湖南汉族人群21个STR基因座的遗传多态性(英文)
邹鹰,郭娟娟,李情鹏,左道宏,刘金山,郭亚东,闫杰,扎拉嘎白乙拉,蔡继峰,兰玲梅
(1.中南大学基础医学院法医学系,湖南长沙 410013;2.中南大学湘雅医学院,湖南长沙 410013;3.内蒙古民族大学附属医院内科,内蒙古通辽 028000)
·综述·
湖南汉族人群21个STR基因座的遗传多态性(英文)
邹鹰1,郭娟娟1,李情鹏2,左道宏2,刘金山3,郭亚东1,闫杰1,扎拉嘎白乙拉1,蔡继峰1,兰玲梅1
(1.中南大学基础医学院法医学系,湖南长沙410013;2.中南大学湘雅医学院,湖南长沙410013;3.内蒙古民族大学附属医院内科,内蒙古通辽028000)
目的调查湖南地区汉族人群21个STR基因座(D3S1358、D13S317、D7S820、D16S539、Penta E、D2S441、TPOX、TH01、D2S1338、CSF1PO、Penta D、D10S1248、D19S433、vWA、D21S11、D18S51、D6S1043、D8S1179、D5S818、D12S391和FGA)的遗传多态性。方法共采集560例湖南汉族健康无关个体血液样本,使用Chelex-100法提取DNA,应用AGCU EX22试剂盒及9700 PCR扩增仪进行复合扩增,扩增产物使用310遗传分析仪进行分离分析。结果共发现248个等位基因,等位基因频率分布在0.001~0.518。除Penta E(P=0.023)外,其余基因座的基因型分布均符合Hardy-Weinberg平衡。21个基因座的累积个人识别率、累积非父排除率、累积匹配率分别为0.999 999 999 999 999 999 999 999 8、0.999 999 998和1.36×10-25。结论21个STR基因座在湖南汉族人群中呈高度多态性。本研究可为法医学个人识别及亲子鉴定提供有价值的数据及理论基础。
法医遗传学;多态现象,遗传;短串联重复序列;湖南;汉族
Introduction
Han nationality is the main nationality with the largestpopulation,notonlyinChinabutalso worldwide.It is generally believed that the ancestors of Han nationality can be traced back to Yan Emperor and Yellow Emperor.The Han people also call themselves“Hua Xia Zi Sun”and“Tang people.”Because of the diversity of gene,region,languageandculture,quiteanumberofdifferent tribes and branches evolved in the formation process of Han nationality.Most historians argue thatthe core of Han nationality was“pre-Qin”and that a unified Han nationality was formed during the QinandHanDynasties.HunanProvince,also called“Xiang Chu”and“Xiao Xiang”,is located at the central south China.With a long history,the province has played an important role in Chinese culture and economy.A trace of human activities was discovered in the Paleolithic Period;100 000 years ago the activities of planting rice came into being.During the early of Western Han Dynasty,Changsha County was established and Hunan gradually became the culture center of the southeast of China.At present,Hunan Province is composed of 54 nationalities besides Han nationality,with a household register population of 65683722(in 2010)[1].
Short tandem repeat(STR),which is occasionally referred to microsatellite sequence,is a kind of DNA sequence inherited by Mendel co-dominance,easy and quick to standardize and genotype,and with high polymorphism[2].STR typing methods have become major detecting means in forensic DNA analysis,gene mapping and genetic linkage analysis of forensic science.In forensic routine casework,Power-Plex®16 System Kit(Promega,Madison,WI),AmpFℓSTR®Identifiler®Plus PCR Amplification Kit and AmpFℓSTR®Sinofiler®PCR Amplification Kit(Applied Biosystems,Foster City,CA)are commonly practiced in the world.For a higher level ofdiscriminatorypower,however,extendedloci may be required in single paternity testing[3]and the paternity testing observed STR mutations[4].In the current study,AGCU EX22 STR kit[5]was used to evaluate the21STRloci(D3S1358,D13S317,D7S820,D16S539,Penta E,D2S441,TPOX,TH01,D2S1338,CSF1PO,Penta D,D10S1248,D19S433,vWA,D21S11,D18S51,D6S1043,D8S1179,D5S818,D12S391 and FGA)performance in population genetics research,human identification,and paternity testing in forensic sciences.
Materials and methods
Samples and DNA extraction
A total of 560 blood samples were collected from unrelated healthy individuals of the Han population,222malesand238females,inHunan Province.The human Genomic DNA was extracted from the blood samples with the method of Chelex-100 extraction.The current study was approved by the Ethics Committee of Central South University Xiangya School of Medicine(Code:201303147),with the written informed consents provided for the collection of the samples and subsequent analyses.
PCR amplification
AGCU EX22 STR kit was applied to the amplification reactions of the 21 autosomal STRs loci following the manufacturer’s instructions.The 10 μL reaction volumes contained 1.0 μL(0.5-2.0 ng)genomic DNA,2.0 μL EX22 Primers,4.0 μL AGCU PCR Reaction buffer,0.4 μL Taq and 0.6 μL ddH2O.Temperature cycling conditions for PCR reactions were as follows:2-min denaturation at 95℃,10 cycles for 30s at 94℃,1min at 60℃,1min at 72℃,20 cycles for 30 s at 90℃,1 min at 58℃,1min at 72℃,and 10-min elongation at 72℃.All amplifications were conducted on a GeneAmp PCR 9700 thermal cycler(Applied Biosystems,Foster City,CA,USA).
DNA typing
A 310 Genetic Analyzer(Applied Biosystems,Foster City,CA,USA)was used to separate and detect the PCR products by capillary electrophoresis.A microliter PCR product or allelic ladder was mixed with 15 μL Hi-Di formamide(Applied Biosystems,Foster City,CA,USA)and 0.5 μL internal lane standard CC5 ILS 600(Promega,Madison,WI,USA),before denatured at 95℃for 3 min,immediately followed by 3-min chilling on ice.Gene-Mapper ID 3.2 software(Applied Biosystems,Foster City,CA,USA)was used to analyze STR alleles by comparison with kit allelic ladders.Standard reference in all experiments was genotyped control DNA from cell line 9947A(Promega,Madison,WI,USA). Statistical analysis
ModifiedPowerStats v1.2softwarepackage(Promega,Madison,WI,USA)was used to calculateallelicfrequencies,polymorphicinformation content(PIC),matching probability(MP),discrimination power(DP),probability of paternity exclusion(PE),observed heterozygosity(Ho)and exact tests of the Hardy-Weinberg equilibrium of the 21 STR loci[6];SHEsis software[7],to calculate the value of D′and r2;and Arlequin v3.11 software[8],to compare the data of Hunan Han popu-lation with the previously published data of other populations. A Neighbor-Joining(N-J)phylogenetic tree based on pairwise Nei’s genetic distance[9]was developed through the phylogeny inference package(Phylip)version 3.695[10].
Results and discussion
The diversity distribution was made of allelic frequencies and forensic statistical parameters of 13 Combined DNA Index System(CODIS)loci and 8 non-Combined DNA Index System(non-CODIS)loci(Penta E,D2S441,D2S1338,Penta D,D10S1248,D19S433,D6S1043 and D12S391)(Table 1&Table 2).The allelic frequencies of 248 alleles at 21 STR loci ranged from 0.001 to 0.518;the genotype frequencies of 910 genotypes,from 0.001 8 to 0.373 2;and the MP,DP,PIC,PE and Ho,from 0.015 to 0.226,0.774 to 0.985,0.554 to 0.909,0.360 to 0.773 and 0.654 to 0.889,respectively.The combined powerof discrimination,combined power of exclusion,and combined matching probability of the 21 STR loci were approximately 0.9999999999999999999999998,0.999999998,and 1.36×10-25,respectively.Observed genotype distributions for each locus showed no deviationsfromHardy-Weinbergequilibriumexcept Penta E(P=0.023).The possible causes were the errors in technology and standard of DNA typing,or random sampling.The D′values ranged from 0.072 to 0.249,whereas the r2values,from 0.001 to 0.005.The value of D′and r2suggested that the alleles were not in linkage disequilibrium.
Theresultswerecomparedwithpreviously published population data at the same STR loci using Arlequin v3.11 software(Table 3).No statistically significant differences(P<0.05)were observed between the current population and Singapore Han[11]populations for all the 13 CODIS STR loci.Statistically significant differences were found between Hunan Han and Anhui Han[12],Zhejiang Han[13],Changzhou Han[14],Shaanxi Han[15],Gansu Tu[16],Hong Kong Han[17],Qinghai Dongxiang[18],Chongqing Han[19],Yunnan Yi[20],Xinjiang Kazak[21],Qinghai Hui[22],Qinghai Salar[23],Shandong Han[24]and Fujian She[25]populations for 2(15),2(15),2(15),3(15),3(15),4(15),4(15),5(14),6(13),12(19),11(15),12(15),14(15)and 19(20)STR loci,respectively(The number within parentheses is the number of common STR loci between two populations).Our results indicated the previous finding[26]thatallelicfrequencydistributionsvariedinthe populationsfromdifferentethnicgroupsinthe same country and even from the same ethnic group who migrated to live in different regions and areas.
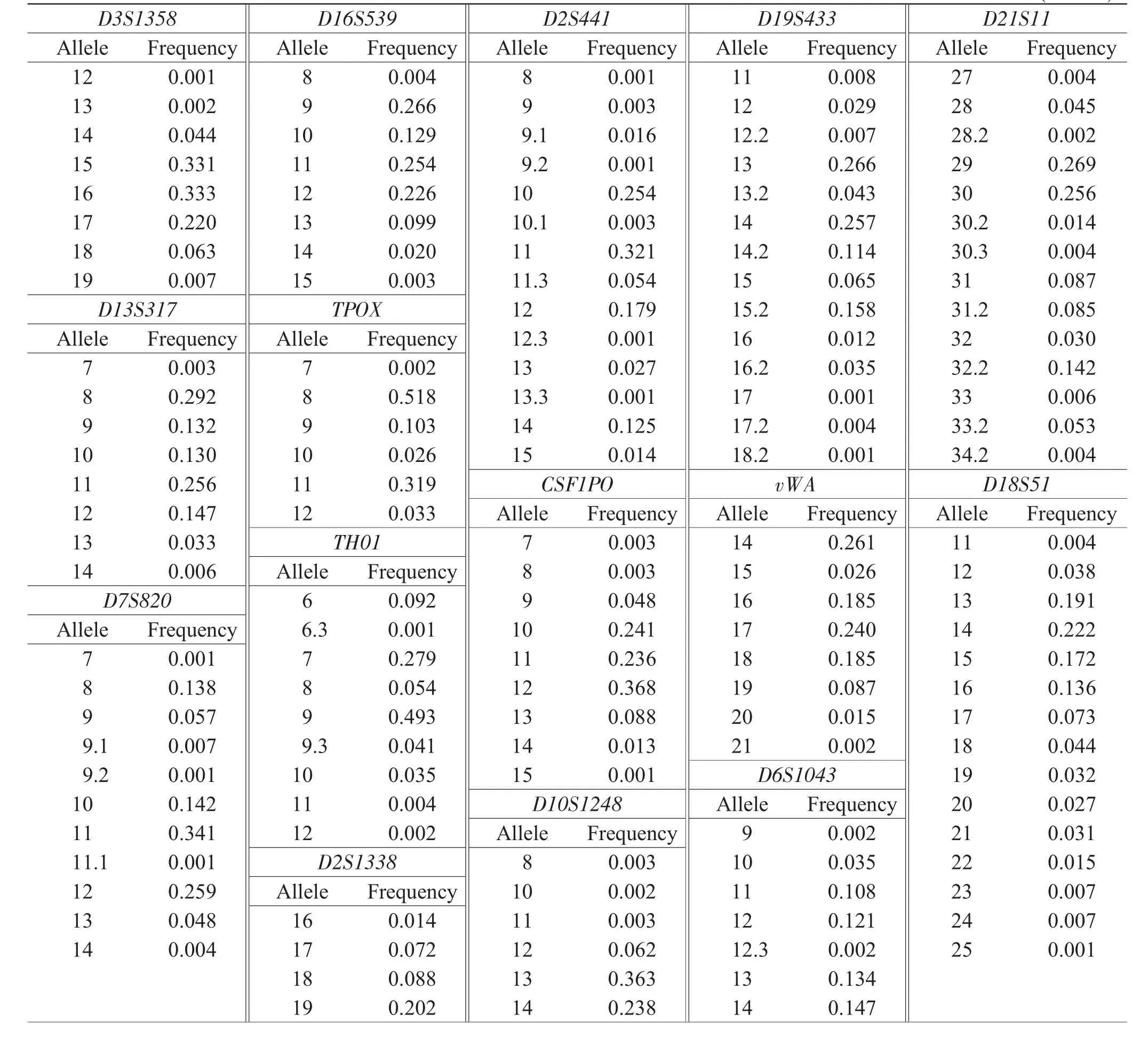
Table 1Allelic frequency distributions of 21 STR loci in Hunan Han population samples(n=560)
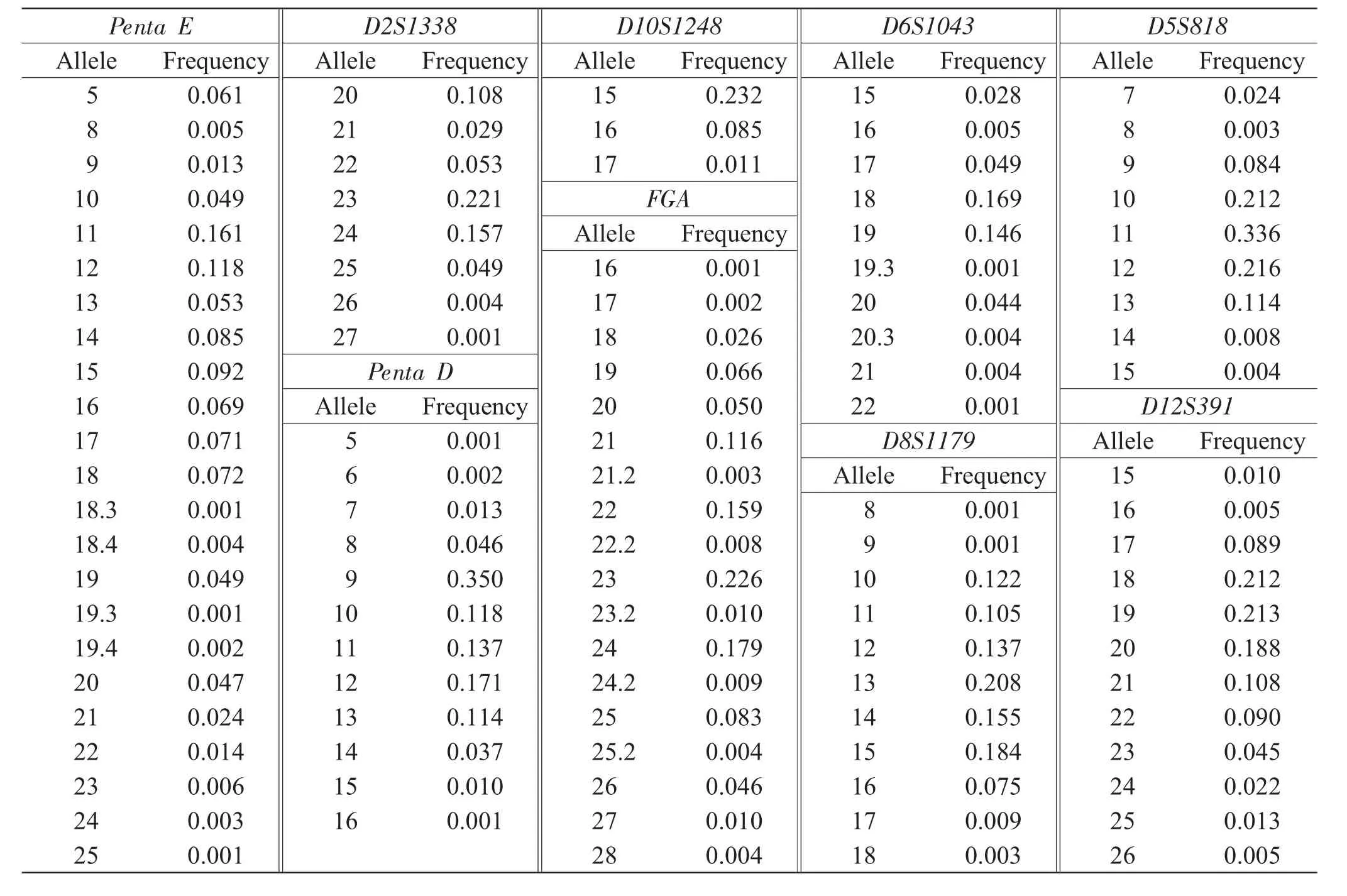
Continued
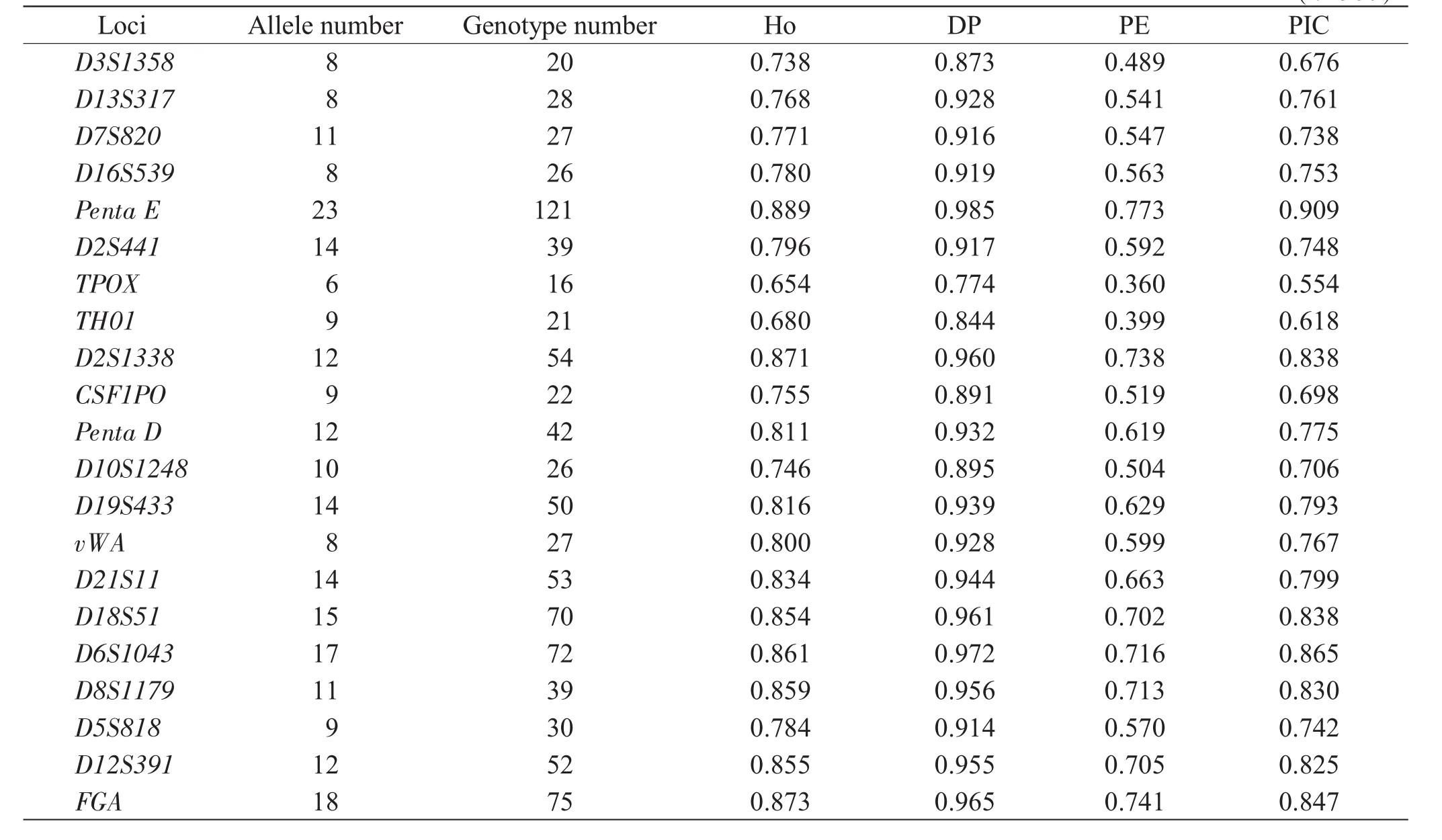
Table 2Genetic parameters of 21 loci in Hunan Han population samples(n=560)
Furthermore,an un-rooted phylogenetic tree was constructed by comparing the allelic frequencies for the 13 CODIS STR loci between Hunan Han population and the other 15 populations[11-25](Fig.1). The analysis showed that Fujian She,Yunnan Yi,Hong Kong Han,Singapore Han,Hunan Han and Zhejiang Han population,who reside in South China or Middle Asian regions,belonged to a cluster. Zhejiang Han population lives more closely to the Hunan Han population,and they have very close relationships.From the West China Gansu Tu,Qinghai Hui,Xinjiang Kazak,Qinghai Dongxiang,Qinghai Salar,Chongqing Han and Shaanxi Han population showed close genetic relationship,while Anhui Han and Shandong Han population from North China belongedtooneclade.Geneexchangebetween populations generally occurs in the same department or near the geographic area[27].The geographical factors play an important role in the formation of genetic polymorphisms,and the heterogeneity level between STR loci in populations from distant geographical distribution is higher[28].
Conclusion
Thedatademonstratethe21STRlociare highly discriminating and polymorphic in the Hunan Han population.The 13 CODIS core STRs and additional 8 new autosomal STRs can obtain more information for routine forensic casework.Additionally,the population data can improve forensic DNA database,enrich Chinese population genetic information resources and provide genetic evidence for the origin of the Hunan Han population.
Acknowledgements
This study was supported by the Natural Science Foundation for the Youth(No.81302621)and the Fundamental Research Funds for the Central Universities of Central South University(2015zzts276).
References:
[1]中华人民共和国国家统计局.2010年第六次全国人口普查主要数据公报[EB/OL].(2014-09-01)[2014-09-12]. http://www.gov.cn/test/2012-04/20/content_2118413.htm.
[2]Chakraborty R,Stivers DN,Su B,et al.The utility of short tandem repeat loci beyond human identification: Implications for development of new DNA typing systems[J].Electrophoresis,1999,20(8):1682-1696.
[3]Poetsch M,Ludcke C,Repenning A,et al.The problem of single parent/child paternity analysis--practical results involving 336 children and 348 unrelated men[J]. Forensic Sci Int,2006,159(2-3):98-103.
[4]Lu D,Liu Q,Wu W,et al.Mutation analysis of 24 short tandem repeats in Chinese Han population[J]. Int J Legal Med,2012,126(2):331-335.
[5]邹凯南,曹禹,夏子芳,等.Expressmarker 22 STR荧光检测试剂盒的法医学应用[J].法医学杂志,2012,28(6):448-450.
[6]Tereba A.Tools for analysis of population statistics[J]. Profiles in DNA,1999,2(3):14-16.
[7]Shi YY,He L.SHEsis,a powerful software plat-form for analyses of linkage disequilibrium,haplotype construction,and genetic association at polymorphism loci[J].Cell Res,2005,15(2):97-98.
[8]ExcoffierL,LavalGandSchneiderS.Arlequin(version 3.0):an integrated softwarepackagefor population genetics data analysis[J].Evol Bioinform Online,2005,1:47-50.
[9]Nei M.Genetic distance between populations[J].Am Nat,1972,949(106):283-292.
[10]Felsenstein J.PHYLIP(phylogeny inference package)version 3.69[Z].Department of Genome Sciences,University of Washington,Seattle,2009.
[11]Syn CK,Chuah SY,Ang HC,et al.Genetic data for the 13 CODIS STR loci in Singapore Chinese[J]. Forensic Sci Int,2005,152(2-3):285-288.
[12]杨玉玲,王冬花,陈玲,等.安徽汉族人群15个STR基因座遗传多态性[J].法医学杂志,2007,23(1):36-38.
[13]Zhu Y,Lu S,Xie Z,et al.Genetic analysis of 15 STR loci in the population of Zhejiang Province(Southeast China)[J].Forensic Sci Int Genet,2009,3(4): e139-e140.
[14]巴华杰,刘冰泉.常州地区汉族人群15个STR基因座遗传多态性[J].中国法医学杂志,2012,27(1):59-61.
[15]Wu YM,Zhang XN,Zhou Y,et al.Genetic polymorphisms of 15 STR loci in Chinese Han population living in Xi'an city of ShaanxiProvince[J]. Forensic Sci Int Genet,2008,2(2):e15-e18.
[16]Zhu B,Yan J,Shen C,et al.Population genetic analysis of 15 STR loci of Chinese Tu ethnic minority group[J].Forensic Sci Int,2008,174(2-3):255-258.
[17]Chan KM,Chiu CT,Tsui P,et al.Population data for the Identifier 15 STR loci in Hong Kong Chinese[J]. Forensic Sci Int,2005,152(2-3):307-309.
[18]Deng Y,Zhu B,Yu X,et al.Genetic polymorphisms of 15 STR loci of Chinese Dongxiang and Salar ethnic minority living in Qinghai Province of China[J].Leg Med(Tokyo),2007,9(1):38-42.
[19]Deng YJ,Yan JW,Yu XG,et al.Genetic analysis of 15 STR loci in Chinese Han population from West China[J].Genomics Proteomics Bioinformatics,2007,5(1):66-69.
[20]Zhu BF,Shen CM,Wu QJ,et al.Population data of 15 STR loci of Chinese Yi ethnic minority group[J]. Leg Med(Tokyo),2008,10(4):220-224.
[21]张丽萍,许晨波,陈慧锦,等.新疆巴里坤哈萨克族人群19个STR基因座的遗传多态性[J].中南大学学报:医学版,2012,37(9):934-938.
[22]Deng YJ,Zhu BF,Shen CM,et al.Genetic polymorphism analysis of 15 STR loci in Chinese Hui ethnic group residing in Qinghai province of China[J]. Mol Biol RPE,2011,38(4):2315-2322.
[23]Zhu J,Shen C,Ma Y,et al.Genetic polymorphisms of 15 STR in Chinese Salar ethnic minority group[J]. Forensic Sci Int,2007,173(2-3):210-213.
[24]张茂修,韩淑毅,高洪梅,等.山东汉族人群19个STR基因座的遗传多态性[J].法医学杂志,2013,29(6):440-443.
[25]Yuan L,Ou Y,Liao Q,et al.Population genetics analysis of 38 STR loci in the She population from Fujian Province of China[J].Leg Med(Tokyo),2014,16(5):314-318.
[26]Meng HT,Han JT,Zhang YD,et al.Diversity study of 12 X-chromosomal STR loci in Hui ethnic from China[J].Electrophoresis,2014,35(14):2001-2007.
[27]Wang HD,Wu D,Feng ZQ,et al.Genetic polymorphisms of 20 short tandem repeat loci from the Han population in Henan,China[J].Electrophoresis,2014,35(10):1509-1514.
[28]Rosser ZH,Zerjal T,Hurles ME,et al.Y-chromosomal diversity in Europe is clinal and influenced primarily by geography,rather than by language[J].Am J Hum Genet,2000,67(6):1526-1543.
(Editor:LI Cheng-tao)
Genetic Polymorphisms of 21 STR Loci in Hunan Province-based Han Population
ZOU Ying1,GUO Juan-juan1,LI Qing-peng2,ZUO Dao-hong2,LIU Jin-shan3,GUO Ya-dong1,YAN Jie1,ZHA Lagabaiyila1,CAI Ji-feng1,LAN Ling-mei1
(1.Department of Forensic Science,School of Basic Medical Sciences,Central South University,Changsha 410013,China;2.Xiangya School of Medicine,Central South University,Changsha 410013,China;3.Department of Internal Medicine,Affiliated Hospital of Inner Mongolia University for Nationalities,Tongliao 028000,China)
Objective To investigate the genetic polymorphisms of 21 short tandem repeat(STR)loci(D3S1358,D13S317,D7S820,D16S539,Penta E,D2S441,TPOX,TH01,D2S1338,CSF1PO,Penta D,D10S1248,D19S433,vWA,D21S11,D18S51,D6S1043,D8S1179,D5S818,D12S391 and FGA).Methods A total of 560 blood samples were collected from unrelated healthy individuals of Han population in Hunan Province.Chelex-100 extraction method was applied to the extraction of genomic DNA,and an AGCU EX22 Kit and 9700 STR amplification was used in amplification reactions.The products were separated and analyzed on 310 Genetic Analyzer.Results A total of 248 alleles were observed,the allelic frequencies ranging from 0.001 to 0.518.Observation of genotype distributions for each locus showed no deviations from Hardy-Weinberg equilibrium except Penta E(P=0.023).The combined power of discrimination,combined power of exclusion,and combined matching probability of the 21 STR loci were approximately 0.999 999 999 999 999 999 999 999 8,0.999 999 998,and 1.36×10-25,respectively. Conclusion The 21 STR loci show high polymorphisms in the Han population,which can provide valuable data and a theoretical basis for forensic individual identification and paternity testing.
forensic genetics;polymorphism,genetic;short tandem repeat sequences;Hunan;Han nationality
LAN Ling-mei,M.S.,experimentalist,major in forensic genetics;E-mail:lanlingmei0731@163.com
(date:2015-07-20)
DF795.2
A
10.3969/j.issn.1004-5619.2016.05.010
1004-5619(2016)05-0356-07
Author:ZOU Ying,M.S.,experimentalist,major in forensic genetics;E-mail:zouhawk@163.com

