农业利用对海南省天然次生林土壤微生物的影响
张逸飞,刘娟娟,孟 磊,邓 欢,姜允斌,张金波,钟文辉,*
1 南京师范大学地理科学学院,江苏省环境演变与生态建设重点实验室, 南京 210046 2 南京化工职业技术学院, 南京 210048 3 海南大学农学院, 海口 570228
农业利用对海南省天然次生林土壤微生物的影响
张逸飞1,2,刘娟娟1,孟 磊3,邓 欢1,姜允斌1,张金波1,钟文辉1,*
1 南京师范大学地理科学学院,江苏省环境演变与生态建设重点实验室, 南京 210046 2 南京化工职业技术学院, 南京 210048 3 海南大学农学院, 海口 570228
为揭示天然次生林经过长期农业利用后土壤微生物学性质以及土壤养分状况的变异,采集了海南省天然次生林土壤以及由天然次生林经农业开垦转变而成的香蕉、桉树和橡胶林区表层土壤样品,利用磷脂脂肪酸(PLFA)分析、变性梯度凝胶电泳(DGGE)、群落水平生理特征(CLPP)分析等方法探究天然次生林经农业利用对土壤微生物生物量、微生物活性、群落多样性和功能多样性的影响。研究结果显示,天然次生林土壤总磷脂脂肪酸显著高于经农业利用的土壤,分别是香蕉和橡胶林土壤的3倍和2倍。平均颜色变化率(AWCD)以及由PLFA、DGGE和CLPP分析获得的土壤微生物群落多样性和功能多样性均显示出天然次生林高于农业利用的土壤。天然次生林土壤与农业利用土壤的微生物群落结构也存在明显的分异。另外,天然次生林土壤pH值、有机碳、总氮、总磷、速效氮和速效钾含量高于经农业利用的土壤。逐步回归分析显示土壤pH值、有机碳和速效氮是影响土壤微生物生物量、微生物活性、群落多样性和功能多样性的主要因素。研究结果表明,天然次生林经农业利用后,由于种植单一树种和农业管理措施可能造成土壤有机质和养分含量下降,导致土壤酸化,对土壤微生物群落造成负面影响。
香蕉;桉树;橡胶;土壤微生物;土壤养分
天然次生林向农业利用转变在发展中国家很普遍[1],在我国热带和亚热带地区,天然次生林被砍伐、焚烧并种植单一作物如桉树、香蕉、橡胶等[2]。天然次生林向农业利用转变可能使土壤生态系统承受巨大压力甚至造成土壤严重退化[3],对天然次生林的农业利用进行生态影响评价显得尤为重要。土壤微生物群落结构、功能多样性、微生物生物量以及活性是研究土地利用方式转变的常用微生物指标,这些指标反映的信息对于了解土壤健康和土壤质量至关重要[4]。已有研究表明,在森林转换为农田或牧场过程中,耕作、施肥、灌溉、放牧、除草等管理措施易扰动土壤微生物并可能改变其多样性[5-6],Jangid等人提出森林转换对土壤微生物多样性的影响取决于管理实践的强度以及植物类型[7],其中植被类型决定了碳输入的质与量并影响土壤理化性质,从而影响土壤微生物群落[8]。在亚热带地区,土地管理措施是影响土壤微生物的主要因素[9]。目前关于管理措施对亚热带地区农田土壤微生物影响的研究较多[10-11]。天然次生林转化为单一树种如香蕉、桉树和橡胶,其耕作强度低于农田土壤,这种转化对土壤微生物群落和功能多样性、微生物生物量和活性将产生什么影响?目前关于这方面的研究报道比较少。
群落水平生理特征(CLPP)分析、磷脂脂肪酸(PLFA)分析和变性梯度凝胶电泳(DGGE)是评价土壤微生物多样性常用方法。CLPP用于表征微生物功能多样性,而PLFA和DGGE用于表征群落多样性。土壤微生物的总体活性可以通过CLPP测定中获得的平均颜色变化率(AWCD)表示[12],总PLFA量被认为是微生物总生物量的一个指标[13]。PLFA分析相对比较敏感但分辨率低;DGGE结合测序方法可以确定微生物种类,但会因PCR(Polymerase Chain Reaction)技术的缺陷而产生偏差。因此,将3种方法结合起来可较为全面地评价土壤微生物学性质,包括微生物生物量、活性、群落多样性和功能多样性。在本研究中,采集位于海南省热带地区的天然次生林和由天然次生林转换而来的香蕉、桉树和橡胶林土壤,通过CLPP,PLFA和DGGE方法研究土壤微生物群落多样性、功能多样性、微生物生物量以及活性,以评价天然次生林向农业利用转变对土壤微生物的影响。
1 研究地区与研究方法
1.1 采样地点描述和土壤取样
采样地位于海南省儋州市的中国热带农业科学院实验场内,天然次生林自原始林在20世纪60年代破坏后发育而来,育林时间约50a。该地区属于热带海洋季风气候,海拔260—455 m,年平均气温23.2 ℃,年降水量1815 mm,年平均蒸发量1628 mm。土壤类型均为花岗岩发育而来的砖红壤。20世纪80年代中期一些地区的天然次生林地转为农用,用于栽种橡胶;20世纪90年代末另一些地区的天然次生林转变为香蕉林和桉树林。天然次生林与农业利用区相距约3 km,香蕉林、桉树林和橡胶林相距1—2 km。天然次生林的优势种主要是美叶菜豆树 (RadermacherafrondosaChun et How)、中平树(Macarangadenticulata) 和橄榄 (Canariumalbum)。桉树林、天然次生林和人工橡胶林下的灌木草本植物主要是矛叶荩草(Arthraxonlanceolatus)、香泽兰(Eupatoriumodoratum)、盾蕨(Neolepisorusovatus)、益智 (AlpiniaoxyphyllaMiq) 和九节木 (Psychotriarubra) 等。天然次生林和桉树林未施肥,橡胶林、香蕉主要以施化肥为主,橡胶林年施肥量为90 kg N/hm2,30 kg P2O5/hm2,40 kg K2O/hm2香蕉年施肥量为528 kg N/hm2, 190 kg P2O5/hm2,1300 kg K2O/hm2。
本研究选择上述天然次生林以及由其转化而来的香蕉林、桉树林和橡胶林土壤进行研究。土壤样品采集于2010年3月。采集土壤样品前,在每种利用方式土地中,选取3个相邻小区,每个小区面积约为100 m2(10 m × 10 m)。在每个小区按照S形采集10 个样点土壤并混合代表该小区。每个样点面积0.25 m2(0.5 m × 0.5 m),用铁锹取表层土壤 (0—20 cm),土壤样品用2 mm筛网过筛。用于PLFA和CLPP测定的土壤样品4 ℃保存,1周内测定;-70 ℃保存用于DNA提取;自然干燥后土壤样品用于化学分析。
1.2 土壤化学分析
土壤化学分析采用常规的方法[14]。土壤pH值采用pH玻璃电极测定,土水质量体积比1∶2.5。土壤有机碳采用重铬酸钾氧化法测定,总氮用凯氏定氮法测定,速效氮采用碱解扩散法测定,总磷采用磷钼蓝法测定,速效磷采用HCl-NH4F萃取再用分光光度法测定,速效钾采用醋酸铵提取再用原子吸收光谱法测定。
1.3 磷脂脂肪酸(PLFA)分析
采用Bligh Dyer方法进行脂类提取和磷酸酯脂肪酸分析[15],土样用15 mL Bligh Dyer提取液 (0.1 mol/L柠檬酸缓冲液∶氯仿∶甲醇=0.8∶1∶2) 提取,提取液用硅酸键合固相抽提柱(SPE-SI)层析,分别用氯仿、丙酮和无水甲醇洗脱,将含磷脂的洗脱液用氮气吹干,然后用碱性甲醇水解和皂化 (甲基化) 得到磷酯脂肪酸甲基酯 (FAME),用Sherlock微生物鉴定系统 (MIDI,Newark,DE) 确定PLFA种类和量,其中PLFA定量采用19∶0FAM内标法。根据结构不同,不同的特征脂肪酸对应不同的微生物,如细菌、真菌,细菌又可分为革兰氏阳性菌、革兰氏阴性菌、放线菌。本研究中分离获得的PLFA见表1。
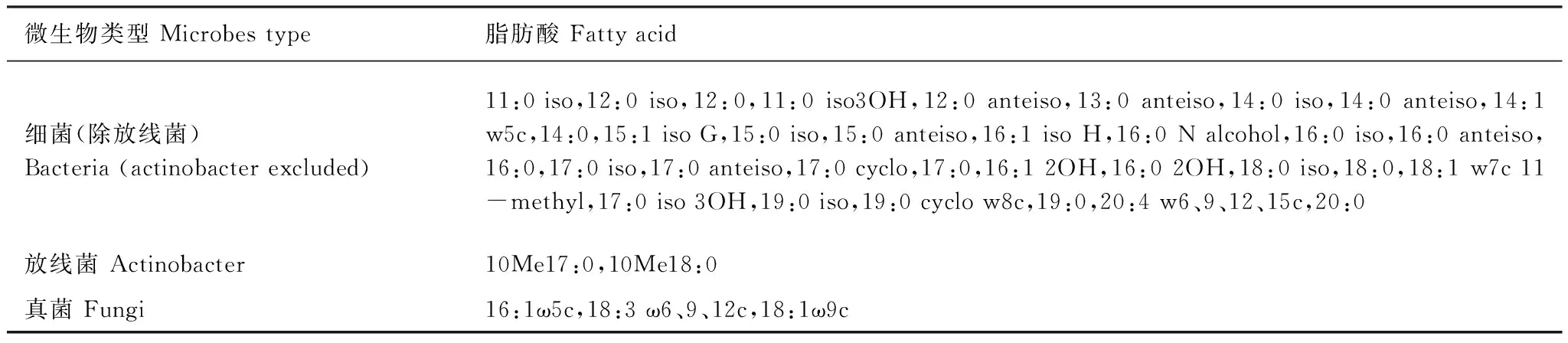
表1 土壤微生物PLFA生物标记物[16-17]Table 1 Phospholipid fatty acids (PLFA) signatures of soil micro-organisms[16-17]
1.4 群落水平生理特征 (CLPP) 分析[18]
将10 g新鲜土壤样品加入到100 mL无菌磷酸盐缓冲液 (0.05 mol/L,pH值 7.0),摇床震荡30 min,取1 mL土壤悬浮液于9 mL无菌磷酸盐缓冲液 (0.05 mol/L,pH值 7.0),混匀,制得10-2土壤悬浮液。取150 μL该10-2土壤悬浮液接种于BIOLOG Eco微平板孔中 (Biolog,Hayward,CA) 置25 °C培养,用自动快速菌类鉴定系统 (Biolog,Hayward,CA) 每隔12 h测定590 nm波长处的吸光值。采用培养96 h数据分析为每孔平均颜色变化率 (AWCD)、Shannon指数 (H′)、主成分 (PCA) 分析。
1.5 DNA提取和PCR扩增
土壤总DNA提取方法依照土壤DNA快速提取试剂盒 (Bio 101)制造商提供的说明书。DNA浓度采用微量紫外分光光度计ND-1000 (NanoDrop Technologies Inc.,Wilmington,DE) 测定,提取的DNA置-70 °C存储。
细菌16S rRNA基因扩增采用引物338F-GC (5′-ACT CCT ACG GGA GGC AGC AG CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG G-3′) 和518R (5′-ATT ACC GCG GCT GCT G-3′)[19]。50 μL反应体系含有:1.25 U聚合酶(TaKaRa Bio Inc., Shiga, Japan),每种引物均为25 pmol,每种核苷酸为10 nmol,5 μL 10×缓冲液和0.1 μmol MgCl2,1 μL提取的DNA为模板。PCR反应在iCycler thermocycler (Bio-Rad Laboratories Inc., CA) 上进行。反应程序为:预变性94 ℃4 min,35个循环包括94 ℃变性30 s,55 ℃退火30 s,72 ℃延伸90 s,最后72 ℃延伸5 min。
真菌18S rRNA基因扩增采用引物NS26 (5′-CTG CCC TAT CAA CTT TCG A-3′) 和518R-GC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GAT TAC CGC GGC TGC TGG-3′)[20]。50 μL反应体系含有:1.25 U聚合酶(TaKaRa Bio Inc., Shiga, Japan),每种引物均为25 pmol,每种核苷酸为10 nmol,5 μL 10×缓冲液和0.1 μmol MgCl2,1 μL提取的DNA为模板。PCR反应在iCycler thermocycler (Bio-Rad Laboratories Inc., CA) 上进行。反应程序为:预变性94 ℃4 min,30个循环包括95 ℃变性30 s,55 ℃退火30 s,72 ℃延伸90 s,最后72 ℃延伸5 min。
1.6 DGGE
本研究采用Dcode (BIO RAD Laboratories, Hercules, CA)系统分析细菌16S rRNA、真菌18S rRNA基因的DGGE (deneaturant gradient gel electrophoresis) 分子指纹图谱。细菌及真菌PCR产物在8%聚丙烯酰胺凝胶中电泳,变性梯度范围35%—70% (其中100%的变性剂含有7 mol/L尿素和40%甲酰胺),电泳缓冲液为0.5×TAE,电泳温度60 ℃。电泳结束,将凝胶置于SYBR Green I染液 (1∶10000体积分数,Invitrogen Probe产品) 中染色30 min,用分子成像器FX (Bio-Rad Laboratories Inc. Hercules, CA) 扫描。DGGE图像用Quantity One软件进行分析。
1.7 数据分析
不同土地利用类型之间的显著差异采用单因素方差分析 (Analysis of Variance,ANOVA) 来确定,显著性水平为P< 0.05。对PLFA、CLPP和DGGE图谱计算Shannon多样性指数 (H′) 并进行主成分分析 (PCA)。采用多元逐步回归分析确定土壤pH、有机碳以及养分含量中影响土壤微生物生物量、活性、微生物群落多样性及功能多样性的主要变量,所有的统计分析采用SPSS 16进行。
2 结果与分析
2.1 土壤pH值和养分含量
天然次生林土壤pH值、有机碳、总氮、总磷、速效氮和速效钾含量显著高于农用利用土壤 (表2)。香蕉林由于施磷肥较多,土壤中速效磷含量最高。橡胶林土壤中pH值、总磷、速效磷和速效钾含量最低。
2.2 DGGE图谱分析
细菌16S rRNA基因和真菌18S rRNA基因的DGGE图谱用来分析不同土地利用类型对细菌群落 (图1a) 和真菌群落的影响 (图1b)。天然次生林、香蕉、桉树林和橡胶林的细菌16S rRNA基因的DGGE平均条

表2 4种不同土地利用类型土壤pH值和养分含量Table 2 Soil pH and nutrient content under the four land use types
同列相同英文小写字母表示差异不显著 (P> 0.05)
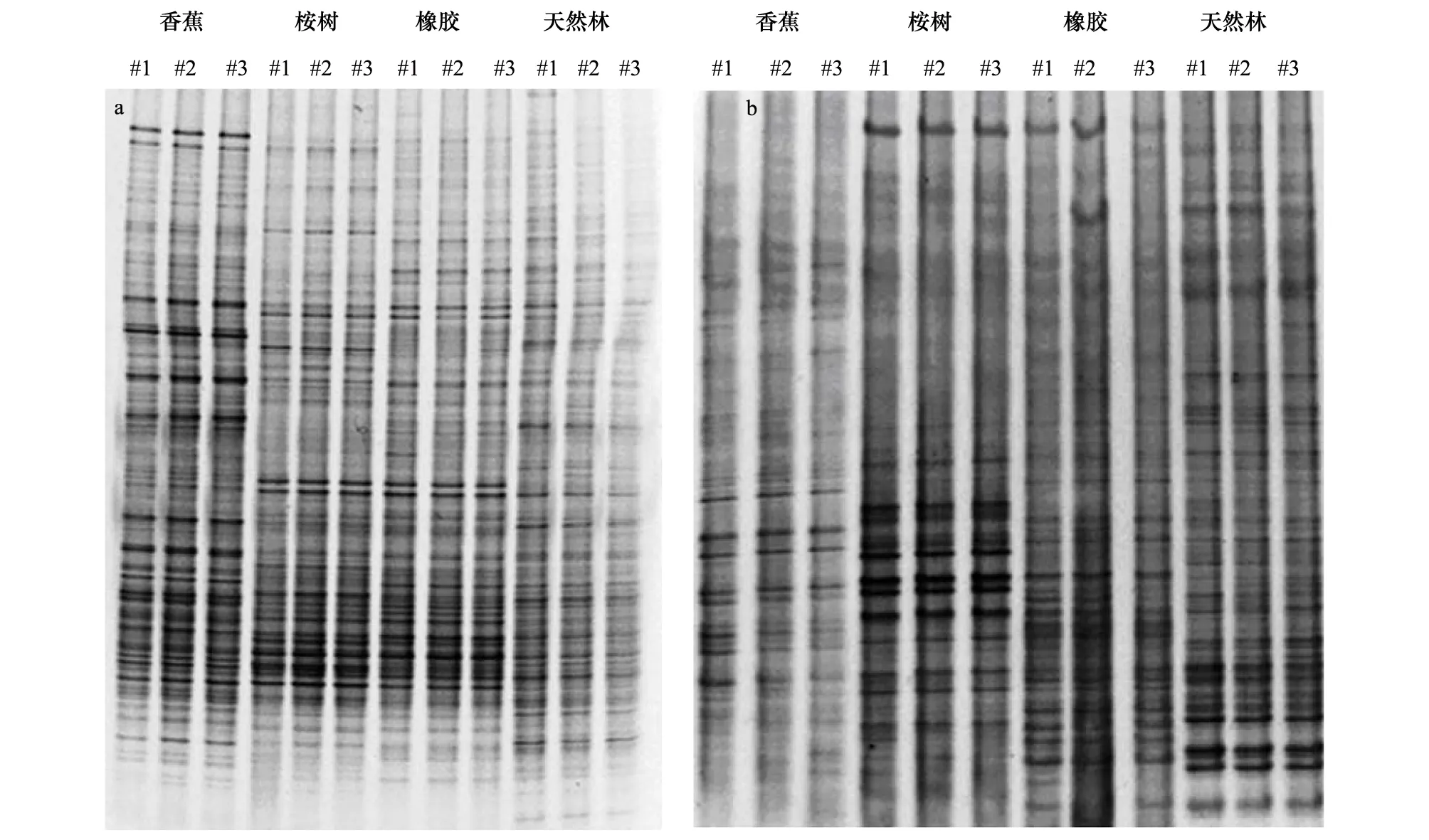
图1 四种利用类型土壤 16S rRNA基因DGGE图谱(a)和4种利用类型土壤18S rRNA基因DGGE图谱(b)Fig.1 The DGGE pattern of 16S rRNA gene fragment from soils under the four land use types (a) The DGGE pattern of 18S rRNA gene fragment from soils under the four land use types (b)#1、#2、#3代表4种利用类型的3个小区土壤样品
带数分别为78、70、76、72条 (以条带亮度大于所在泳道条带总亮度1%的条带计);真菌18S rRNA基因的DGGE平均条带数分别为39、30、26、34条。天然次生林土壤16S rRNA 基因的H′最高,香蕉、橡胶林明显较低 (图2)。18S rRNA基因的H′也表现出相似的模式:天然次生林>橡胶林>香蕉>桉树林。其中,天然次生林显著高于桉树和香蕉林 (P<0.05)。PCA分析(图3)显示天然次生林,橡胶林、香蕉林和桉树林在PC1和PC2方向上存在明显分异,表明土地利用方式是影响细菌和真菌群落结构的重要因素。
2.3 磷脂脂肪酸多样性分析
从4种不同利用类型土壤中共检测出58种脂肪酸,其中含量大于1%的有38种(表1)。土地利用类型显著影响土壤微生物PLFA量(表3)。天然次生林中总磷脂脂肪酸以及细菌、革兰氏阳性菌、革兰氏阴性菌、放线菌和真菌的磷脂脂肪酸最高,香蕉林土壤中上述指标含量最少。

图2 4种利用类型土壤中微生物群落多样性和功能多样性指数(H′) Fig.2 The Shannon diversity (H′) of soil microbial diversity and functional diversity from the four land use types. Means with the same letter are not significantly different具有相同字母者表示差异不显著 (P > 0.05,邓肯测验,n=3),短竖代表的标准误差
4种土地利用方式下PLFA图谱分析表明,香蕉林H′显著低于其他3种土地利用类型 (图2) (P<0.05)。对4种土地利用类型的PLFA分析结果进行PCA分析显示,其PC1的方差贡献率为63%,PC2的方差贡献率为13% (图3)。4种土地利用类型在PC2方向上彼此分离。包括真菌和放线菌在内的26种磷脂脂肪酸沿着沿PC1 (图3) 有较高方差贡献率 (> 0.60),11种磷脂脂肪酸沿PC2有较高方差贡献率 (> 0.60) (图4)。
2.4 CLPP多样性分析
不同土地利用方式对土壤微生物活性和功能多样性存在显著影响。天然次生林土壤的AWCD显著高于农用利用土壤,而桉树林、橡胶林和香蕉土壤之间没有显著差异 (表3)。对4种土地利用类型的CLPP图谱进行PCA分析的结果显示 (图3),天然次生林和农业利用区分异明显,桉树林与橡胶林无明显分异,但都与橡胶林存在明显分异。BIOLOG Eco微平板的碳源可分为6类,由于胺类和酚类混合物的碳源数目较少,各为2种,故分别与氨基酸类和聚合物类合并成4大类,分别为聚合物、碳水化合物、羟酸和胺/氨基酸[21]。吐温80,吐温40,α-环糊精和糖原等4种聚合物沿PC1和PC2具有较高的方差贡献率 (> 0.60,图5),7种碳水合物沿PC1和PC2具有方差贡献率 (> 0.60),4种胺/氨基酸、2种羧酸沿PC1和PC2具有较高的方差贡献率 (> 0.60)。
2.5 逐步回归分析
逐步回归分析表明,土壤微生物活性、生物量和多样性受土壤pH值和养分含量的影响显著 (表4)。速效氮显著影响细菌DGGE和PLFA的H′指数。总PLFA,细菌、放线菌的PLFA,CLPP的H′指数与速效磷和土壤pH显著相关。AWCD、真菌DGGE图谱和真菌PLFA的H′指数与有机碳显著相关。
表3 4种土地利用类型下的磷脂脂肪酸 (PLFA) 总量,细菌、革兰氏阳性菌、革兰氏阴性菌、真菌、放线菌的磷脂脂肪酸总量和每孔平均颜色变化率 (AWCD)
Table 3 The amounts of total PLFAs, bacterial, Gram-positive bacterial, Gram-negative bacterial, fungal and actinobacterial PLFAs and the average color development (AWCD) under four land use types

土地利用类型Landusetype天然次生林Naturalsecondaryforest香蕉Banana桉树Eucalyptus橡胶Rubber总磷脂脂肪酸TotalPLFA/(nmol/g干重)48.90a15.92c40.33b24.54bc细菌磷脂脂肪酸BacterialPLFA/(nmol/g干重)41.14a13.19b36.20a19.60b革兰氏阳性菌/革兰氏阴性菌G+/G-2.11b3.15a1.50c3.02a真菌磷脂脂肪酸FungalPLFA/(nmol/g干重)7.76a2.73c4.13b4.94b放线菌磷脂脂肪酸ActinobacterialPLFA/(nmol/g干重)2.33a1.09c1.17c1.74b平均颜色变化率AWCD1.02a0.69b0.75b0.88ab
同一行内英文小写字母相同表示差异不显著(P> 0.05)

图4 土壤中单独磷脂脂肪酸沿主成分1和2的负荷Fig.4 Loadings of the individual PLFAs from the PCA of the PLFAs data along principal components 1 and 2
3 讨论
本研究中,综合采用PLFA,DGGE和CLPP分析等方法较为全面地评价了天然次生林与由天然次生林转换而来的香蕉、桉树和橡胶林土壤微生物群落多样性、功能多样性、活性和微生物生物量的差异。结果表明,与天然次生林土壤相比,农业利用之后土壤微生物活性(AWCD)、微生物生物量(PLFA)和功能多样性(CLPP)显著降低,微生物群落多样性也降低,且天然次生林土壤与农业利用土壤之间、不同农业利用土壤之间微生物群落结构有显著差异。

图5 CLPP测定过程中的31种碳源利用率沿主成分1和2负荷 Fig.5 Loadings of the 31 carbon sources from the PCA of the CLPP along principal components 1 and 2具有相同字母者表示差异不显著 (P > 0.05,邓肯测验,n=3),短竖代表的标准误差
天然次生林的农业利用对土壤微生物群落的负面影响很大程度上归因于管理措施。管理过程中的人为扰动和植被类型都可能对土壤微生物群落产生负面影响[22]。经农业利用的土壤比天然次生林土壤受到更多的人为干扰,如耕作和清扫凋落物会导致土壤碳和养分的减少[23]。pH值对于微生物的多样性、活性和生物量具有重要影响[24-25],通常偏离中性条件对微生物群落产生胁迫,在本研究中,橡胶林和香蕉林土壤比天然次生林土壤pH值低,可能是由于氮肥输入增强硝化作用导致土壤酸化[26];另外,不同土地利用方式下,凋落物和根系分泌物有机碳输入的差异造成土壤有机碳的数量及质量的差异,并进而导致不同土壤pH,因此施肥和碳输入可能是本研究中经农业利用的林区pH值显著低于天然次生林的主要原因,这与Deekor等人的研究结果相似[27]。
以往的研究表明,天然次生林土壤转换为栽种香蕉后,碳输入的减少并导致土壤有机碳减少[28],而橡胶树和桉树凋落物中木质素与氮的比例较高,因而凋落物的可分解性较差[29-30]。因此本研究中3种经农业利用的土壤有机碳的减少也可能是由于碳输入的减少。土壤有机质为生物反应和生命活动提供了良好基质,因此经农业利用的土壤中有机碳含量的减少导致了微生物生物量、活性以及群落多样性和功能多样性的降低[8,31]。
表4 由4种利用类型土壤中微生物指标及其相关变量得到的的逐步回归分析 Table 4 The microbial indicators or indices and their correlated variables obtained by stepwise regression analysis in the soils under four land use types
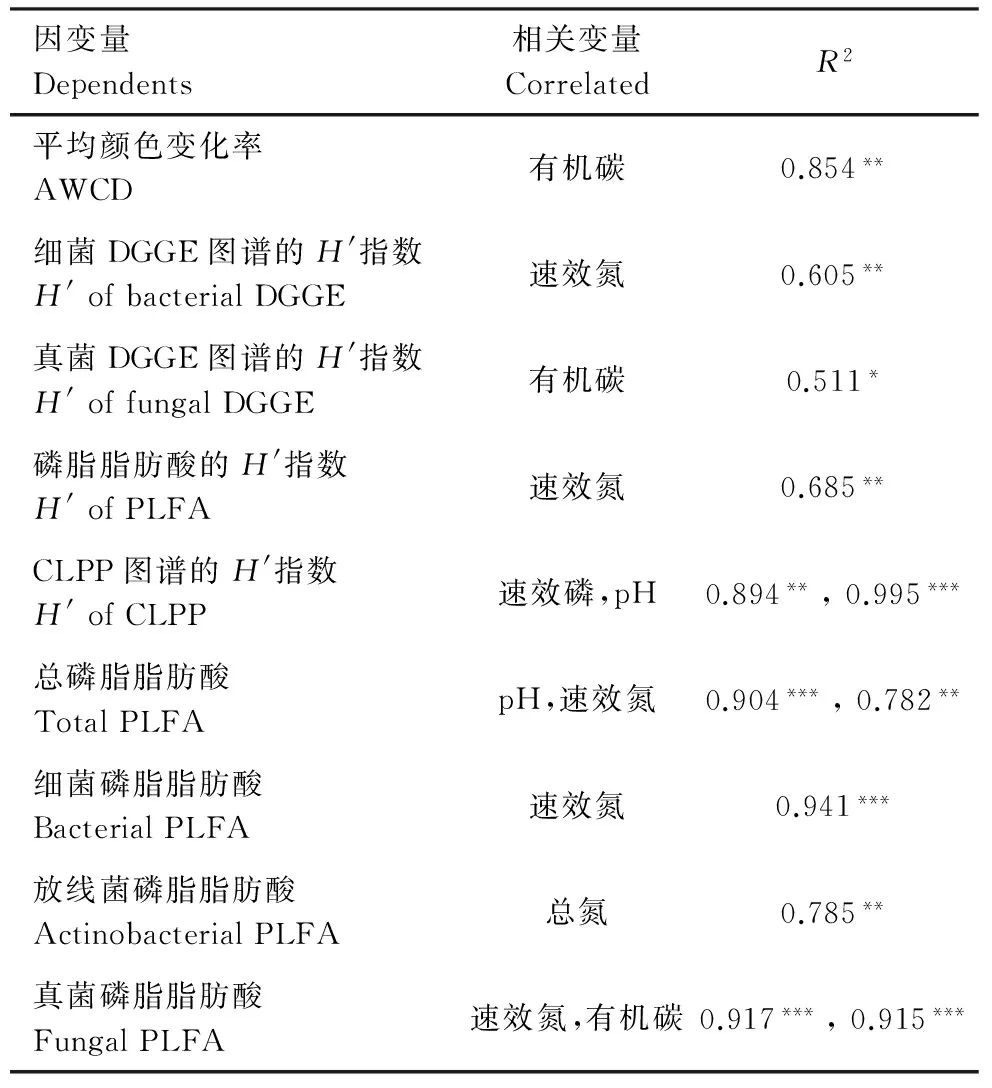
因变量Dependents相关变量CorrelatedR2平均颜色变化率AWCD有机碳0.854**细菌DGGE图谱的H'指数H'ofbacterialDGGE速效氮0.605**真菌DGGE图谱的H'指数H'offungalDGGE有机碳0.511*磷脂脂肪酸的H'指数H'ofPLFA速效氮0.685**CLPP图谱的H'指数H'ofCLPP速效磷,pH0.894**,0.995***总磷脂脂肪酸TotalPLFApH,速效氮0.904***,0.782**细菌磷脂脂肪酸BacterialPLFA速效氮0.941***放线菌磷脂脂肪酸ActinobacterialPLFA总氮0.785**真菌磷脂脂肪酸FungalPLFA速效氮,有机碳0.917***,0.915***
微生物群落多样性和功能多样性还与植物的类型有关[32],天然次生林比单一树种拥有更加多样的植物,这会使得次生林碳输入的种类更加多样,有助于提高土壤中微生物的多样性和功能多样性。CLPP图谱显示出的微生物群落结构在不同利用方式土壤间的差异,很有可能由于不同植被下的碳输入种类不同,导致了土壤微生物利用碳源的差异。另外,逐步回归分析表明,细菌多样性和包括革兰氏阳性菌、革兰氏阴性菌、放线菌在内的细菌PLFA含量在很大程度上受速效氮影响,而真菌多样性及生物量主要受到有机碳的影响,以往的研究结果显示细菌生物量增长却更依赖氮[33],真菌生物量的增加更多的依赖有机碳[34,35]。通常细菌生物体内C/N比约为3—6,而真菌生物体中的C/N比约为5—15[36],本研究的结果能与已有的研究结论相互印证。
本研究显示经农业利用后土壤中真菌和放线菌PLFA总量低于天然次生林。以往的研究也显示农业活动如耕作、土壤压实会破坏放线菌菌丝体以及真菌菌丝体从而导致生物量的减少[37-38]。表明真菌和放线菌对土地利用变化较为敏感。革兰氏阳性菌与阴性菌的比例变化可以反映土壤微生物群落所受到的胁迫[25]。本研究中香蕉、橡胶林土壤中革兰氏阳性菌与阴性菌的比例明显高于次生林土壤。与革兰氏阳性菌相比,革兰氏阴性细菌外膜存在额外的稳定层以抵抗渗透压[39],不施肥的天然次生林土壤的N、P、K含量最高,其含盐量较高,可能造成革兰氏阳性菌与阴性菌的比例较低。另一方面,本研究中显示桉树林土壤中革兰氏阳性菌与阴性菌的比例明显低于次生林及其它农业利用土壤,这可能是桉树的落叶产生的抑菌作用引起的,杨东升等人的研究表明桉叶浸提物对部分革兰氏阳性菌有抑菌作用, 而对革兰氏阴性菌无抑菌作用[40]。
[1] Smith C K, de Assis Oliveira F, Gholz Hl, Baima A. Soil carbon stocks after forest conversion to tree plantations in lowland Amazonia, Brazil. Forest Ecology and Management, 2002, 164(1/3): 257- 263.
[2] Zhang P C, Shao G F, Zhao G, Le Master D C, Parker G R, Dunning J B Jr, Li Q L. China′s forest policy for the 21st century. Science, 2000, 288(5474): 2135- 2136.
[3] Venkatesan S, Senthurpandian V K. Comparison of enzyme activity with depth under tea plantations and forested sites in south India. Geoderma, 2006, 137(1/2): 212- 216.
[4] Tilman D, Reich P B, Knops J M H. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature, 2006, 441(7093): 629- 632.
[5] Levine U Y, Teal T K, Robertson G P, Schmidt T M. Agriculture′s impact on microbial diversity and associated fluxes of carbon dioxide and methane. The ISME Journal, 2011, 5(10): 1683- 1691.
[6] Upchurch R, Chiu C Y, Everett K, Dyszynski G, Coleman D C, Whitman W B. Differences in the composition and diversity of bacterial communities from agricultural and forest soils. Soil Biology and Biochemistry, 2008, 40(6): 1294- 1305.
[7] Jangid K, Williams M A, Franzluebbers A J, Sanderlin J S, Reeves J H, Jenkins M B, Endale D M, Coleman D C, Whitman W B. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biology and Biochemistry, 2008, 40(11): 2843- 2853.
[8] Deng H, Zhang B, Yin R, Wang H L, Mitchell S M, Griffiths B S, Daniell T J. Long-term effect of re-vegetation on the microbial community of a severely eroded soil in sub-tropical China. Plant and Soil, 2010, 328(1/2): 447- 458.
[9] Bossio D A, Girvan M S, Verchot L, Bullimore J, Borelli T, Albrecht A, Scow K M, Ball A S, Pretty J N, Osborn A M. Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microbial Ecology, 2005, 49(1): 50- 62.
[10] Zhong W H, Gu T, Wang W, Zhang B, Lin X G, Huang Q R, Shen W S. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant and Soil, 2010, 326(1- 2): 511- 522.
[11] Chu H Y, Lin X G, Fujii T, Morimoto S, Yagi K, Hu J L, Zhang J B. Soil microbial biomass, dehydrogenase activity, bacterial community structure in response to long-term fertilizer management. Soil Biology and Biochemistry, 2007, 39(11): 2971- 2976.
[12] Loranger-Merciris G, Barthes L, Gastine A, Leadley P. Rapid effects of plant species diversity and identity on soil microbial communities in experimental grassland ecosystems. Soil Biology and Biochemistry, 2006, 38(8): 2336- 2343.
[13] Thoms C, Gattinger A, Jacob M, Thomas F M, Gleixner G. Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biology and Biochemistry, 2010, 42(9): 1558- 1565.
[14] 鲁如坤. 土壤农业化学分析方法. 北京: 中国农业科技出版社, 2000.
[15] Bligh E G, Dyer W J. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 1959, 37(8): 911- 917.
[16] Zelles L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biology and Fertility of Soils, 1999, 29(2): 111- 129.
[17] Buyer J S, Teasdale J R, Roberts D P, Zasada I A, Maul J E. Factors affecting soil microbial community structure in tomato cropping systems. Soil Biology and Biochemistry, 2010, 42(5): 831- 841.
[18] Campbell C D, Grayston S J, Hirst D J. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. Journal of Microbiological Methods, 1997, 30(1): 33- 41.
[19] Muyzer G, de Waal E, Uitterlinden A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology, 1993, 59(3): 695- 700.
[20] Schabereiter-Gurtner C, Piar G, Lubitz W, Rölleke S. Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA sequence analysis. Journal of Microbiological Methods, 2001, 47(3): 345- 354.
[21] Choi K H, Dobbs F C. Comparison of two kinds of Biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. Journal of Microbiological Methods, 1999, 36(3): 203- 213.
[22] Steenwerth K L, Jackson L E, Calderon F J, Stromberg M R, Scow K M. Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biology and Biochemistry, 2002, 34(11): 1599- 1611.
[23] Doran J W, Zeiss M R. Soil health and sustainability: managing the biotic component of soil quality. Applied Soil Ecology, 2000, 15(1): 3- 11.
[24] Bååth E, Anderson T H. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biology and Biochemistry, 2003, 35(7): 955- 963.
[25] Deng H, Li X F, Cheng W D, Zhu Y G. Resistance and resilience of Cu-polluted soil after Cu perturbation, tested by a wide range of soil microbial parameters. FEMS Microbiology Ecology, 2009, 70(2): 293- 304.
[26] Brown T T, Koenig R T, Huggins D R, Harsh J B, Rossi R E. Lime effects on soil acidity, crop yield, and aluminum chemistry in direct-seeded cropping systems. Soil Science Society of America Journal, 2008, 72(3): 634- 640.
[27] Deekor T N, Iwara A I, Ogundele F O, Amiolemen S O, Ita A E. Changes in soil properties under different land use covers in parts of Odukpani, Cross River State, Nigeria. Journal of Environment and Ecology, 2012, 3(1): 86- 99.
[28] Powers J S. Changes in soil carbon and nitrogen after contrasting land-use transitions in northeastern Costa Rica. Ecosystems, 2004, 7(2): 134- 146.
[29] Guo L B, Sims R E H. Litter decomposition and nutrient release via litter decomposition in New Zealand eucalypt short rotation forests. Agriculture, Ecosystems & Environment, 1999, 75(1/2): 133- 140.
[30] Maulin S P, Reddy G V, Banerjee R, Ravindra Babu P, Kothari I L. Microbial degradation of banana waste under solid state bioprocessing using two lignocellulolytic fungi (Phylostictaspp. MPS-001 andAspergillusspp. MSP-002). Process Biochemistry, 2005, 40(1): 445- 451.
[31] Yin R, Deng H, Wang H L, Zhang B. Vegetation type affects soil enzyme activities and microbial functional diversity following re-vegetation of a severely eroded red soil in sub-tropical China. CATENA, 2014, 115: 96- 103.
[32] Fierer N, Jackson R B. The diversity and biogeography of soil bacterial communities. Proceeding of the National Academy of Science of the United States of America, 2006, 103(3): 626- 631.
[33] Carney K M, Hungate B A, Drake B G, Megonigal J P. Altered soil microbial community at elevated CO2leads to loss of soil carbon. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(12): 4990- 4995.
[34] de Vries F T, Hoffland E, van Eekeren N, Brussaard L, Bloem J. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biology and Biochemistry, 2006, 38(8): 2092- 2103.
[35] Güsewell S, Gessner M O. N: P ratios influence litter decomposition and colonization by fungi and bacteria in microcosms. Functional Ecology, 2009, 23(1): 211- 219.
[36] Strickland M S, Rousk J. Considering fungal: bacterial dominance in soils-methods, controls, and ecosystem implications. Soil Biology and Biochemistry, 2010, 42(9): 1385- 1395.
[37] Vargas-Gil S, Pedelini R, Oddino C, Zuza M, Marinelli A, March G J. The role of potential biocontrol agents in the management of peanut root rot in Argentina. Journal of Plant Pathology, 2008, 90(1): 35- 41.
[38] Perez-Brandán C, Arzeno J L, Huidobro J, Grümberg B, Conforto C, Hilton S, Bending G D, Meriles J M, Vargas-Gil S. Long-term effect of tillage systems on soil microbiological, chemical and physical parameters and the incidence of charcoal rot byMacrophominaphaseolina(Tassi) Goid in soybean. Crop Protection, 2012, 40: 73- 82.
[39] Silhavy T J, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology, 2010, 2(5): a000414.
[40] 杨东升, 张青苗, 谢晓红, 罗先群. 海南桉叶浸提物的抗菌及降解双酚A研究. 化学与生物工程, 2007, 24(11): 35- 37.
Agricultural use of natural secondary forests affects soil microorganisms in Hainan Province, China
ZHANG Yifei1,2, LIU Juanjuan1, MENG Lei3, DENG Huan1, JIANG Yunbin1, ZHANG Jinbo1, ZHONG Wenhui1,*
1JiangsuKeyLaboratoryofEnvironmentalChange&EcologicalConstruction,SchoolofGeographyScience,NanjingNormalUniversity,Nanjing210046,China
2NanjingCollegeofChemicalTechnology,Nanjing210048,China
3CollageofAgriculture,HainanUniversity,Haikou570228,China
Natural forests tend to be converted into agricultural lands in developing countries for economic development. In the subtropical regions of China, natural secondary forests are generally converted to banana, rubber, and eucalyptus plantations. Unlike natural forests, agricultural lands are mainly characterized by mono-plantations, tillage, fertilization, and litter removal. These practices may have unfavorable consequences on the soil ecosystem. Moreover, the soil in the subtropical regions of China is classied as Ultisol, which is vulnerable to erosion and degradation under improper soil management. Yet, knowledge about the effects of the conversion of natural forests to agricultural lands on soil quality in these regions remains scarce. Soil microorganisms are critical for organic matter conversion and nutrient cycling, in addition to being sensitive to environmental changes. Thus, soil microbial parameters such as microbial biomass, activity, biodiversity, and composition are considered as reliable indicators of soil quality. To understand the effect of forest conversion to agricultural lands on soil microbial parameters, as well as soil chemical properties, we collected soil samples from a well-conserved natural secondary forest, in addition to transformed banana, eucalyptus, and rubber forests. Phospholipid fatty acid (PLFA) analysis, denaturing gradient gel electrophoresis (DGGE), and community-level physiological profiles (CLPP) were used to investigate various microbial parameters, including microbial biomass, activity, community diversity, and functional diversity. Microbial diversity was expressed as Shannon diversity (H′). Principal component analysis (PCA) was performed to analyze the soil microbial structure based on PLFA, CLPP, and DGGE data. In addition, soil chemical properties were determined, including pH, organic carbon, available nitrogen, total phosphorus, available phosphorus, and available potassium. Stepwise multiple regression analysis was used to determine the main soil properties that influenced soil microbial parameters. The results showed that the natural secondary forest had the highest total PLFA, which was 3 and 2 times higher than that recorded for the banana and rubber forests, respectively. The average color development (AWCD), community diversity, and functional diversity determined from the PLFA, DGGE, and CLPP profiles were also highest in the natural secondary forest. Soil microbial community structure differed between the natural secondary forest and agricultural lands, as well as between the vegetation types. In addition, soil pH, organic carbon, total nitrogen, total phosphorus, available nitrogen, and available potassium were higher in the natural secondary forest compared to the agricultural lands. The stepwise analysis showed that soil pH and available phosphorus affected H′ and CLPP values, while organic carbon affected AWCD,H′ of fungal DGGE, and fungal biomass values. Total nitrogen and available nitrogen generally affected microbial biomass (total, bacterial, and fungal PLFAs) and community diversity (H′ of DGGE and PLFA data). Our results indicate that the conversion of natural secondary forests to agricultural lands leads to soil acidification and a significant decrease in soil organic carbon and nutrient content. Furthermore, conversion decreases microbial biomass, activity, diversity, and functional diversity, causing shifts in soil microbial community structure. The adverse effects of this conversion on soil chemical and microbial properties may be due to the agricultural management practices being followed at local sites.
banana; eucalyptus; rubber; soil microorganisms; soil nutrient
国家自然科学基金资助项目(41271255,41222005); 江苏省高校自然科学研究重大项目(12KJA170001); 江苏省高校优势学科建设工程项目(164320H101)
2013- 10- 19;
日期:2015- 04- 14
10.5846/stxb201310192525
*通讯作者Corresponding author.E-mail: zhongwenhui@njnu.edu.cn
张逸飞,刘娟娟,孟磊,邓欢,姜允斌,张金波.钟文辉.农业利用对海南省天然次生林土壤微生物的影响.生态学报,2015,35(21):6983- 6992.
Zhang Y F, Liu J J, Meng L, Deng H, Jiang Y B, Zhang J B, Zhong W H. Agricultural use of natural secondary forests affects soil microorganisms in Hainan Province, China.Acta Ecologica Sinica,2015,35(21):6983- 6992.

