绵羊GDF9基因mRNA、DNA和调控区序列克隆及其在11个品种中遗传多态性检测
潘章源,贺小云,刘秋月,胡文萍,王翔宇,郭晓飞,曹晓涵,狄 冉,储明星
(中国农业科学院北京畜牧兽医研究所,农业部畜禽遗传资源与种质创新重点实验室,北京 100193)
绵羊GDF9基因mRNA、DNA和调控区序列克隆及其在11个品种中遗传多态性检测
潘章源,贺小云,刘秋月,胡文萍,王翔宇,郭晓飞,曹晓涵,狄冉*,储明星*
(中国农业科学院北京畜牧兽医研究所,农业部畜禽遗传资源与种质创新重点实验室,北京 100193)
本研究旨在克隆绵羊生长分化因子9(Growth differentiation factor 9,GDF9)基因mRNA、DNA和调控区全序列,并检测其在11个绵羊品种中的遗传多态性,探究GDF9基因与绵羊多羔性状的关系。首先应用RACE和PCR技术克隆GDF9基因全长序列,其次利用SNaPshot分型技术检测11个绵羊品种GDF9基因遗传多态性。结果显示,克隆获得GDF9基因mRNA全长1 852 bp(GenBank序列号:KR063137),编码区两侧翼分别具有5′-UTR 58 bp(1~58 nt)和3′-UTR 432 bp(1 421~1 852 nt)。进一步克隆获得长为2 898 bp的DNA序列和2 304 bp的调控区序列,分析发现调控区序列比数据库序列(NC_019462.1)多两个长片段。序列比对发现了已知突变G260A(G1)和调控区新突变-2078C>G。分型结果显示,11个绵羊品种均不含FecGE、FecGH、FecGT、FecGF、FecGV突变,而G260A和-2078C>G广泛存在于除草原型藏羊外的各绵羊品种中,G260A表现3种基因型(AA、BB和AB),首次在小尾寒羊和山谷型藏羊中检测到BB型突变纯合子。-2078C>G也存在3种基因型,基因型结果表明该位点与G260A完全连锁。关联分析结果显示,小尾寒羊AB型产羔数显著高于AA型(P<0.05)。本研究完善了绵羊GDF9 mRNA、DNA和调控区全长序列,为进一步研究GDF9基因功能奠定了基础。同时在不同绵羊品种中发现了G260A和-2078C>G突变,其中G260A作为潜在的有效遗传标记可用于提高绵羊的产羔数。
绵羊;GDF9基因;mRNA;调控区;遗传多态性
产羔数是绵羊最重要的繁殖性状。作为转化生长因子β超家族的一员,GDF9(Growth differentiation factor 9)通过旁分泌方式对卵泡的生长和分化起着重要作用[1-6],并在提高绵羊产羔数中扮演着重要角色。绵羊GDF9基因位于5号染色体[7],其包含2个外显子和1个内含子,编码456个氨基酸,但其成熟蛋白仅由135个氨基酸残基组成[8]。GDF9广泛表达于湖羊的各个组织(下丘脑、垂体、卵巢、子宫、输卵管、心、肝、脾、肺、肾)[9]。对于GDF9多态性与产羔数的关联性研究,早在2004年,J.P.Hanrahan等[10]发现GDF9FecGH(G8)突变与爱尔兰Cambridge和Belclare绵羊的高繁殖力密切相关。随后发现了多个与绵羊产羔数相关的GDF9突变,包括在巴西Santa Ines绵羊中发现的FecGE突变[11-12];在冰岛Thoka绵羊中发现的FecGT突变[13];在伊朗Moghani、Ghezel绵羊中发现的G1突变[14-15];在中国小尾寒羊中发现的G729T突变[16];以及最近在挪威White 和Finnish绵羊中证实与产羔数有关的FecGF(G7)突变[17-18]。由此可见GDF9基因对绵羊多羔性状起着重要作用,然而通过查询多个基因数据库,发现绵羊GDF9 基因mRNA序列可能不完整,因为最长一条mRNA(NM_001142888)从起始密码子开始,且末端无ploy(A)尾,其转录起始位点和终止位点仍然未知;而DNA和调控区也无试验验证性序列,仅为绵羊De novo基因组序列[19],基因组在拼接过程可能出现一些错误,还需通过多方面验证。同时针对该基因的多态性研究主要集中在外显子区,对于调控区鲜见报道。
本研究应用RACE和PCR技术扩增绵羊GDF9 mRNA、DNA和调控区序列全长,并检测其遗传多态性,分析其与产羔数的关联性。为寻找与绵羊产羔数相关的分子遗传标记提供遗传学依据。
1 材料与方法
1.1材料
选择实验室前期采集保存于RNA Later(Qiagen,Hilden,Germany)中的小尾寒羊卵巢组织,用于提取总RNA。384只母羊血样来自11个绵羊品种,其中小尾寒羊(92只)采自山东郓城县,策勒黑羊(30只)采自新疆策勒县,湖羊(36只)采自江苏徐州市,多赛特羊(25只)采自内蒙古呼和浩特市,澳洲美利奴羊(30只)采自内蒙古克什克腾旗,乌珠穆沁羊(29只)采自内蒙古正蓝旗,滩羊(35只)采自宁夏盐池县,巴音布鲁克羊(30只)采自新疆和静县,山谷型藏羊(30只)采自西藏贡嘎县,草原型藏羊(30只)采自西藏当雄县,欧拉羊(17只)采自青海黄南藏族自治州,所有血样都将用于GDF9基因遗传多态性检测。以上11个绵羊品种中,小尾寒羊、策勒黑羊、湖羊、澳洲美利奴属于多羔品种,而多赛特羊、乌珠穆沁羊、滩羊、巴音布鲁克羊、山谷型藏羊、草原型藏羊、欧拉羊为单羔品种,同时小尾寒羊具有产羔记录。
1.2RNA和DNA提取
使用Trizol Reagent (Invitrogen,USA)提取卵巢组织总RNA,溶解于RNase free ddH2O中,-80℃保存。使用血液DNA提取试剂盒(天根,北京)提取血样DNA,ddH2O溶解,置于-20 ℃备用。
1.3引物设计与合成
根据GenBank公布的绵羊GDF9基因序列(NM_001142888和NC_019462.1),利用 NCBI Primer-BLAST软件分别设计RACE GSP引物、DNA扩增引物和调控区扩增引物(表1),以上引物均由生工生物工程(上海)股份有限公司合成。
表1GDF9基因RACE、DNA和调控区扩增引物
Table 1Primers used for amplification of RACE,DNA,regulatory region ofGDF9 gene

引物Primer引物序列(5'→3')Primersequence退火温度/℃Annealingtemperature目的片段/bpAmplifiedDNAfragment5'GSP-1GATTACGCCAAGCTTCATGGTGTGAACCGGAGAGCCATAC6512445'GSP-2GATTACGCCAAGCTTGCACTCTCCTGGTCTCTGCGGTGAC6510123'GSP-1GATTACGCCAAGCTTCACTGTTCGGCTCTTCACCCCCTGT6513133'GSP-2GATTACGCCAAGCTTTGTAAGATCGTCCCGTCACCGCAGA65719GDF9-E1-FGDF9-E1-RATGGGGAAATGTGTTCCTTGCTTCCCTCCACCCATTAACC61470GDF9-I1-FGDF9-I1-RTGAGGCTGAGACTTGGTCCTCAGCAGATCCACTGATGGAA581420GDF9-E2-FGDF9-E2-RGGGGAGAAAAGGGACAGAAGTCAATTAAAACCGCACACAGA611493TK-FTK-RCTTGCTGAAGTAGTGCGGGAAGGTAGAGGTGGCGTCTGTTGGATTT562357
1.4RACE扩增GDF9全序列
选择小尾寒羊卵巢RNA,根据SMARTer RACE 5′/3′ Kit (Clontech Laboratories,Inc.USA)说明书合成cDNA第一条链。以第一条链为模板,利用上述设计好的GSP引物(5′GSP-2和3′GSP-2产物全覆盖GDF9 CDS序列),按照试剂盒操作说明,通过巢式PCR方法分别扩增获得5′和3′RACE PCR 产物,经0.5%琼脂糖凝胶电泳和DNA凝胶回收试剂盒(BBI,Canada)回收纯化,连接至pMD-19T载体,提取质粒并送生工生物工程(上海)股份有限公司测序。
1.5GDF9基因DNA和调控区序列克隆及其多态性检测
利用3对DNA扩增引物(GDF9-E1、I1和E2,表1)和1对调控区扩增引物(TK,表1),根据TaKaRa ExTaq(TaKaRa,Dalian,China)试剂盒说明书,对10只小尾寒羊样本基因组DNA进行PCR扩增,扩增产物经过DNA凝胶回收试剂盒(BBI,Canada)进行回收和纯化,连接至pMD-19T载体,提取质粒并送生工生物工程(上海)股份有限公司测序。比对分析10只小尾寒羊序列,寻找SNP位点。
1.6SNaPshot分型
针对绵羊GDF9基因新发现的SNP位点和现有文献报道的5个SNPs位点(FecGE、FecGH、FecGT、FecGF、FecGV),设计PCR扩增引物和SNaPshot分型引物(表2)。SNaPshot是由美国应用生物公司(ABI)开发的一种基于荧光标记单碱基延伸原理的分型技术,其可以快速地对多个位点进行分型,目前已经得到广泛的使用[20-22]。按照SNaPshot分型操作说明,使用扩增引物进行PCR,扩增后的产物通过使用SAP酶(Fermentas)和ExoⅠ酶(New England Biolabs)消化清除里面的残余引物和dNTP;以纯化后的PCR产物为底物,使用SNaPshot分型引物进行延伸反应;延伸产物通过CIP酶(New England Biolabs)纯化后,直接使用3730XL测序仪(ABI)检测基因型。
1.7数据分析
对SNP分型结果统计基因型频率、等位基因频率。考虑到胎次对产羔数有影响,本研究配合固定效应模型[16]进行最小二乘分析,比较小尾寒羊产羔数在不同基因型之间的差异,利用 SPSS 15.0 软件(SPSS Inc,Chicago,IL,USA)的广义线性模型(GLM)过程完成。
表2SNaPshot扩增和分型引物
Table 2The amplification and genotyping primers of SNaPshot

位点Site变异Variation扩增引物AmplificationprimerSNaPshot分型引物SNaPshotgenotypingprimerG260A(G1)[G260A]F:GTTGGAATCTGAGGCTGAGACR:GTGTTGTAGAGGTGGCGTCTGCAGCCAGATGACAGAGCTTTGC-2078C>G[C(-2078)G]F:CGCCGCCAACCCGAGTCCTTR:GCGTCCGATCTACCGGAAGTTTTTTTTTTCGCGCTGTCTCGGGGACCCCTGFecGvFecGEFecGF(G7)FecGH(G8)FecGT[C943T][T1034G][G1111A][C1184T][A1279C]F:CTGAACGACACAAGTGCTCAR:AGGAGTCTGTTAACGACAGGTTGCTGAGGGTGTAAGATCGTCCTTTTTTTTTTTTTTTTTTTTCT-GAGTGAATACTTCAAACAGTGCTGAAGTGGGACAACTGGATTTTTTTTTTTTGCGGTCGGACATCGGTATGGCTTTTTTTTTTTTTTTTTTTACCTGC-CAAGTATAGCCCTTTG
2 结 果
2.1RACE扩增GDF9 mRNA完整序列
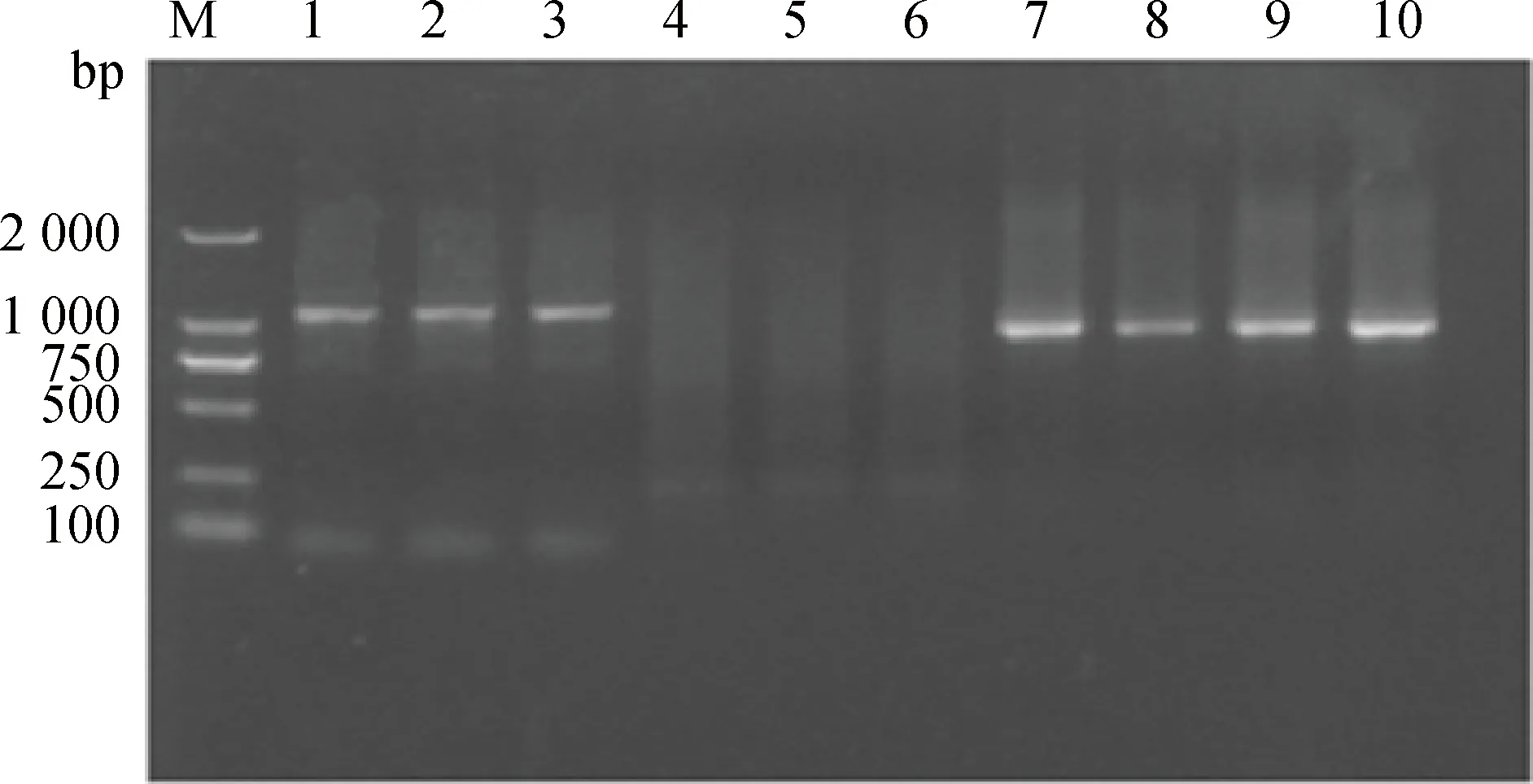
M.DNA相对分子质量标准;1~3.5′GSP-2产物(1 095 bp);4~6.空白对照(水);7~10.3′GSP-2产物(929 bp)M.DL2000 DNA marker;1-3.Products of 5′GSP-2 (1 095 bp);4-6.Blank (water);7-10.Products of 3′GSP-2(929 bp)图1 GDF9 5′RACE片段和3′RACE片段扩增产物电泳Fig.1 Electrophoresis of 5′RACE mRNA and 3′RACE mRNA fragments of GDF9 gene
通过5′RACE 和3′RACE,获得了5′ 端长为1 095 bp和3′ 端长为929 bp的片段(图1)。序列拼接和比对结果表明,GDF9 mRNA全长1 852 bp(GenBank序列号:KR063137),包含2个外显子,编码区为1 368 bp,编码456个氨基酸。与GenBank绵羊GDF9 mRNA序列(NM_001142888)相比,5′ 端多58 bp,3′ 端多185 bp(图2)。GDF9基因编码区两侧翼分别具有5′-UTR 58 bp(1~58 nt)和3′-UTR 432 bp(1 421~1 852 nt)。本研究仅扩增出单一条带,表明GDF9基因在小尾寒羊卵巢中仅存在一种剪接体。
2.2GDF9基因DNA全序列扩增及其多态性分析
利用3对引物分别获得长为470、1 420和1 493 bp的片段(图3),测序结果表明均为目的条带。对3个产物片段进行拼接,获得一条长为2 898 bp的DNA序列,序列分析表明其与NC_019462.1数据库序列一致。进一步分析比对10只小尾寒羊序列,发现外显子1存在一个错义突变(G260A)(图4),该突变导致精氨酸变成组氨酸(CGC>CAC),此突变为已知突变G1[10]。
2.3GDF9基因5′调控区扩增及其多态性分析
首先在小尾寒羊中通过调控区扩增引物,获得了长为2 636 bp的扩增片段(图5),覆盖起始密码子上游调控区2 304 bp,比预测PCR产物(2 357 bp)长279 bp。为了进一步确认该结果,本研究在不同绵羊品种DNA中进行PCR扩增,结果均一致。序列分析显示,测序序列和数据库序列部分一致,从起始密码子ATG到上游-1 848 bp基本一致,但在-1 848~-2 336 bp间本研究测序结果比NC_019462.1多两个长片段:1 10和1 69 bp(图6)。10只小尾寒羊序列比对发现,在-2 078 bp位点存在C>G突变(图7)。
2.4GDF9在11个绵羊品种中遗传多态性分布
对本研究发现的2个位点以及GDF9已知的功能位点进行SNP分型,发现11个绵羊品种均不含FecGE、FecGH、FecGT、FecGF、FecGV突变,G260A和-2078C>G突变广泛存在于除了草原型藏羊外的其他各绵羊品种中。其中,首次发现山谷型藏羊、欧拉羊、策勒黑羊、澳洲美利奴羊和乌珠穆沁羊携带G260A突变(表3)。G260A包含AA、BB和AB 3种基因型,其中BB型仅存在于小尾寒羊和山谷型藏羊中。哈迪-温伯格平衡检验发现该位点各基因型频率在各群体中处于非平衡状态。进一步分析发现,-2078C>G突变在基因型上表现出与G260A完全连锁,在基因型频率和等位基因频率上二者表现完全一致。通过分析小尾寒羊产羔数与基因型的关联性(表4),发现BB突变纯合子个体能繁殖,且产单羔,而AB杂合型的产羔数显著高于AA型(P<0.05)。

图2 GDF9 mRNA序列与NM_001142888序列比对Fig.2 Sequence alignment between GDF9 mRNA and NM_001142888

M.DNA相对分子质量标准;1~5.GDF9-E1产物(470 bp);6~10.GDF9-I1产物(1 420 bp);11~15.GDF9-E2产物(1 493 bp)M.DL2000 DNA marker;1-5.Products of GDF9-E1 (470 bp);6-10.Products of GDF9-I1 (1 420 bp);11-15.Products of GDF9-E2 (1 493 bp)图3 GDF9 外显子1、外显子2、内含子1 PCR扩增产物电泳Fig.3 Electrophoresis of exon 1,exon 2,intron 1 of GDF9 gene

图4 G260A基因型测序峰Fig.4 Sequencing profiles of G260A site
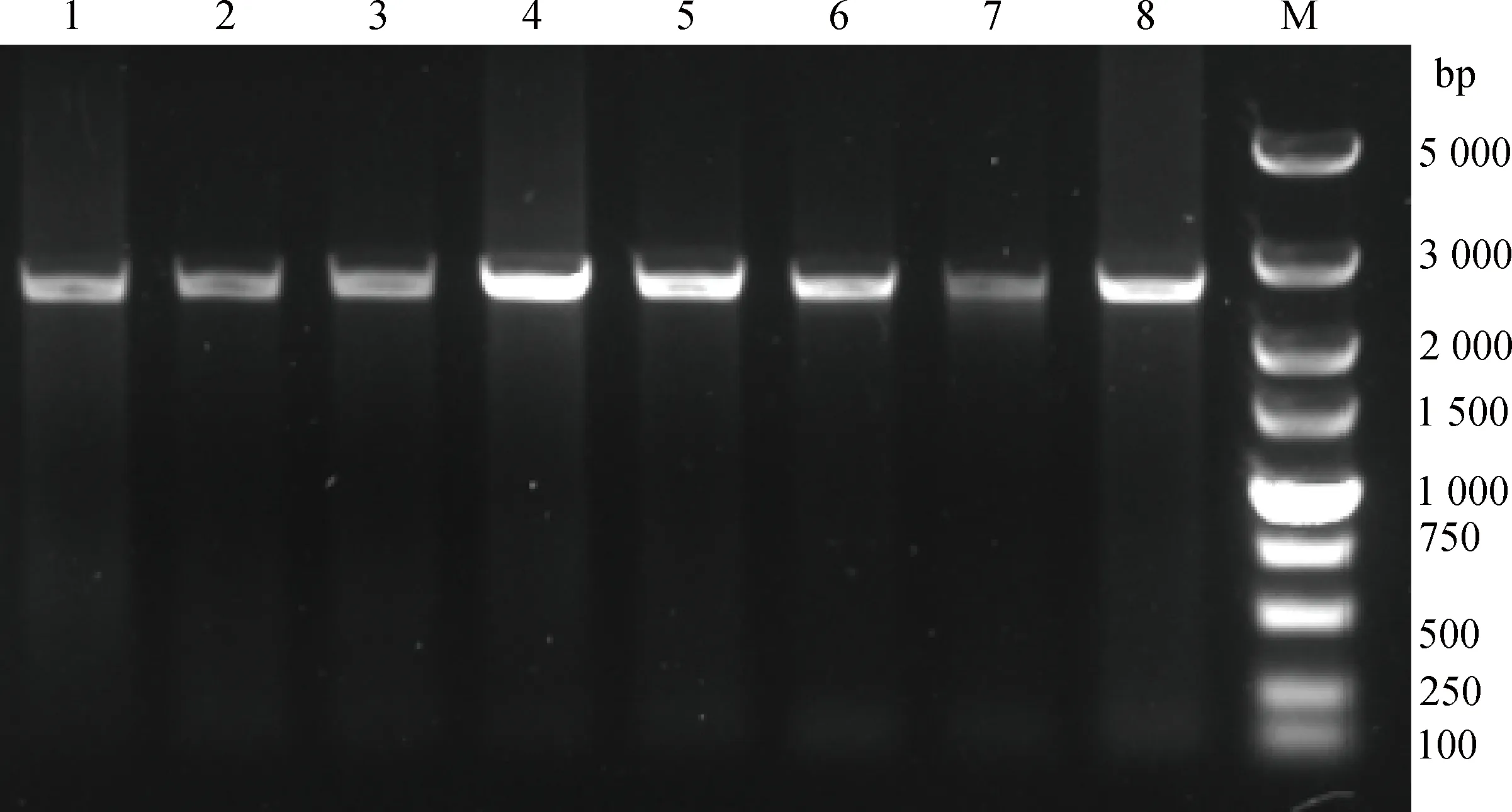
M.DNA相对分子质量标准;1~8.TK产物M.DL5000 DNA marker;1-8.Products of TK (2 636 bp)图5 GDF9调控区扩增产物电泳Fig.5 Electrophoresis of regulatory region of GDF9 gene

图6 GDF9基因调控区与序列比对Fig.6 Sequence alignment between GDF9 regulatory region and NC_019462.1

图7 调控区突变位点-2078C>G测序峰Fig.7 Sequencing profiles of -2078C>G site
表3不同绵羊品种GDF9基因G260A和-2078C>G位点基因型频率及等位基因频率
Table 3Allele and genotype frequencies of G260A and -2078C>G mutations ofGDF9 gene in different sheep breeds

品种Breed个体数NumberG260A基因型频率Genotypefrequency等位基因频率AllelefrequencyAAABBBAB小尾寒羊Small-tailHan920.935(86)0.054(5)0.011(1)0.9620.038策勒黑羊CeleBlack300.833(25)0.167(5)0.000(0)0.9170.083湖羊Hu360.917(33)0.083(3)0.000(0)0.9580.042多赛特羊Dorset250.920(23)0.080(2)0.000(0)0.9600.040澳洲美利奴羊AustralianMerino300.733(22)0.267(8)0.000(0)0.8660.134山谷型藏羊ValleyTibetan300.733(22)0.233(7)0.033(1)0.850.150草原型藏羊PrairieTibetan301.000(30)0.000(0)0.000(0)1.0000.000巴音布鲁克羊Bayinbuluke300.800(24)0.200(6)0.000(0)0.9000.100乌珠穆沁Ujumqin290.897(26)0.103(3)0.000(0)0.9480.052欧拉羊Oula170.882(15)0.118(2)0.000(0)0.9410.059滩羊Tan350.914(32)0.086(3)0.000(0)0.9570.043品种Breed个体数Number-2078C>G基因型频率Genotypefrequency等位基因频率AllelefrequencyCCCDDDCD小尾寒羊Small-tailHan920.935(86)0.054(5)0.011(1)0.9620.038策勒黑羊CeleBlack300.833(25)0.167(5)0.000(0)0.9170.083湖羊Hu360.917(33)0.083(3)0.000(0)0.9580.042多赛特羊Dorset250.920(23)0.080(2)0.000(0)0.9600.040澳洲美利奴羊AustralianMerino300.733(22)0.267(8)0.000(0)0.8660.134山谷型藏羊ValleyTibetan300.733(22)0.233(7)0.033(1)0.8500.150草原型藏羊PrairieTibetan301.000(30)0.000(0)0.000(0)1.0000.000巴音布鲁克羊Bayinbuluke300.800(24)0.200(6)0.000(0)0.9000.100乌珠穆沁Ujumqin290.897(26)0.103(3)0.000(0)0.9480.052欧拉羊Oula170.882(15)0.118(2)0.000(0)0.9410.059滩羊Tan350.914(32)0.086(3)0.000(0)0.9570.043
表4小尾寒羊GDF9基因G260A突变不同基因型个体的产羔数
Table 4The litter size of individuals with different genotypes ofGDF9 G260A mutation in Small-tail Han sheep (mean士SD)

突变位点Mutationsite基因型Genotype个体数Number产羔数LittersizeG260AAA862.16±0.318aAB52.80±0.248b
同列小写字母不同表示差异显著(P<0.05)
Data marked with the different superscripts within the same rank differ significantly (P<0.05)
3 讨 论
本研究克隆了绵羊GDF9 mRNA序列,结果与前人研究一致[8],GDF9包含2个外显子,一个内含子,编码区长1 368 bp,但相比前人研究,本研究拓展了该基因的5′-UTR 58 bp和3′-UTR 185 bp,确定其转录起始位点位于-58 bp,这对研究GDF9基因表达调控机制具有重要作用。进一步克隆获得2 898 bp的DNA序列和2 304 bp的调控区序列,序列比对发现DNA序列与NCBI数据库序列一致,但是调控区序列出现了不一致的结果,在本研究的绵羊品种中扩增产物长度均为2 636 bp,比NCBI数据的序列NC_019462.1多2个长片段,由于NC_019462.1为Texel羊的序列,是否在Texel羊中存在缺失还有待进一步的证明。
目前GDF9基因在各绵羊品种中的遗传多态性被广泛研究,本研究发现的一个突变为G260A,该突变在2004年由J.P.Hanrahan等[10]已经对其报道,并命名为G1突变,但未将其作为效应位点。然而随后的研究发现,G260A突变杂合子可增加Moghani和Ghezel母羊的产羔数,且突变纯合子母羊表现出正常的生殖能力[14],在97个个体中,5只G260A位点野生型的个体产双羔(6.3%);13只G260A位点突变杂合型个体中7只产双羔(53.8%);4只G260A位点突变纯合型个体都能繁殖,且产单羔。最近伊朗的Afshari、Baluchi、Makui和Mehraban绵羊也表现出杂合子产羔数显著高于野生型[15],这表明G260A确实对绵羊繁殖力具有一定效应。当前已经在全世界多个绵羊品种中检测到G260A位点,包括Cambridge、Belclare[10],Moghani,Ghezel,Afshari,Baluchi,Makui,Mehraban[14,15],Garole,Bonpala[23-24],Chios,Karagouniki[25],Araucano Creole[26],德国肉用美利奴羊、巴音布鲁克羊、小尾寒羊、湖羊、洼地绵羊、多赛特、特克塞尔、杜泊羊[27-29],其中仅在Moghani、Ghezel、Baluchi、Makui和Chios绵羊中检测到突变纯合子[14-15,25]。本研究首次在山谷型藏羊、欧拉羊、策勒黑羊、澳洲美利奴和乌珠穆沁羊中检测到该位点,且在小尾寒羊和山谷型藏羊中检测到突变纯合子BB型。该突变位点主要以杂合子形式存在,这可能与之前报道的纯合子突变不利于其繁殖有关[14-15],小尾寒羊BB纯合子表现出产单羔印证了这一点。本研究发现该位点虽然在多羔品种和单羔品种中不存在基因型分布差异,但是在小尾寒羊中G260A突变AB型个体产羔数显著高于AA型(P<0.05),说明该位点可能仅在小尾寒羊中具有一定效应,或在小尾寒羊中该位点与致因位点存在搭车效应,且在本研究中AB型个体仅为5只,该位点是否为致因位点还待进一步的研究。综上所述,虽然G260A不能确认为致因位点,但该位点对一些绵羊多羔具有一定的效应,用于指导小尾寒羊多羔育种具有一定的意义,可作为潜在的有效遗传标记。
本研究同时将GDF9基因多态性研究聚焦在调控区,并发现调控区新突变-2078C>G,且该位点和G260A位点完全连锁。G260A虽改变了第87位氨基酸,但该位置并不涉及成熟蛋白的功能活性[10]。而最近的一些研究表明,调控区的突变将会直接影响基因的表达水平[30-32],新发现的-2078C>G突变位于5′调控区,可能对GDF9基因表达具有调控作用,G260A突变的效应是否为-2078C>G引起的,这值得进一步深入研究。
4 结 论
克隆获得绵羊GDF9基因mRNA全长序列1 852 bp、DNA序列2 898 bp和调控区序列2 304 bp,并发现了已知突变G260A(G1)和调控区新突变-2078C>G。11个绵羊品种均不含FecGE、FecGH、FecGT、FecGF、FecGV突变,但G260A和-2078C>G广泛存在于除草原型藏羊外的各绵羊品种中。G260A和-2078C>G均存在3种基因型,-2078C>G在基因型上表现出与G260A完全连锁。在小尾寒羊中,G260A位点AB型个体产羔数显著高于AA型(P<0.05)。
[1]GALLOWAY S M,MCNATTY K P,CAMBRIDGE L M,et al.Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner[J].NatGenet,2000,25(3):279-283.
[2]DAVIS G H.Major genes affecting ovulation rate in sheep[J].GenetSelEvol,2005,37(Suppl 1):S11-S23.
[3]KAIVO-OJA N,BONDESTAM J,KAMARAI-NEN M,et al.Growth differentiation factor-9 induces Smad2 activation and inhibin B production in cultured human granulosa-luteal cells[J].JClinEndocrinolMetab,2003,88(2):755-762.
[4]ELVIN J A,CLARK A T,WANG P,et al.Paracrine actions of growth differentiation factor-9 in the mammalian ovary[J].MolEndocrinol,1999,13(6):1035-1048.
[5]MCGRATH S A,ESQUELA A F,LEE S J.Oocyte-specific expression of growth/differentiation factor-9[J].MolEndocrinol,1995,9(1):131-136.
[6]DONG J,ALBERTINI D F,NISHIMORI K,et al.Growth differentiation factor-9 is required during early ovarian folliculogenesis[J].Nature,1996,383:531-535.
[7]SADIGHI M,BODENSTEINER K J,BEATTIE A E,et al.Genetic mapping of ovine growth differentiation factor 9 (GDF9) to sheep chromosome 5[J].AnimGenet,2002,33(3):244-245.
[8]BODENSTEINER K J,CLAY C M,MOELLER C L,et al.Molecular cloning of the ovine growth/differentiation factor-9 gene and expression of growth/differentiation factor-9 in ovine and bovine ovaries[J].BiolReprod,1999,60(2):381-386.
[9]胡冬利,李齐发,徐业芬,等.湖羊生长分化因子9 (GDF9)基因组织表达特征、mRNA 表达水平与SNPs分析[J].农业生物技术学报,2010,18(3):533-538.
HU D L,LI Q F,XU Y F,et al.The tissue expression profile,mRNA expression level and SNPs analysis on GDF9 gene in Hu sheep[J].JournalofAgriculturalBiotechnology,2010,18(3):533-538.(in Chinese)
[10]HANRAHAN J P,GREGAN S M,MULSANT P,et al.Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovisaries)[J].BiolReprod,2004,70(4):900-909.
[11]MELO E O,SILVA B D M,CASTRO E A,et al.A novel mutation in the Growth and Differentiation Factor 9 (GDF9) gene is associated,in homozygosis,with increased ovulation rate in Santa Ines sheep[J].BiolReprod,2008,78:141,371.
[12]SILVA B D M,CASTRO E A,SOUZA C J H,et al.A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep[J].AnimGenet,2011,42(1):89-92.
[13]NICOL L,BISHOP S C,PONG-WONG R,et al.Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep[J].Reproduction,2009,138(6):921-933.
[14]BARZEGARI A,ATASHPAZ S,GHABILI K,et al.Polymorphisms in GDF9 and BMP15 associated with fertility and ovulation rate in Moghani and Ghezel sheep in Iran[J].ReprodDomestAnim,2010,45(4):666-669.
[15]JAVANMARD A,AZADZADEH N,ESMAILIZADEH A K.Mutations in bone morphogenetic protein 15 and growth differentiation factor 9 genes are associated with increased litter size in fat-tailed sheep breeds[J].VetResCommun,2011,35(3):157-167.
[16]CHU M X,YANG J,FENG T,et al.GDF9 as a candidate gene for prolificacy of Small Tail Han sheep[J].MolBiolRep,2011,38(8):5199-5204.
[17]VAGE D I,HUSDAL M,KENT M P,et al.A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep[J].BMCGenet,2013,14:1.
[18]MULLEN M P,HANRAHAN J P.Direct evidence on the contribution of a missense mutation in GDF9 to variation in ovulation rate of Finnsheep[J].PLoSOne,2014,9(4):e95251.
[19]JIANG Y,XIE M,CHEN W,et al.The sheep genome illuminates biology of the rumen and lipid metabolism[J].Science,2014,344(6188):1168-1173.
[20]TOUATI A,BLOUIN Y,SIRAND-PUGNET P,et al.Molecular epidemiology of mycoplasma pneumoniae:genotyping using single nucleotide polymorphisms and SNaPshot technology[J].JClinMicrobiol,2015,53(10):3182-3194.
[21]DANIEL R,SANTOS C,PHILLIPS C,et al.A SNaPshot of next generation sequencing for forensic SNP analysis[J].ForensicSciIntGenet,2015,14:50-60.
[22]YANG L,SUN H Y,CHEN D Z,et al.Application of multiplex SNaPshot assay in measurement of PLAC4 RNA-SNP allelic ratio for noninvasive prenatal detection of trisomy 21[J].PrenatalDiag,2014,34(2):139-144.
[23]POLLEY S,DE S,BRAHMA B,et al.Polymorphism of BMPR1B,BMP15 and GDF9 fecundity genes in prolific Garole sheep[J].TropAnimHealthPro,2010,42(5):985-993.
[24]ROY J,POLLEY S,DE S,et al.Polymorphism of fecundity genes (FecB,FecX,and FecG) in the Indian Bonpala sheep[J].AnimBiotechnol,2011,22(3):151-162.
[25]LIANDRIS E,KOMINAKIS A,ANDREADOU M,et al.Associations between single nucleotide polymorphisms of GDF9 and BMP15 genes and litter size in two dairy sheep breeds of Greece[J].SmallRuminantRes,2012,107(1):16-21.
[26]PAZ E,QUINONES J,BRAVO S,et al.Identification of G1 and G8 polymorphisms of GDF9 gene in Araucano creole sheep[J].ArchMedVet,2014,46(2):327-331.
[27]左北瑶,钱宏光,刘佳森,等.德国肉用美利奴羊BMPR-IB、BMP15和GDF9基因10个突变位点的多态性检测分析[J].南京农业大学学报,2012,35(3):114-120.
ZUO B Y,QIAN H G,LIU J S,et al.Detection of the 10 mutations of BMPR-IB,BMP15 and GDF9 gene in German Mutton Merino sheep[J].JournalofNanjingAgriculturalUniversity,2012,35(3):114-120.(in Chinese)
[28]ZUO B Y,QIAN H G,WANG Z Y,et al.A study on BMPR-IB genes of Bayanbulak sheep[J].AsianAustralJAnim,2013,26(1):36-42.
[29]古丽格娜,艾买提·买买提,於建国,等.7种绵羊和4种山羊GDF9基因G1突变检测[J].中国草食动物科学,2015,35(4):1-4.
GULIGENA,AIMAITI·MAIMAITI,YU J G,et al.Detection of the G1 mutation of the GDF9 gene in seven sheep and four goat breeds[J].ChinaHerbivoresScience,2015,35(4):1-4.(in Chinese)
[30]PARK C K,LEE S H,KIM J Y,et al.Expression level of hTERT is regulated by somatic mutation and common single nucleotide polymorphism at promoter region in glioblastoma[J].Oncotarget,2014,5(10):3399-3407.
[31]MAHAJAN M,YADAV S K.Gain of function mutation in tobacco MADS box promoter switch on the expression of flowering class B genes converting sepals to petals[J].MolBiolRep,2014,41(2):705-712.
[32]HUANG J,DANG R,TORIGOE D,et al.Genetic variation in the GDNF promoter affects its expression and modifies the severity of Hirschsprung’s disease (HSCR) in rats carrying Ednrb mutations[J].Gene,2016,575(1):144-148.
(编辑郭云雁)
Cloning and Genetic Polymorphism Analysis of mRNA,DNA and Regulatory Region of OvineGDF9 Gene in 11 Breeds
PAN Zhang-yuan,HE Xiao-yun,LIU Qiu-yue,HU Wen-ping,WANG Xiang-yu,GUO Xiao-fei,CAO Xiao-han,DI Ran*,CHU Ming-xing*
(KeyLaboratoryofFarmAnimalGeneticResourcesandGermplasmInnovationofMinistryofAgriculture,InstituteofAnimalScience,ChineseAcademyofAgriculturalSciences,Beijing100193,China)
The aim of this study was to clone the mRNA,DNA and regulatory region sequence of ovineGDF9 gene,detect its genetic polymorphism in 11 sheep breeds,and search for the molecular genetic markers related to sheep litter size.The RACE and PCR technologies were used to clone full-length sequence of ovineGDF9 gene,and SNaPshot was performed to detect the polymorphisms ofGDF9 gene in 11 breeds.A 1 852 bp full-length mRNA of ovineGDF9 gene (GenBank No.:KR063137) was obtained,which contains 58 bp 5′-UTR and 432 bp 3′-UTR.In addition,2 898 bp of DNA sequence and 2 304 bp of regulatory region sequence were amplified.Compared with the sequence of NC_019462.1,the result had 2 more fragments in regulatory region.The genetic polymorphism analysis showed that a known mutation G260A (G1) and a novel mutation -2078C>G within regulatory region were identified.The genotyping results showed that 11 sheep breeds were free ofFecGE,FecGH,FecGT,FecGF,FecGVmutations,while the G260A and -2078C>G were widespread in all sheep except Prairie Tibetan sheep.G260A contained 3 genotypes AA,BB and AB,but the BB genotype only presented in Small-tail Han sheep and Valley Tibetan sheep.-2078C>G also included 3 genotypes,in which the genotype and allele frequencies were exactly the same with G260A.It seems that -2078C>G was complete linkage with G260A.Association analysis in Small-tail Han sheep showed that the litter size of individuals with AB genotype was significantly higher than that of AA genotype (P<0.05).We improved the mRNA,DNA and regulatory region sequences of ovineGDF9,which provided a foundation for further functional study ofGDF9 gene.Meanwhile,G260A and -2078C>G mutations were found in different sheep breeds,and G260A site could be a potential genetic marker for improving litter size in sheep.
sheep;GDF9 gene;mRNA;regulatory region;genetic polymorphism
10.11843/j.issn.0366-6964.2016.08.005
2015-12-14
中国农业科学院科技创新工程(ASTIP-IAS13);国家肉羊产业技术体系专项(CARS-39);国家自然科学基金项目(31472078);国家转基因科技重大专项(2016ZX08009-003-006;2016ZX08010-005-003);内蒙古自治区战略性新兴产业发展专项资金计划
潘章源(1986-),男,江西赣州人,博士,主要从事动物遗传育种与繁殖研究,E-mail:pzq170450077@163.com
狄 冉,副研究员,E-mail:dirangirl@163.com;储明星,研究员,E-mail:mxchu@263.net
S826;S821.2
A
0366-6964(2016)08-1555-10

