[77] Banasaz M,Norin E,Holma R,et al.Increased enterocyte production in gnotobiotic rats mono-associated with Lactobacillus rhamnosus GG [J].Appl Environ Microbiol,2002,68(6): 3031-3034.
[78] Alam M,Midtvedt T,Uribe A.Differential cell kinetics in the ileum and colon of germfree rats [J].Scand J Gastroenterol,1994,29(5): 445-451.
[79] Husebye E,Hellstrom PM,Midtvedt T.Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex [J].Dig Dis Sci,1994,39(5): 946-956.
[80] Hooper LV,Gordon JI.Commensal host-bacterial relationships in the gut [J].Science,2001,292(5519): 1115-1118.
[81] Isaacson R,Kim HB.The intestinal microbiome of the pig [J].Anim Health Res Rev,2012,13(1): 100-109.
[82] Frese SA,Parker K,Calvert C,et al.Diet shapes the gut microbiome of pigs during nursing and weaning [J].Microbiome,2015,3: 28.
[83] Petri D,Hill JE,Van Kessel AG.Microbial succession in the gastrointestinal tract (GIT) of the preweaned pig [J].Livestock Science,2010,133(1-3): 107-109.
[84] Bian G,Ma S,Zhu Z,et al.Age,introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model [J].Environ Microbiol,2016,18(5): 1566-1577.
[85] Rist VT,Weiss E,Eklund M,et al.Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: a review [J].Animal,2013,7(7): 1067-1078.
[86] Pieper R,Vahjen W,Zentek J.Dietary fibre and crude protein: impact on gastrointestinal microbial fermentation characteristics and host response [J].Animal Prod Sci,2015,55(12): 1367-1375.
[87] Tian L,Bruggeman G,van den Berg M,et al.Effects of pectin on fermentation characteristics,carbohydrate utilization,and microbial community composition in the gastrointestinal tract of weaning pigs [J].Mol Nutr Food Res,2017,61(1): 1600186
[88] Cao KF,Zhang HH,Han HH,et al.Effect of dietary protein sources on the small intestine microbiome of weaned piglets based on high-throughput sequencing [J].Lett Appl Microbiol,2016,62(5): 392-398.
[89] 刘忠臣.不同来源脂肪对仔猪的营养效应及对E.coli攻毒的保护作用研究 [D].四川农业大学,2011.
[90] Ren W,Duan J,Yin J,et al.Dietary L-glutamine supplementation modulates microbial community and activates innate immunity in the mouse intestine [J].Amino Acids,2014,46(10): 2403-2413.
[91] Ren W,Chen S,Yin J,et al.Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity [J].J Nutr,2014,144(6): 988-995.
[92] Hojberg O,Canibe N,Poulsen HD,et al.Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets [J].Appl Environ Microbiol,2005,71(5): 2267-2277.
[93] Looft T,Johnson TA,Allen HK,et al.In-feed antibiotic effects on the swine intestinal microbiome [J].Proc Natl Acad Sci U S A,2012,109(5): 1691-1696.
[94] Levesque CL,Hooda S,Swanson KS,et al.Alterations in ileal mucosa bacteria related to diet complexity and growth performance in young pigs [J].PLoS One,2014,9(9): e108472.
[95] Zhang D,Ji H,Liu H,et al.Changes in the diversity and composition of gut microbiota of weaned piglets after oral administration of Lactobacillus or an antibiotic [J].Appl Microbiol Biotechnol,2016,100(23): 10081-10093.
[96] Schokker D,Zhang J,Zhang LL,et al.Early-life environmental variation affects intestinal microbiota and immune development in new-born piglets [J].PLoS One,2014,9(6): e100040.
[97] 胡远亮.利用分子生物技术研究益生菌对断奶仔猪生长及粪便菌群的影响 [D].华中农业大学,2014.
[98] Mair C,Plitzner C,Domig KJ,et al.Impact of inulin and a multispecies probiotic formulation on performance,microbial ecology and concomitant fermentation patterns in newly weaned piglets [J].J Anim Physiol Anim Nutr (Berl),2010,94(5): e164-e177.
[99] Ren W,Wang P,Yan J,et al.Melatonin alleviates weanling stress in mice: involvement of intestinal microbiota [J].J Pineal Res,2017.doi: 10.1111/jpi.12448.[Epub ahead of print]
[100] Yi JQ,Piao XS,Li ZC,et al.The effects of enzyme complex on performance,intestinal health and nutrient digestibility of weaned pigs [J].Asian-Australas J Anim Sci,2013,26(8): 1181-1188.
[101] Ahmed ST,Hwang JA,Hoon J,et al.Comparison of single and blend acidifiers as alternative to antibiotics on growth performance,fecal microflora,and humoral immunity in weaned piglets [J].Asian-Australas J Anim Sci,2014,27(1): 93-100.
[102] 吴超,张莉,吴跃明,等.中草药添加剂对早期断奶仔猪生长性能和肠道菌群的影响 [J].中国畜牧杂志,2010(03): 31-35.
Currentunderstandingoftheintestinalmicrobiotaofpiglets
XIA Yao-yao1,2,REN Wen-kai1,2*,HUANG Rui-lin1,ZENG Ben-hua3,WEI Hong3,YIN Yu-long1*
(1.Key Laboratory of Agro-Ecological Processes in Subtropical Region,National Engineering Laboratory for Pollution Control and Waste Utilization in Livestock and Poultry Production,Hunan Provincial Engineering Research Center for Healthy Livestock and Poultry Production,Scientific Observation and Experimental Station of Animal Nutrition and Feed Science in South-Central China,Ministry of Agriculture,Institute of Subtropical Agriculture,Chinese Academy of Sciences,Changsha 410125,China; 2.University of Chinese Academy of Sciences,Beijing 100049; 3.Department of Laboratory Animal Science,College of Basic Medicine Science,Third Military Medical University,Chongqing 400038)
The role of intestinal microbiota in mammals and humans are gaining increasing attention.The health of piglets requires a dynamically balanced intestinal microbiota.However,there are temporal and spatial changes in the distribution and composition of microbiota from the esophagus to the rectum during the life cycle of the pig.The intestinal microbiota has various beneficial functions in pig,like nutrient metabolism,intestinal mucosal barrier,immune responses and pathogen infection.A variety of factors play an important role in the formation and stabilization of intestinal microbiota,including the ways of delivery (vaginal or caesarean),the diet during infancy (breast milk or formula feeds) and the usage of antibiotics or antibiotic-like molecules.In this review,we mainly discussed the relationship between intestinal microbiota and the intestinal health of piglets from the aspects of the composition and colonization of intestinal microbiota,as well as the functions and influencing factors of intestinal microbiota,so as to further understanding the importance of intestinal microbiota in intestinal function of piglets.
Intestinal microbiota; Intestinal health; Piglets
REN Wen-kai.E-mail: renwenkai19@126.com; YIN Yu-long.E-mail: yinyulong@isa.ac.cn
国家自然科学基金(No.31330075,No.31301989)。
夏耀耀(1994-),男,硕士研究生,研究方向为单胃动物营养及分子生物学。E-mail:1016812759@qq.com
任文凯,男,博士,主要从事营养与免疫方面研究。E-mail:renwenkai19@126.com;
印遇龙,男,博士,研究员,博士生导师,主要从事猪氨基酸营养代谢与调控研究。E-mail:yinyulong@isa.ac.cn
Q95-33
A
1005-4847(2017) 06-0681-08
10.3969/j.issn.1005-4847.2017.06.018
2017-10-19

