畜禽养殖过程抗生素使用与耐药病原菌及其抗性基因赋存的研究进展
隋倩雯,张俊亚,魏源送,,*,陈梅雪,董红敏,熊继海
1. 中国科学院生态环境研究中心 环境模拟与污染控制国家重点联合实验室,北京 100085 2. 中国农业科学院农业环境与可持续发展研究所,北京 100081 3. 江西省科学院能源研究所,南昌 330096
畜禽养殖过程抗生素使用与耐药病原菌及其抗性基因赋存的研究进展
隋倩雯1,张俊亚1,魏源送1,3,*,陈梅雪1,董红敏2,熊继海3
1. 中国科学院生态环境研究中心 环境模拟与污染控制国家重点联合实验室,北京 100085 2. 中国农业科学院农业环境与可持续发展研究所,北京 100081 3. 江西省科学院能源研究所,南昌 330096
兽用抗生素在提高畜禽生产性能、防治疾病方面发挥着重要作用,目前全球超过一半以上抗生素用于畜禽养殖,畜禽养殖源耐药病原菌、抗性基因及其传播风险愈益得到人们的重视。我国是畜禽养殖和抗生素使用大国,但兽用抗生素使用、病原菌耐药水平及其抗性基因类型等数据却较为缺乏,不利于今后畜禽养殖源耐药病原菌及其传播风险的控制。因此,本文通过文献调研,对我国和主要发达国家的兽用抗生素使用情况、畜禽养殖源耐药病原菌及其携带的抗性基因、基因移动元件以及向环境传播的途径进行分析、总结,以期为规范合理用药、降低耐药病原菌及其抗性基因传播风险,建立从畜禽养殖场至公共环境全过程的抗性污染控制链条提供借鉴。
兽用抗生素;重金属;耐药病原菌;抗生素抗性基因;基因移动元件
Received 22 September 2015 accepted 21 October 2015
世界卫生组织指出抗生素抗性是21世纪人类面临最大的挑战,但有关畜禽养殖业抗生素抗性的数据非常缺乏,应监督和促进畜禽养殖业的合理用药,加强抗生素使用与抗性传播的研究[1]。自1949年美国科学家发现饲料中添加金霉素可使猪、鸡增重,节约饲料以来[2],兽用抗生素使用已有60年历史。兽用抗生素按照用途分为3类,分别为促生长类(animal growth promoters)、预防类(prophylactic use)和治疗类(therapeutic use)[3],随着畜禽业的集约化、规模化发展,为提高动物生产性能、防治疾病,兽用抗生素发挥着重要作用。据统计全球兽用抗生素用量是人类用量的2倍[4]。全球平均牛、鸡、猪在单位动物产品生产过程中抗生素消耗量分别为45、148和172 mg·kg-1[5]。2010年兽用抗生素用量最大的国家依次为中国(23%)、美国(13%)、巴西(9%)、印度(3%)和德国(3%),中国高居全球兽用抗生素消耗最大国家[5]。
抗生素以饲料添加、口服、注射等方式在畜禽养殖业使用,其中饲料添加剂抗生素用量最大。除抗生素之外,重金属如铜、锌、砷等矿物元素也作为饲料添加剂大量使用[6]。Chantziaras等[7]的研究表明兽用抗生素(包括氯霉素类、磺胺类、链霉素、四环素、氨基青霉素类、庆大霉素、第三代头孢菌素等)用量与畜禽养殖源分离的埃希氏大肠杆菌(Escherichia coli, E. coli )的耐药水平具有正相关关系。由于兽用抗生素引起细菌耐药性,欧盟国家已在1997年开始逐步禁止促生长类抗生素的使用,在随后的5年中畜禽养殖源分离的耐药菌尤其鸡源分离的粪肠球菌(Enterococcus faecium )对安普霉素、维吉尼亚霉素、泰乐菌素、卑霉素的耐药率大幅降低[8]。不同于人用抗生素的使用与耐药性监测,畜禽养殖过程的相关数据非常缺乏。
畜禽养殖源赋存大量病原菌,如肠杆菌科细菌、肠球菌、葡萄球菌等[9]。在抗生素的选择压力和重金属的协同选择作用下[10-11],动物肠道、粪便、废水存储大量耐药病原菌[12-13],其携带的抗性基因由移动基因元件介导具有水平转移功能[14],存在较大的疫病传播和公共健康风险[15]。Martínez等[16]将移动基因元件介导且宿主细菌为人类病原菌的抗性基因排序为一级风险,与其他类型抗性基因相比对人类健康威胁最大。
由于畜禽粪便、粪水的还田利用或排放,畜禽源耐药病原菌从养殖场传播至外界环境(土壤、河流)[17-18],存在抗性污染的传播风险,尤其畜禽源耐药菌对环境抗性水平提高的贡献以及对公共健康安全的威胁,目前仍未有定论,缺乏系统性研究与评估[19]。此外,有研究表明人类肠道四环素抗性基因丰度最高,极有可能来自于兽用抗生素的使用以及抗性基因沿食物链的传播[20]。有研究从蔬菜分离得到来自于动物粪便的耐药病原菌及抗性基因[9, 21]。耐药病原菌及其携带的抗性基因沿食物链传播,可能是畜禽养殖抗性基因向人类传播的途径之一,然而该传播机制尚不明确。
因此,本文通过文献调研,总结归纳了我国及主要发达国家兽用抗生素以及重金属使用情况,畜禽养殖源分离的病原菌的耐药水平及其携带的抗性基因,以及由移动基因元件介导的抗性基因类型,并探讨了畜禽养殖场通过粪便的土地利用引起环境中耐药病原菌传播的可能。本文着重从畜禽养殖抗生素使用到动物与环境中耐药病原菌及其抗性基因为主线进行总结、讨论,并对今后的研究重点和方向提出建议和展望,以期为合理使用兽用抗生素与重金属、降低环境抗生素抗性传播提供借鉴。
1 我国和主要发达国家兽用抗生素和重金属的使用状况(The veterinary antibiotics and heave metal use in China and other developed countries)
中国是抗生素生产和消费大国。据统计2013年我国抗生素生产总量24.8万t,国内消耗16.2万t,磺胺类、四环素类、氟喹诺酮类、大环内脂类、β-内酰胺类及其他分别占总用量的5%、7%、17%、26%、21%和24%,消耗的抗生素中48%用于人类,52%用于动物[22]。我国兽用抗生素的生产主要集中于山东、河南、河北、江苏、四川等畜禽养殖大省[23]。饲料添加是兽用抗生素的重要组成部分。Coates等[24]研究发现饲料抗生素对无菌动物无明显生长促进作用,其作用机理在于抑制肠道有害微生物生长,减少对营养成分的竞争;还可使肠绒毛和肠壁变薄,进而提高饲料的养分代谢率。我国农业部在《饲料药物添加剂使用规范》中将饲料添加抗生素分为2类,一类可在饲料中长时间添加使用,主要用于促进动物生长;另一类是通过混饲给药的饲料添加剂,用于防治动物疾病,规定了用量、用法和休药期。第一类促生长抗生素包括聚醚离子载体类6种、多肽类5种、四环素类2种,另外包括砷制剂2种;而第二类防治动物疾病的抗生素包括大环内脂类3种、氨基糖苷类4种、磺胺类3种、喹诺酮类2种、林可酰胺类1种等[25]。根据我国兽用抗生素的使用规范和文献中其他种类兽用抗生素的使用情况,将其分类总结于表1。
作为允许饲料抗生素使用的国家之一,美国存在饲料抗生素普遍使用的现象。2011年美国食品和药物管理局(Food and Drug Administration, FDA)报告指出,美国每年消耗的抗生素超过一半用于动物生产而非用于人类,其中兽用抗生素按消耗量排序依次为四环素类(5 643 t)、离子载体类(4 123 t)、青霉素类(880 t)、大环内脂类(583 t)、磺胺类(371 t)、氨基糖苷类(214 t)、林可酰胺类(190 t)、头孢菌素类(27 t)以及其他类(1 510 t)[26],其中用量最大的2类兽用抗生素与我国《饲料药物添加剂使用规范》中第一类饲料添加剂种类相似。至今美国食品和药物管理局仍允许抗生素作为生长促进和疾病预防目的使用,仅以自愿方式减少抗生素的使用[27]。美国712个猪场的调研发现,使用最多的兽用抗生素分别为四环素、卡巴多司、杆菌肽、泰乐菌素、安普菌素和林可霉素等种类,92.2%的抗生素是连续添加,而按饲养阶段分类抗生素用量依次为保育猪>育成猪>育肥猪,但仅仅有12%的畜禽养殖场不使用抗生素饲料添加剂[28]。在不同饲养阶段,抗生素的使用种类也存在差异。Apley等[29]统计了美国规模化猪场饲料抗生素使用情况,17种兽用抗生素中,保育猪较多使用金霉素、土霉素、替米考星,而育成猪/育肥猪较多使用金霉素、泰乐菌素和土霉素。实际饲养过程中,饲料抗生素在不同动物种类、饲养阶段、添加量各有不同,详见表2。例如,通常在生猪保育阶段猪饲料中抗生素添加量高于其他饲养阶段,即断奶仔猪抗生素用量最大;而不同动物相比,猪的饲料抗生素添加量高于牛和鸡,以单位动物产品估算,动物抗生素用量依次是猪>鸡>牛[5]。
自1997年禁用安普霉素作为动物促生长剂(animal growth promoter)后,欧盟国家开始逐步扩大动物促生长剂类抗生素的禁用范围,到2006年全面禁用动物促生长剂类抗生素。虽然随之发现治疗类抗生素使用量出现增加趋势,但兽用抗生素总用量降低了50%[27]。据统计,2012年欧盟及欧洲经济区(26国)的兽用抗生素用量合计8 046.4 t,依次为德国(1 714 t)、西班牙(1 694.7 t)、意大利(1 543 t)、法国(778.4 t)和波兰(518.3 t);折合单位动物产品抗生素用量排序依次为塞浦路斯、意大利、匈牙利、西班牙和德国,分别为396.5、341、245.5、242和204.8 mg·(population correction unit, PCU)-1;不同种类兽用抗生素用量所占比例依次为四环素类(44.2%)、青霉素类(18.3%)、磺胺类(14.5%)、大环内脂类(9.1%)等[30],与我国和美国消耗的兽用抗生素类型较为相似。
如表3所示,重金属(如铜、锌、砷)通常作为矿物元素广泛应用于畜禽养殖,是重要的饲料添加剂,用于提高饲料转化率、促进动物生长、提高生产性能。我国和美国都允许饲料中添加重金属[41-44],虽然欧盟国家已禁止促生长类抗生素的使用,但重金属作为饲料添加剂仍被允许使用[6]。
不被吸收的重金属以及代谢不完全的抗生素随粪便、尿液排泄出来[31]。众多研究表明养殖场抗生素、重金属的排放与动物饲养过程抗生素的摄入有关,猪粪和污水中的抗生素和重金属主要源于饲料添加[32-33]。关于畜禽养殖场抗生素的排放已有广泛研究,对养殖场周边土壤、水体环境造成较大影响[31-32, 34-35]。
2 畜禽养殖源耐药病原菌及其携带的抗性基因(Antibiotic resistance pathogen and the carried antibiotic resistance genes in animal production)
畜禽养殖场是病原菌及抗性基因的重要蓄积库[45-46],但目前畜禽养殖源耐药病原菌及其携带的抗性基因研究极为不足。畜禽养殖环境病原菌种类多样,其中包含大量病原菌(如葡萄球菌、肠球菌、沙门氏菌等)。Ferreira等[5]测试了猪场废水中总大肠菌群(total coliform, TC)、E. coli 数量分别为1.2×106、2.8×105MPN·(100 mL)-1。Resende等[9]测定牛场粪水中肠球菌和肠杆菌科细菌(Enterobactetiaceae )数量分别为3.71×105和4.42×108CFU·mL-1。Brooks等[45]采用定量PCR方法测试了育肥猪场废水中沙门氏菌、弯曲杆菌数量分别为7.24×103、1.41×104copies·mL-1,丰度分别为8.51×10-7、1.66×10-6copies/16S rRNA。Tulayakul等[33]测试了猪场废水中沙门氏菌的血清型检出频率最高的依次为Rissen、Anatum、Kedougou、Stanley、Typhimurium、Paratyphi B var. Java。

表1 兽用抗生素的种类、抑菌方式及抑菌类型Table 1 Type and antimicrobial mode of veterinary antibiotics
注:a分类依据《饲料药物添加剂使用规范》[25],*表示《饲料药物添加剂使用规范》规定的促生长类抗生素;**《饲料药物添加剂使用规范》规定的防治动物疾病类抗生素;***表示文献中常用兽用抗生素;G+表示革兰氏阳性菌;G-表示革兰氏阴性菌。
Notes:aClassification according to “Norms for Application of Feed Additives”[25]; * denotes animal growth promoting antibiotics defined in “Norms for Application of Feed Additives”; ** denotes disease control antibiotics defined in “Norms for Application of Feed Additives”; *** denotes other veterinary antibiotics from references; G+denotes Gram-positive bacteria; G-denotes Gram-negative bacteria.
表2 饲料抗生素的添加量
Table 2 Usage of antibiotics in feed for animal production

抗生素Antibiotics金霉素(Chlortetracycline)土霉素(Oxytetracycline)替米考星(Tilmicosin)泰乐菌素(Tylosin)金霉素(Chlortetracycline)金霉素a(Chlortetracyclinea)磺胺甲嘧啶a(Sulfamerazinea)青霉素a(Penicillina)卡巴多司(Carbadox)莫能霉素钠b,c(Monensinsodiumb,c)维吉尼亚霉素b(Virginiamycinb)磷酸泰乐菌素c(Tylosinphosphatec)金霉素(Chlortetracycline)莫能霉素钠d(Monensinsodiumd)泰乐菌素d(Tylosind)动物种类与饲养阶段Animaltypeandgrowthstage保育猪Nurserypig育成/育肥猪Finisher/fatteningpig保育猪Nurserypig育成/育肥猪Finisher/fatteningpig保育猪Nurserypig育成/育肥猪Finisher/fatteningpig保育猪Nurserypig育成/育肥猪Finisher/fatteningpig哺乳猪Lactatingsow妊娠猪Gestatingsow育肥(日龄:18周)Fattening,ageat18weeks保育猪Nurserypig肉鸡Boiler肉牛Beefcattle肉牛(12~15个月龄)Beefcattle,ageat12-15months使用量/(g·t-1)Consumption/(g·t-1)促生长Growthpromotion疾病预防Diseaseprevention疾病治疗Diseasetreatment2504004001004004005045045050400400363181363-36336340401002040100-330--550-100-100-50-1050110-15-20-22-29.9-11-文献References[29][36][37][38][39][40][41]
注:aASP250饲料添加剂包含金霉素、磺胺甲嘧啶和青霉素;b莫能霉素钠与维吉尼亚霉素结合使用;c莫能霉素钠与磷酸泰乐菌素结合使用;d莫能霉素钠与泰乐菌素结合使用;-无有效数据。
Notes:aASP250 feed addictive in comprise of chlortetracycline, sulfamerazine, penicillin;bMonensin sodium and virginiamycin combined use;cMonensin sodium and tylosin phosphate combined use;dMonensin sodium and tylosin combined use; - not available.
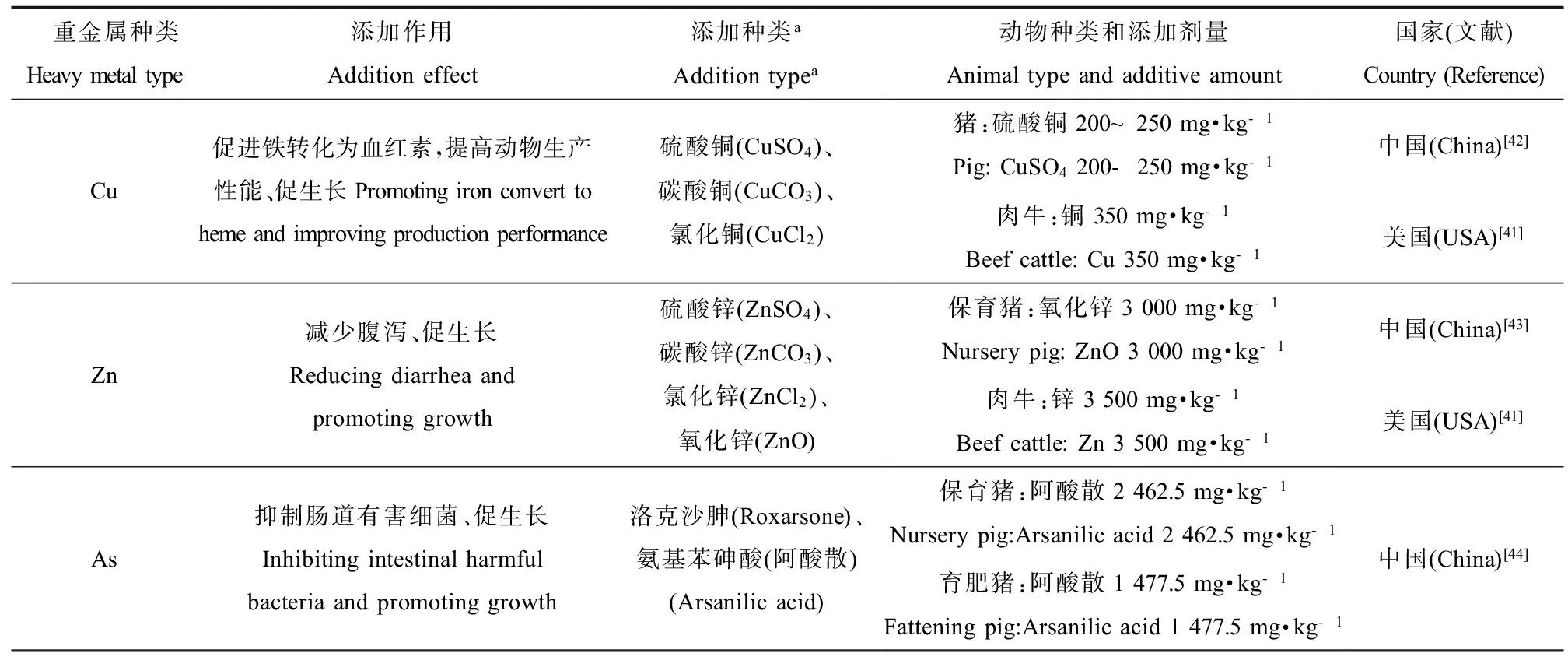
表3 饲料中重金属添加种类、作用及使用量Table 3 Category, function and dosage of heavy metals in feed summarized from literature
抗生素对动物肠道和粪便细菌具有选择压力,改变了细菌菌群结构。Danzeisen等[39]的研究结果表明,莫能霉素(monensin)与维吉尼亚霉素(virginiamycin)或与泰乐菌素(tylosin)结合使用会显著改变肉鸡盲肠菌群结构,使E. coli 数量提高。Looft等[37]的研究表明,饲料中添加ASP250(金霉素、磺胺甲嘧啶、青霉素的混合物,见表3)使猪粪中变形菌门(Proteobacteria )的比例从1%提高到11%,其中E. coli 比例增加20至100倍。
除细菌菌群结构改变以外,抗生素还对动物肠道、粪便细菌的耐药性具有选择压力,提高了细菌耐药水平。研究表明肉牛饲喂金霉素会显著增加粪便中E. coli 和肠球菌的耐药率[40]。氨苄西林用于仔猪饲养,口服或注射(20 mg·kg-1body weight)都会显著提高粪便中肠杆菌科细菌的耐药性,注射抗生素的耐药率从0.9%~12%升高至26%,口服抗生素的耐药率提高至49%;口服抗生素会使粪便中blaTEM拷贝数从104~106copies·g-1升高至107~109copies·g-1,而注射对粪便中抗性基因的影响不显著[47]。Looft等[37]的研究发现,饲料中添加金霉素、磺胺甲嘧啶、青霉素显著提高了猪粪中四环素外排泵、A类β -内酰胺酶、磺胺抗性、氨基糖苷磷酸转移酶、两类外排泵的抗性基因丰度。病原菌或潜在病原菌的耐药性及携带的抗性基因在动物肠道、粪便、废水中分布详见表4。
目前我国和部分发达国家的相关职能部门定期开展动物源细菌的耐药性测试。例如,我国农业部开展了对畜禽养殖场、屠宰场的动物肠道、动物产品的动物源细菌耐药性监测,2013年涵盖了15个畜禽养殖大省,细菌种类包括大肠杆菌、肠球菌、沙门氏菌、金黄色葡萄球菌、空肠弯曲杆菌、猪多杀巴氏杆菌和副猪嗜血杆菌;耐受药物主要包括青霉素、链霉素、红霉素、泰乐菌素、四环素、万古霉素等主要抗生素种类[48],但我国动物源细菌耐药性数据却鲜有公开报道。美国疾病控制中心、农业部、食品和药物管理局定期联合发布全国耐药性检验报告(National Antimicrobial Resistance Monitoring System, NARMS),其中包括动物源细菌耐药性检验结果,并给出相关耐药菌比例,如沙门氏菌由人类、屠宰鸡、屠宰猪分离得到的多重耐药率分别为9%、8%和16%[49]。
肠球菌和金黄色葡萄球菌是重要监控的畜禽源革兰氏阳性菌类型。动物饲喂泰乐菌素显著提高了粪便中大环内脂耐药菌及其抗性基因的数量[50]。而畜禽粪便中分离得到的大环内脂-林可酰胺-链阳霉素B(macrolide-lincosamide-streptogramin B, MLSB)多重耐药菌可能与饲喂泰乐菌素有关[41],其中粪肠球菌(Enterococcus faecalis )具有MLSB多重耐药性是质粒pAMβ1携带的抗性基因erm B介导[12]。耐万古霉素肠球菌(vancomycin-resistant enterococci, VRE)可能与饲料中添加的安普霉素有关[51],万古霉素是治疗肠球菌和金黄色葡萄球菌等革兰氏阳性菌引发疾病的“最后抗生素”,美国疾病防控中心将VRE归为“具有严重威胁”的耐药菌,而动物源分离的VRE由van A介导,具有万古霉素和替考拉宁抗性[52],因此VRE传播存在很大的疫病防治隐患。葡萄球菌也是重要的畜禽养殖源致病微生物,Neyra等[53]研究了屠宰场工人携带的金黄色葡萄球菌(Staphylococcus aureus )的耐药性,发现由mec A编码的甲氧西林耐药率为14.3%,多重耐药率为37.1%,主要耐受红霉素、头孢西丁、环丙沙星等抗生素,表明耐药菌及抗性基因可能通过动物屠宰以及动物产品的途径进行传播。
大肠埃希氏菌(大肠杆菌)、沙门氏菌、志贺氏菌等肠杆菌科(Enterobacteriaceae )革兰氏阴性细菌也是重要的动物源致病菌,同时是食品源微生物的重要检测与控制指标。β-内酰胺类抗生素包括青霉素类和头孢菌素类,应用广泛。然而β-内酰胺酶可由肠杆菌科细菌产生使抗生素失活。头孢噻呋是用作动物疾病治疗的第三代头孢菌素,主要用于治疗奶牛乳腺炎、动物呼吸道和肠道疾病[54-55],然而研究表明牛和猪在使用头孢噻呋后粪便分离的大肠杆菌、沙门氏菌具有人用三代头孢菌素(如头孢曲松钠)的抗性[13, 56]。Wu等[57]发现猪粪分离的E. coli 中普遍存在磺胺类抗性基因,其中sul 2(44%)检出率高于sul 1(29%),sul 基因由不兼容质粒(Inc)介导,转移接合率为82%。

表4 畜禽养殖源致病性耐药菌种类与分布Table 4 Antibiotic resistant pathogens and the distributions from animal source
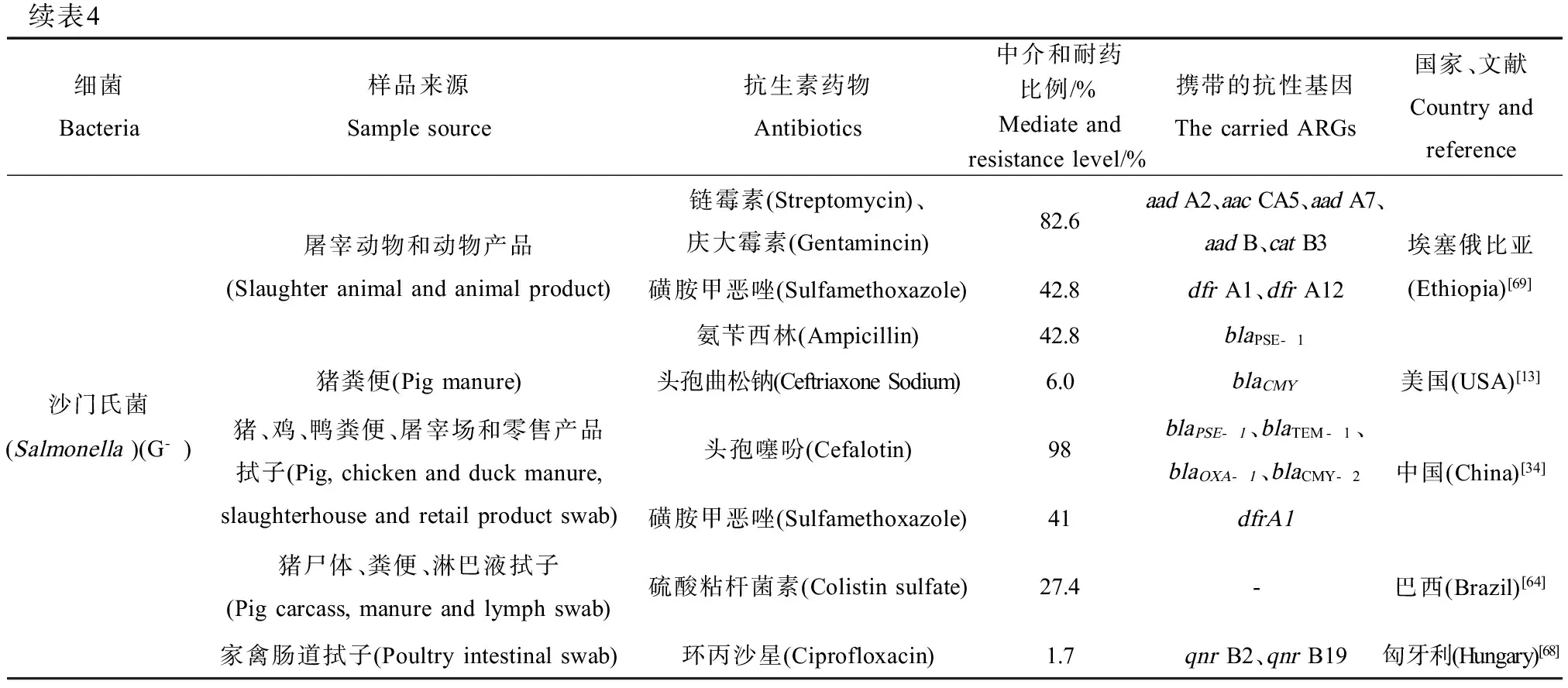
续表4细菌Bacteria样品来源Samplesource抗生素药物Antibiotics中介和耐药比例/%Mediateandresistancelevel/%携带的抗性基因ThecarriedARGs国家、文献Countryandreference沙门氏菌(Salmonella)(G-)屠宰动物和动物产品(Slaughteranimalandanimalproduct)猪粪便(Pigmanure)猪、鸡、鸭粪便、屠宰场和零售产品拭子(Pig,chickenandduckmanure,slaughterhouseandretailproductswab)猪尸体、粪便、淋巴液拭子(Pigcarcass,manureandlymphswab)家禽肠道拭子(Poultryintestinalswab)链霉素(Streptomycin)、庆大霉素(Gentamincin)82.6aadA2、aacCA5、aadA7、aadB、catB3磺胺甲恶唑(Sulfamethoxazole)42.8dfrA1、dfrA12氨苄西林(Ampicillin)42.8blaPSE-1头孢曲松钠(CeftriaxoneSodium)6.0blaCMY头孢噻吩(Cefalotin)98blaPSE-1、blaTEM-1、blaOXA-1、blaCMY-2磺胺甲恶唑(Sulfamethoxazole)41dfrA1硫酸粘杆菌素(Colistinsulfate)27.4-环丙沙星(Ciprofloxacin)1.7qnrB2、qnrB19埃塞俄比亚(Ethiopia)[69]美国(USA)[13]中国(China)[34]巴西(Brazil)[64]匈牙利(Hungary)[68]
注:a-用于采样的灭菌棉拭子,b-无有效数据,c具有甲氧西林耐药性的金黄色葡萄球菌,d庆大霉素和安普霉素交叉抗性。
Notes:a- sterile swab for sampling,b-not available,cwith methicillin- resistant Staphylococcus aureus (MRSA),dcross- resistance to gentamicin and apramycin.
3 畜禽养殖源中移动基因元件介导的抗生素抗性基因(Antibiotic resistance genes mediated by mobile genetic elements in animal production)
Martínez等[16]指出移动基因元件介导的抗性基因具有较高的传播风险。表5总结了由畜禽养殖源分离得到的移动基因元件介导的抗性基因。细菌依靠基因移动元件(mobile genetic element, MGE)包括接合质粒、转座元件(转座子和插入序列)和整合子发生基因的水平转移[14]。Looft等[37]研究发现,饲料抗生素(金霉素、磺胺甲嘧啶、青霉素)显著提高了未添加抗生素(如氨基糖苷类)的抗性基因和多药剂外排泵(multi-drug efflux)的丰度,这可能由于多种抗性基因位于同一移动原件上,由抗生素的协同选择(co-selection)所造成。Chen等[41]指出饲喂泰乐菌素对牛粪中四环素抗性基因(tet A、tet G和核糖体保护类四环素抗性基因)具有协同选择的作用。Kanwar等[62]发现,与饲料中不添加金霉素相比,添加金霉素会显著增加牛粪中blaCMY-2和blaCTX-M的丰度,可能存在金霉素对头孢菌素类抗性基因的协同选择作用。虽然农业部及药品监管部门限制了兽用抗生素的使用种类,但其协同选择作用可能引起人用抗生素(如第三代头孢菌素、万古霉素)的抗性水平提高。然而,协同选择不仅发生在抗生素之间,Seiler等[6]指出重金属对抗生素抗性基因具有协同选择作用。协同选择的原因之一为某种抗性基因编码的酶或蛋白具有提高细胞耐受多种抑菌物质(如抗生素或重金属)的能力,如多重药剂外排泵(multi drug efflux pumps),可以将毒性物质迅速排出细胞外[11, 14]。另一方面,2种或多种抗性功能的基因相互邻近并在同一个移动基因元件上[70],如猪粪中分离的质粒pMC2,携带了大环内脂、四环素等抗生素抗性基因和汞、铬等重金属抗性基因,具有很强的移动和接合能力[71]。由于重金属与抗生素的协同选择作用,增加了抗生素抗性传播的控制难度和抗性基因研究的复杂程度,而由移动基因元件介导的抗性基因是相关研究的重点。
Binh等[72]的研究表明,猪粪中分离的接合质粒包括IncP-1、pHV216-like、IncN、IncW,其中IncN比例最高(34%),携带阿莫西林、磺胺嘧啶的抗性基因分别为bla TEM、sul 1、sul 2、sul 3。
整合子是重要的基因移动元件,在猪粪及其施用的土壤中广泛存在。Agersø等[73]的研究表明,33%猪粪分离的革兰氏阴性菌、17%猪粪分离的革兰氏阳性菌、5%土壤分离的革兰氏阴性菌、12%土壤分离的革兰氏阳性菌携带一类整合子,其中土壤中携带一类整合子的菌属分别为假单胞菌、产碱杆菌、棒状杆菌、节杆菌;猪粪中携带一类整合子的菌属分别为E. coli 、肠杆菌、节杆菌等。
Zhang等[74]研究了不同养殖来源E. coli 中由整合子介导抗性基因的分布特征,发现只有鸡场分离的E. coli 携带二类整合子(int I2),int I2携带的基因盒长度为2 400 bp,介导的抗性基因序列dfr A1-sat 1-aad A1-orf X,转移接合率为69.2%;int I1携带介导的抗性基因序列分别为aad A23B、aad A2、arr -3-dfr16等,基因盒长度为1 009~2 000 bp小于int I2携带的基因长度,转移接合率74.8%。

表5 畜禽养殖源移动基因元件介导的抗性基因Table 5 ARGs mediated by mobile gene elements in animal source
注:a质粒上携带转座子和插入序列,- 无有效数据。
Notes:aThe plasmid contained transposon and insertion sequence, - not available.
诸多研究表明,肠球菌携带的铜抗性基因(tcr B和cue O)与四环素、大环内脂、万古霉素、氨苄西林的抗性基因具有协同转移(co-transfer)的特点,这可能因为它们位于同一基因移动元件上[75-76]。Amachawadi等[77]的研究表明,肉牛饲料中矿物元素铜添加量的增加(10 mg·kg-1提高到100 mg·kg-1)显著提高了粪便中耐铜屎肠球菌(Enterococcus faecium )丰度(P <0.05),抗铜基因tcr B由接合质粒介导并同时携带四环素和大环内脂类抗性基因tet M和erm B,与粪肠球菌(Enterococcus faecalis )种间水平转移率为2.0×10-5。Cavaco等[78]发现由猪场和肉牛场分离的耐甲氧西林金黄色葡萄球菌(MRSA)同时携带抗锌表型和抗锌基因(czr C),表明饲料中添加锌可能促进MRSA的出现,促进其传播。由基因移动元件介导的抗生素抗性基因及其对重金属响应的研究还较为不足,由于重金属对抗生素抗性基因的协同选择作用,在重金属饲料添加使用方面应更为谨慎。
4 抗生素耐药性从畜禽养殖场向环境的传播(Antibiotic resistance spread from animal farm to environment)
抗性基因及基因移动元件从畜禽养殖场向外界传播途径如图1所示,主要传播途径包括畜禽养殖人员的暴露,畜禽产品的造成食品加工人员的暴露,以及畜禽粪污还田利用造成环境和人群的暴露。畜禽粪便、肉质产品中赋存的病原菌耐药水平及其抗性基因类型已在表4进行了总结,且畜禽养殖人员和食品加工者在抗生素抗性中的暴露主要是对小范围人群的影响。然而畜禽粪污还田利用使粪污中携带的耐药病原菌以及基因移动元件暴露于土壤、径流,使抗性污染从养殖场传播至外界环境,存在较大的疾病传播和公共健康隐患。然而该传播机制尚不明确[19],今后有待深入研究。

图1 抗性基因和移动基因元件从畜禽(以禽类为例)养殖场向公共环境的传播[81]
畜禽废弃物中赋存的病原菌在还田过程仍会长时间存活,不同细菌在土壤中的赋存时间不同。Jacobsen等[82]指出养殖场及周边环境是沙门氏菌的多发环境,沙门氏菌主要通过粪便、废水的还田利用向土壤环境传播,而沙门氏菌在土壤中可存活数周甚至数月。Piorkowski等[83]的研究结果表明,牛粪水还田后E. coli 消减1 log需要约40~50 d,消减2 log则需要约60~76 d。Wang等[84]的研究表明,E. coli O157:H7在土壤中平均存活时间为2.1~3.6 d,而沙质土、低pH值、低有机碳含量的土壤中E. coli O157:H7赋存时间较短。
畜禽养殖废弃物还田利用可能引起耐药病原菌进入土壤环境,使土壤中细菌耐药性和抗性基因丰度提高。Pourcher等[17]将恩诺沙星饲喂的鸡粪施用于农田土壤,发现在前35天肠杆菌科细菌丰度有所提高之后逐渐恢复本底水平,而耐恩诺沙星E. coli 由鸡粪引入,可在土壤中存活89 d以上。Cook等[85]研究了鸡粪还田后肠球菌的演变,发现肠球菌在土壤中90%消减时间需要7.41 d,施肥后148 d土壤中肠球菌仍维持在2~3 log CFU·g-1,土壤中抗性基因tet W、sul 1、str B在施肥148 d后仍无法恢复本底水平。Bech等[86]研究发现大肠杆菌和四环素耐药菌会在粪水还田的土壤中持续存活46~49 d。
畜禽养殖废弃物的排放或土地利用导致周边环境(土壤、河流)病原菌耐药水平的提高。West等[18]指出邻近畜禽养殖场的河流虽然常规水质指标满足标准,但粪大肠杆菌具有较高的多重耐药菌率(41.6%),比其他河流高25.1%,所携带的tet B、tet C由质粒介导可以与鼠伤寒沙门氏菌(Salmonella tyhimurium )间发生接合转移。Chee-Sanford等[88]发现猪场氧化塘下游250 m仍可测得四环素抗性基因tet M,养殖场四环素耐药菌主要为肠球菌、葡萄球菌、罗伊氏乳杆菌(Lactobacillus reuteri )。Chen等[41]研究了福建闽江流域E. coli 的耐药性,畜禽粪便可能是该流域抗生素耐药率高的重要因素,分离的大肠杆菌中41%携带一类整合子,整合子的基因盒携带aad A1、drf A1、drf A27、arr -3等抗性基因。
畜禽养殖废水浇灌蔬菜引起蔬菜携带抗性基因和耐药菌的研究较少,然而该途径可能是畜禽养殖源抗性基因进入食物链的途径之一,但相关研究非常缺乏。Yang等[88]对施用鸡粪种植的蔬菜内生菌进行了耐药性测试,发现在芹菜、小白菜、黄瓜中头孢氨苄耐药菌的比例分别为16.9%~86.33%、21.76%~91.31%和0.21%~0.44%,蔬菜内生菌具有抗生素抗性的原因可能是耐药菌通过土壤进入植物,或者由于土壤中抗性基因被植物吸收,这需要进一步研究。Hofmann等[89]采用FISH-CLSM研究病原菌在植物根际的寄生位置,发现植物表皮细菌及内生菌中存在沙门氏菌等病原菌,与粪便相比,废水农田利用存在更大的病原菌传播风险。Reuland等[90]检测了荷兰零售蔬菜中超广谱β-内酰胺酶(extended beta-lactamase, ESBL)肠杆菌科细菌,发现6%的蔬菜检测到ESBL肠杆菌科细菌,其携带的抗性基因包括blaCTX-M-15、blaCTX-M-14、blaCTX-M-1、blaSHV-12,作者推测这可能与畜禽粪便的还田利用有关,进而引起了抗性基因沿食物链的传播。Marti等[91]发现,施用猪粪的蔬菜表皮抗性基因的检出率较高,包括IncP ori V、sul 2、tet (BT)、erm A/F、qnr B、blaPSE和blaOXA-20等抗性基因,指出直接食用蔬菜是人类接触土壤耐药菌和抗性基因的途径之一。
5 结语与展望(Conclusion and outlook)
(1)我国是兽用抗生素生产和使用大国,尽管已出台兽用抗生素种类与用量的相关规定,但抗生素的实际使用水平及其对耐药性影响仍鲜有公开报道。更为严格的抗生素使用规范是大势所趋,全面、系统的抗生素使用与耐药性数据有利于标准的修订。
(2)畜禽粪便、废水是病原菌和抗性基因的重要蓄积库。应深入研究病原菌具有的耐药表型和携带的抗性基因,建立抗生素使用与病原菌耐药性、抗性基因的联系,有利于合理用药,从而降低病原菌的耐药水平。
(3)应深入研究由基因移动元件介导的抗性基因,尤其在重金属的协同选择作用下,加强其在病原菌之间发生的水平转移效率、抗性传播风险的机制研究,从诱导肠道、粪便中的耐药病原菌入手,深入研究饲料中重金属的添加阈值。
(4)畜禽养殖粪便、废水从养殖场通过还田利用、灌溉等途径进入环境,由移动基因元件介导且宿主为人类病原菌的抗性基因存在较大的公共健康威胁,然而从畜禽养殖场到环境的抗性传播机制尚不明确,应深入研究建立从畜禽养殖场至公共环境全过程的抗性污染控制链条。
[1] WHO. Antimicrobial Resistance: Global Report on Surveillance [R]. WHO, 2014
[2] 刘金旭. 配合饲料中的抗生素[J]. 国外畜牧科技, 1986(2): 47
[3] Barton M D. Antibiotic use in animal feed and its impact on human health [J]. Nutrition Research Reviews, 2000, 13(2): 279-299
[4] Aarestrup F. Sustainable farming: Get pigs off antibiotics [J]. Nature, 2012, 486(7404): 465-466
[5] Van Boeckel T P, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals [C]. Proceedings of the National Academy of Sciences, 2015, 112(18): 5649-5654
[6] Seiler C, Berendonk T U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture [J]. Frontiers in Microbiology, 2012, 3: 399
[7] Chantziaras I, Boyen F, Callens B,et al. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries [J]. Journal of Antimicrobial Chemotherapy, 2014, 69(3): 827-834
[8] Wegener H C. Ending the use of antimicrobial growth promoters is making a difference [J]. ASM News, 2006, 69(9): 443-448
[9] Resende J A, Silva V L, de Oliveira T L R, et al. Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure [J]. Bioresource Technology, 2014, 153: 284-291
[10] Walczak J J, Xu S. Manure as a source of antibiotic-resistant Escherichia coli and Enterococci : A case study of a Wisconsin , USA family dairy farm [J]. Water Air Soil Pollution, 2011, 219(1-4): 579-589
[11] Baker-Austin C, Wright M S, Stepanauskas R, et al. Co-selection of antibiotic and metal resistance [J]. Trends in Microbiology, 2006, 14(4): 176-182
[12] Marosevic D, Cervinkova D, Vlkova H, et al. In vivo spread of macrolide-lincosamide-streptogramin B (MLSB) resistance-A model study in chickens [J]. Veterinary Microbiology, 2014, 171(3-4): 388-396
[13] Lutz E , McCarty M J, Mollenkopf D F, et al. Ceftiofur use in finishing swine barns and the recovery of fecal Escherichia coli or Salmonella spp. resistant to ceftriaxone [J]. Foodborne Pathogens and Disease, 2011, 8(11): 1229-1234
[14] Bennett P M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria [J]. British Journal of Pharmacology, 2008,153(s): S347-S357
[15] 张俊亚, 魏源送, 陈梅雪, 等. 畜禽粪便生物处理与土地利用全过程中抗生素和重金属抗性基因的迁移转化研究进展[J]. 环境科学学报, 2015, 35(4): 935-946
Zhang J Y, Wei Y S, Chen M X, et al. Fate of antibiotic and heavy metal resistance genes during the whole biological treatment and soil application process of livestock manure: A review [J]. Acta Scientiae Circumstantiae, 2015, 35(4): 935-946 (in Chinese)
[16] Martínez J L, Coque T M, Baquero F. What is a resistance gene? Ranking risk in resistomes [J]. Nature Reviews Microbiology, 2014, 13(2): 116-123
[17] Pourcher A M, Jadas-Hécart A, Cotinet P, et al. Effect of land application of manure from enrofloxacin-treated chickens on ciprofloxacin resistance of Enterobacteriaceae in soil [J]. Science of the Total Environment, 2014, 482-483(3): 269-275
[18] West B M, Liggit P, Clemans D L, et al. Antibiotic resistance, gene transfer, and water quality patterns observed in waterways near cafo farms and wastewater treatment facilities [J]. Water Air Soil Pollution, 2011, 217(1-4): 473-489
[19] Berendonk T U, Manaia C M, Merlin C, et al. Tackling antibiotic resistance: The environmental framework [J]. Nature Reviews Microbiology, 2015, 13: 310-317
[20] Hu Y, Yang X, Qin J, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota [J]. Nature Communications, 2013, 4: 2151
[21] 隋倩雯, 张俊亚, 魏源送, 等. 畜禽养殖废水生物处理与农田利用过程抗生素抗性基因的转归特征研究进展[J]. 环境科学学报(已接受), 2015
Sui Q W, Zhang J Y, Wei Y S, et al. Fate of antibiotic resistance genes in the total process of biological treatment and land application of animal wastewater: An overview [J]. Acta Scientiae Circumstantiae, 2015 (accepted) (in Chinese)
[22] Zhang Q Q, Ying G G, Pan C G, et al. A comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modelling, and linkage to bacterial resistance [J]. Environmental Science & Technology, 2015, 49: 6772-6782
[23] 中华人民共和国农业部. 中国动物卫生状况报告[R]. 北京: 中国农业出版社, 2009
[24] Coates M E, Davies M K, Kon S K. The effect of antibiotics on the intestine of the chick [J]. The British Journal of Nutrition, 1955, 9(1): 110-119
[25] 中华人民共和国农业部. 饲料药物添加剂使用规范[S]. 北京: 中华人民共和国农业部, 2001
[26] United States Food and Drug Administration (US FDA). Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals [R]. Washington DC: FDA, 2011
[27] Maron D F, Smith T J S, Nachman K E. Restrictions on antimicrobial use in food animal production: An international regulatory and economic survey [J]. Globalization and Health, 2013, 9: 48
[28] Dewey C, Cox B, Straw B, et al. Use of antimicrobials in swine feeds in the United States [J]. Swine Health and Production, 1999, 7(1): 19-25
[29] Apley M D, Bush E J, Morrison R B, et al. Use estimates of in-feed antimicrobials in swine production in the United States [J]. Foodborne Pathogens and Disease, 2012, 9(3): 272-279
[30] European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 25 EU/EEA Countries in Third ESVAC Report [R]. London: European Medicines Agency, 2013
[31] 王瑞, 魏源送. 畜禽粪便中残留四环素类抗生素和重金属的污染特征及其控制[J]. 农业环境科学学报, 2013, 32(9): 1705-1719
Wang R, Wei Y S. Pollution and control of tetracyclines and heavy metals residues in animal manure [J]. Journal of Agro-Environment Science, 2013, 32(9): 1705-1719 (in Chinese)
[32] Zhou L J, Ying G G, Liu S, et al. Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China [J]. Science of the Total Environment, 2013, 444: 183-195
[33] Tulayakul P, Boonsoongnern A, Kasemsuwan S, et al. Comparative study of heavy metal and pathogenic bacterial contamination in sludge and manure in biogas and non-biogas swine farms [J]. Journal of Environmental Sciences, 2011, 23(6): 991-997
[34] Li J, Shao B, Shen J,et al. Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots [J]. Environmental Science & Technology, 2013, 47(6): 2892-2897
[35] Zhou L J, Ying G G, Zhang R Q, et al. Use patterns, excretion masses and contamination profiles of antibiotics in a typical swine farm, South China [J]. Environmental Science: Processes & Impacts, 2013, 15(4): 802-813
[36] Duriez P, Topp E. Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm [J]. Applied and Environmental Microbiology, 2007, 73(17): 5486-5493
[37] Looft T, Johnson T A, Allen H K, et al. From the cover: In-feed antibiotic effects on the swine intestinal microbiome [J]. Proceedings of the National Academy of Sciences, 2012, 109(5): 1691-1696
[38] Allen H K, Looft T, Bayles D O, et al. Antibiotics in feed induce prophages in swine fecal microbiomes [J]. MBio, 2011, 2(6):1-9
[39] Danzeisen J L, Kim H B, Isaacson R E, et al. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment [J]. PLoS One, 2011, 6(11): e27949
[40] Platt T M, Loneragan G H, Scott H M, et al. Antimicrobial susceptibility of enteric bacteria recovered from feedlot cattle administered chlortetracycline in feed [J]. American Journal of Veterinary Research, 2008, 69(8): 988-996
[41] Chen J, Fluharty F L, St-Pierre N, et al. Technical note: Occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin [J]. Journal of Animal Science, 2008, 86(9): 2385-2391
[42] 赵飞燕, 汪以真. 我国饲料安全的现状与对策[J]. 中国饲料, 2004(4): 37-39
[43] 彭点懿, 何健, 杨玉峰. 氧化锌在断奶仔猪配合饲料中的应用及其抗腹泻的可能机制[R]. 中国畜牧杂志, 2011, 47(20): 66-70
[44] 西南大学. 重金属与金霉素对无公害肉猪生产和环境的影响研究: 中国[P/OL]. (2008-08-06).
[45] Brooks J P, Adeli A, McLaughlin M R. Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems [R]. Water Research, 2014, 57: 96-103
[46] Zhu Y G, Johnson T, Su J Q, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3435-3440
[47] Bibbal D, Dupouy V, Ferré J P, et al. Impact of three ampicillin dosage regimens on selection of ampicillin resistance in Enterobacteriaceae and excretion of blaTEMgenes in swine feces [J]. Applied and Environmental Microbiology, 2007, 73(15): 4785-4790
[48] 中华人民共和国农业部. 2013年动物源细菌耐药性监测计划[R]. 北京: 中华人民共和国农业部, 2013
[49] United States Food and Drug Administration (US FDA). National antimicrobial resistance monitoring system [R]. Washington DC: US The Centers for Disease Control and Prevention, 2009
[50] Jacob M E, Fox J T, Narayanan S K, et al. Effects of feeding wet corn distillers grains with solubles with or without monensin and tylosin on the prevalence and antimicrobial susceptibilities of fecal foodborne pathogenic and commensal bacteria in feedlot cattle [J]. Journal of Animal Science, 2008, 86(5): 1182-1190
[51] Kühn I, Iversen A, Finn M, et al. Occurrence and relatedness of vancomycin-resistant Enterococci in animals, humans, and the environment in different European regions [J]. Applied and Environmental Microbiology, 2005, 71(9): 5383-5390
[52] Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant Enterococci by PCR [J]. Journal of Clinical Microbiology, 1995, 33(1): 24-27
[53] Neyra R C, Frisancho J A, Rinsky J L, et al. Multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA) [J]. Environmental Health Perspectives, 2014, 122(5): 471-477
[54] Dahmen S, Métayer V, Gay E, et al. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France [J]. Veterinary Microbiology, 2013, 162(2-4): 793-799
[55] Sato T, Okubo T, Usui M, et al. Association of veterinary third-generation cephalosporin use with the risk of emergence of extended-spectrum-cephalosporin resistance in Escherichia coli from dairy cattle in Japan [J]. PLoS One, 2014, 9(4): e96101
[56] Jiang X, Yang H, Dettman B, et al. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves [J]. Foodborne Pathogens and Disease, 2006, 3(4): 355-365
[57] Wu N, Qiao M, Zhang B, et al. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China [J]. Environmental Science & Technology, 2010, 44(18): 6933-6939
[58] Jaglic Z, Vlkova H, Bardon J, et al. Distribution, characterization and genetic bases of erythromycin resistance in Staphylococci and Enterococci originating from livestock [J]. Zoonoses Public Health, 2012, 59(3): 202-211
[59] Kotzamanidis C, Zdragas A, Kourelis A, et al. Characterization of vanA-type Enterococcus faecium isolates from urban and hospital wastewater and pigs [J]. Journal of Applied Microbiology, 2009, 107(3): 997-1005
[60] Chénier M R, Juteau P. Fate of chlortetracycline- and tylosin-resistant bacteria in an aerobic thermophilic sequencing batch reactor treating swine waste [J]. Microbial Ecology, 2009, 58(1): 86-97
[61] Nemeghaire S, Argudín M, Haesebrouck F, et al. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines [J]. BMC Veterinary Research, 2014, 10(1): 153
[62] Kanwar N, Scott H M, Norby B, et al. Impact of treatment strategies on cephalosporin and tetracycline resistance gene quantities in the bovine fecal metagenome [J]. Scietific Report, 2014, 4: 5100
[63] Graves A K, Liwimbi L, Israel D W. Distribution of ten antibiotic resistance genes in E . coli isolates from swine manure, lagoon effluent and soil collected from a lagoon waste application field [J]. Folia Microbiologica, 2011, 56(2): 131-137
[64] Morales A S, Fragoso de Araújo J, de Moura Gomes V T, et al. Colistin resistance in Escherichia coli and Salmonella enterica strains isolated from swine in Brazil [J]. The Scientific World Journal, 2012, 2012: 109795
[65] Jensen V F, Jakobsen L, Emborg H D, et al. Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli [J]. Journal of Antimicrobial Chemotherapy, 2006, 58(1): 101-107
[66] Horton R A, Randall L P, Snary E L, et al. Fecal carriage and shedding density of CTX-M extended-spectrum β -lactamase-producing Escherichia coli in cattle, chickens, and pigs: Implications for environmental contamination and food production [J]. Applied and Environmental Microbiology, 2011, 77(11): 3715-3719
[67] Endimiani A, Rossano A, Kunz D, et al. First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland [J]. Diagnostic Microbiology and Infectious Disease, 2012, 73(1): 31-38
[68] Jones-Dias D, Manageiro V, Francisco A P, et al. Assessing the molecular basis of transferable quinolone resistance in Escherichia coli and Salmonella spp. from food-producing animals and food products [J]. Veterinary Microbiology, 2013, 167(3-4): 523-531
[69] Molla B, Miko A, Pries K, et al. Class 1 integrons and resistance gene cassettes among multidrug resistant Salmonella serovars isolated from slaughter animals and foods of animal origin in Ethiopia [J]. Acta Tropica, 2007, 103(2): 142-149
[70] Chapman J S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance [J]. International Biodeterioration and Biodegradation, 2003, 51(4): 271-276
[71] Rahube T O, Yost C K. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application [J]. Journal of Applied Microbiology, 2012, 112(6): 1123-1133
[72] Binh C T T, Heuer H, Kaupenjohann M, et al. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids [J]. FEMS Microbiology Ecology, 2008, 66(1): 25-37
[73] Agersø Y, Sandvang D. Class 1 integrons and tetracycline resistance genes in Alcaligenes , Arthrobacter , and Pseudomonas spp. isolated from pigsties and manured soil [J]. Applied and Environmental Microbiology, 2005, 71(12): 7941-7947
[74] Zhang X Y, Ding L J, Yue J. Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in Northeastern China [J]. Microbial Drug Resistance, 2009, 15(4): 323-328
[75] Silveira E, Freitas A R, Antunes P, et al. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages [J]. Journal of Antimicrobial Chemotherapy, 2014, 69(4): 899-906
[76] Hasman H, Aarestrup F M. The tcrB gene conferring transferable copper resistance in Enterococcus faecium : Occurrence, transferability, and linkage to macrolide and glycopeptide resistance [J]. Antimicrobial Agents and Chemotherapy, 2002, 46(5): 1410-1416
[77] Amachawadi R G, Scott H M, Alvarado C, et al. Occurrence of the transferable copper resistance gene tcrB among fecal Enterococci of U.S. feedlot cattle fed copper-supplemented diets [J]. Applied and Environmental Microbiology, 2013, 79(14): 4369-4375
[78] Cavaco L M, Hasman H, Aarestrup F M. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance [J]. Veterinary Microbiology, 2011, 150(3-4): 344-348
[79] Chen B, Zheng W, Yu Y, et al. Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River, Fujian Province, China [J]. Applied and Environmental Microbiology, 2011, 77(1): 148-155
[80] Smet A, Rasschaert G, Martel A, et al. In situ ESBL conjugation from avian to human Escherichia coli during cefotaxime administration [J]. Journal of Applied Microbiology, 2011, 110(2): 541-549
[81] You Y, Silbergeld E K. Learning from agriculture: Understanding low-dose antimicrobials as drivers of resistome expansion [J]. Frontiers in Microbiology, 2014, 5: 1-10
[82] Jacobsen C S, Bech T B. Soil survival of Salmonella and transfer to freshwater and fresh produce [J]. Food Research International, 2012, 45(2): 557-566
[83] Piorkowski G S, Bezanson G S, Jamieson R C, et al. Effect of hillslope position and manure application rates on the persistence of fecal source tracking indicators in an agricultural soil [J]. Journal of Environmental Quality, 2014, 43(2): 450-458
[84] Wang H, Zhang T, Wei G, et al. Survival of Escherichia coli O157: H7 in soils under different land use types [J]. Environmental Science and Pollution Research International, 2014, 21(1): 518-524
[85] Cook K L, Netthisinghe A M P, Gilfillen R A. Detection of pathogens, indicators, and antibiotic resistance genes following land application of poultry litter [J]. Journal of Environmental Quality, 2014, 10(6): 42104
[86] Bech T B, Rosenbom A E, Kjaer J, et al. Factors influencing the survival and leaching of tetracycline-resistant bacteria and Escherichia coli through structured agricultural fields [J]. Agriculture, Ecosystems and Environment, 2014, 195: 10-17
[87] Chee-Sanford J C, Amniov R I, Krapac I J, et al. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities [J]. Applied and Environmental Microbiology, 2001, 67(4): 1494-1502
[88] Yang Q, Ren S, Niu T, et al. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables [J]. Environmental Science and Pollution Research, 2014, 21(2): 1231-1241
[89] Hofmann A, Fischer D, Hartmann A, et al. Colonization of plants by human pathogenic bacteria in the course of organic vegetable production [J]. Frontiers in Microbiology, 2014, 5: 1-11
[90] Reuland E A, Al Naiemi N, Raadsen S A, et al. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables [J]. European Journal of Clinical Microbiology & Infectious Diseases, 2014, 33: 1843-1846
[91] Marti R, Scott A, Tien Y C, et al. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest [J]. Applied and Environmental Microbiology, 2013, 79(18): 5701-5709
◆
Veterinary Antibiotics Use, Occurrence of Antibiotic Resistance Pathogen and Its Antibiotic Resistance Genes in Animal Production: An Overview
Sui Qianwen1, Zhang Junya1, Wei Yuansong1,3,*, Chen Meixue1, Dong Hongmin2, Xiong Jihai3
1. State Key Joint Laboratory of Environmental Simulation and Pollution Control, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China 2. Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agricultural Sciences, Beijing 100081, China 3. Institute of Energy, Jiangxi Academy of Sciences, Nanchang 330096, China
Veterinary antibiotics play an important role in animal growth promotion and disease control in food-animal production. Over a half of total antibiotics is used in animal industry. Antibiotic resistance bacteria (ARB), antibiotic resistance genes (ARGs) and their spread risk caused by the animal industry have drawn widely attention. China is the biggest country of animal industry and veterinary antibiotics consumption in the world, but many gaps exist about veterinary antibiotics use, antibiotic resistance level of pathogens and their carried ARGs, which are difficult for the control of antibiotic resistance pathogens and its spread risk. Through literature review, the veterinary antibiotic use of China and other developed countries were summarized, the AR level of pathogenic bacteria and the carried ARGs, mobile genetic elements, as well as the spread risk of AR pathogenic bacteria to environment were analyzed. Therefore the purposes of this paper are to provide support for improving rational drug use and reducing the spread risk of antibiotic resistance pathogens, and establishing the linkage of antibiotic resistance pollution from animal production to public environment.
veterinary antibiotics; heavy metal; antibiotic resistance pathogenic bacteria; antibiotic resistance genes; mobile genetic element
公益性行业(农业)科研专项经费课题(No. 201303091);国家“水体污染控制与治理”科技重大专项课题(2012ZX07202-005;2015ZX07203-007);江西省科学院省院协同创新团队(2014-SYXTCX-02);国家自然科学基金(No. 41501513);国家自然科学基金(No. 21577161)
隋倩雯(1986-),女,博士后,研究方向为畜禽养殖废水生物处理及抗性基因减控,E-mail:suiqianwen@163.com;
*通讯作者(Corresponding author), E-mail: yswei@rcees.ac.cn
10.7524/AJE.1673-5897.20150922001
2015-09-22录用日期:2015-10-21
1673-5897(2015)5-020-15
X171.5
A
魏源送(1969—),男,环境工程博士,研究员,研究方向污水处理与再生利用、污泥处理与资源化、抗生素抗性基因的转归与控制,已在国内外刊物发表学术论文130余篇。
隋倩雯, 张俊亚,魏源送,等.畜禽养殖过程抗生素使用与耐药病原菌及其抗性基因赋存的研究进展[J]. 生态毒理学报,2015, 10(5): 20-34
Sui Q W, Zhang J Y, Wei Y S, et al. Veterinary antibiotics use, occurrence of antibiotic resistance pathogen and its antibiotic resistance genes in animal production: An overview[J]. Asian Journal of Ecotoxicology, 2015, 10(5): 20-34 (in Chinese)

