非小细胞肺癌铂类化疗药物基因组学研究
崔佳佳,尹继业
肺癌是目前全世界面临的最严重公共健康问题之一。其病理类型主要有小细胞肺癌和非小细胞肺癌(non-small-cell lung cancer,NSCLC)2种。其中,NSCLC占85%左右[1]。到目前为止,基于铂类的联合化疗仍然是NSCLC主要的治疗方案,尤其对于中晚期肿瘤患者。但在临床实践中,不同患者的化疗疗效存在巨大差异。部分患者对铂类药物敏感,有些患者则有原发或者继发性耐药。药物基因组学的研究表明,遗传因素是导致药物反应个体差异的决定性因素[2]。随着研究的深入,人们同样也发现遗传变异是影响NSCLC铂类化疗疗效的重要因素之一。
铂类药物是一种含铂的重金属配合物,进入细胞后可以与DNA链上的碱基形成交叉联结,从而起到破坏肿瘤细胞DNA结构和功能的作用,在临床广泛应用于实体瘤的治疗[3]。它在体内的具体过程如图1所示,其疗效主要受DNA损伤与修复、药物代谢与解毒、细胞周期和凋亡、药物转运体与膜受体等通路的影响。因此,上述通路基因的遗传变异是NSCLC铂类化疗药物基因组学研究的重点。
1 DNA损伤与修复通路
DNA损伤与修复通路是影响铂类化疗效果最重要的机制。因此,该通路的基因变异目前得到了最广泛的研究。与铂类化疗有关的DNA修复途径主要有核酸剪切修复(nucleotide excision repair,NER)、碱基剪切修复(base excision repair,BER)、错配修复(mismatch repair,MMR)和双链断裂(double-strand break,DSB)修复 4种。
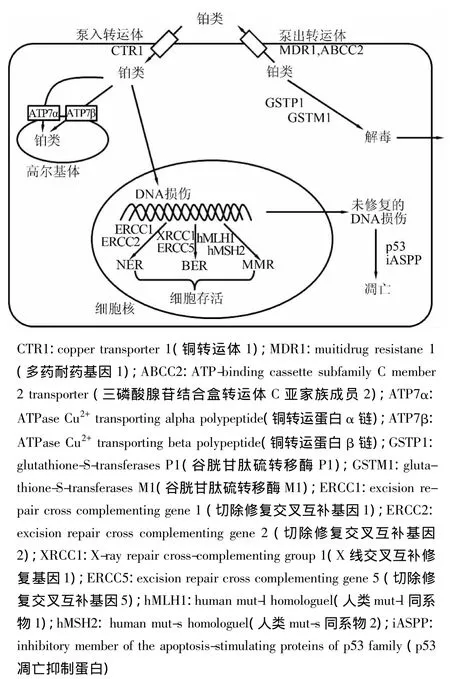
图1 铂类药物的体内代谢过程
NER是细胞修复铂类造成DNA破坏的主要途径,而ERCC1是NER途径中研究最多的基因。目前对于ERCC1多态性的研究主要集中于C354T和C8092A。Li等[4]对89例NSCLC 的研究表明,ERCC1 C354T CC基因型的敏感性比TC、TT基因型的低,但其差异没有达到统计学意义。而Kalikaki等[5]在119例NSCLC患者中的研究,则发现TT基因型患者的敏感性比 CC、CT基因型低。Isla等[6]同样也在62例接受顺铂和多西他赛联合化疗的NSCLC患者中研究了该位点,结果研究表明该多态性与铂类化疗敏感性无关。而对于C8092A多态性的研究来自于Takenaka等[7]研究,他们发现C8092A多态性与ERCC1蛋白表达和铂类化疗敏感性无关。由于这些研究的样本量太少,因此部分学者对它们进行了荟萃分析。比如Yu等[8]对1 252例NSCLC患者的荟萃分析表明,ERCC1 C354T和C8092A多态性与铂类化疗敏感性无显著性相关。Yin等[9]的研究也得到同样的结果。NER通路另一个被广泛研究的基因是ERCC2,主要集中于Asp312Asn和Lys751 Gln 2个位点。Booton等[10]对 108例 NSCLC患者的研究表明,野生型患者的铂类化疗敏感性(40%)比Asp312Asn/Lys751Gln(35.3%)和Asn312Asn/Gln751Gln(22.2%)基因型高。而Kim等[11]和 Provencio等[12]研究显示,Asp312Asp/Lys751Lys基因多态性与铂类化疗敏感性无显著相关性。针对中国人群的研究也存在类似的结果。Li等[13]对496例NSCLC患者的研究证明,Asp312Asn和Lys751Gln突变型患者的敏感性比野生型患者低。Wu等[14]对353例NSCLC患者的研究则未发现Asp312Asn和Lys751Gln突变与铂类化疗敏感性显著相关。荟萃分析的结果也不一致。Qin等[15]包含2 383例NSCLC患者的结果表明在白种人中,携带突变型等位基因的患者比野生型患者化疗敏感性要高,而在亚洲人群中这个结果则正好相反。而Yin等[9]和Qiu等[16]的研究结果则表示,ERCC2多态性与铂类化疗敏感性之间无显著性关联。除Asp312Asp/Lys751Lys基因之外,NER途径的核糖核苷酸还原酶M1(ribonucleotide reductase M1,RRM1)的 C-37A[17],ERCC5 的rs751402、rs2094258 和 rs2296147[18],XPC 的 Lys939Gln和Ala499Val也得到了研究[19]。但这些研究样本量较少,还需要重复性研究。
BER通路研究最多的基因是XRCC1,其Arg399 Gln位点在中国人群被广泛研究。大量研究表明,Arg399Gln突变型患者对铂类化疗敏感性较野生型患者高。比如Zhao等[20]对147例NSCLC患者的研究表明,突变型等位基因携带者的化疗敏感性明显比野生型高。Zhang等[21]对375例NSCLC患者的研究表明,AA基因型的化疗敏感性、无疾病生存期和总体生存率比其他基因型患者显著增加。对这些研究的荟萃分析也得到类似结论。AA基因型患者的铂类化疗敏感性(45.2%)高于AG(29.9%)、GG(30.7%)基因型患者[22]。BER通路另一个广泛研究的位点是 XRCC3的 Thr241Met,包含2 828例和2 201例NSCLC患者的荟萃分析发现,在白种人中突变型患者的化疗敏感性显著高于野生型,而亚洲人群中不存在这种相关性[23-24]。因此,XRCC3 Thr241Met位点的临床意义存在种族特异性。此外,XRCC1 Arg194Trp和ERCC5 His46His也有报道。Sun等[25]对82例NSCLC患者的研究表明,XRCC1 Arg194Trp的CT基因型患者铂类化疗敏感率(48.6%)要显著高于CC(20.5%)基因型,还显示ERCC5 His46His CT(36.1%)、CC(35.3%)基因型患者铂类化疗敏感率要高于TT(13.5%)基因型。
除以上广泛研究的2条通路之外,其他DNA修复通路的基因多态性也有报道。Cui等[26]对438例NSCLC患者的研究表明,亚甲基四氢叶酸还原酶(methylenetetrahydrofolate reductase,MTHFR)C677T TT基因型患者的铂类化疗敏感率要高于CT、CC基因型;来自于Zhu等[27]的荟萃分析结果也表明,MTHFR C677T TT基因型对铂类化疗更有效。其他的研究还包括 hMSH2 IVS12-6T>C、hMLH1 T-1151A[28]、胞苷脱氨酶(cytidine deaminase,CDA)Lys27Gln[29]、连接酶 4(ligase Ⅳ,LIG4)Thr9Ile[30]等(表 1)。
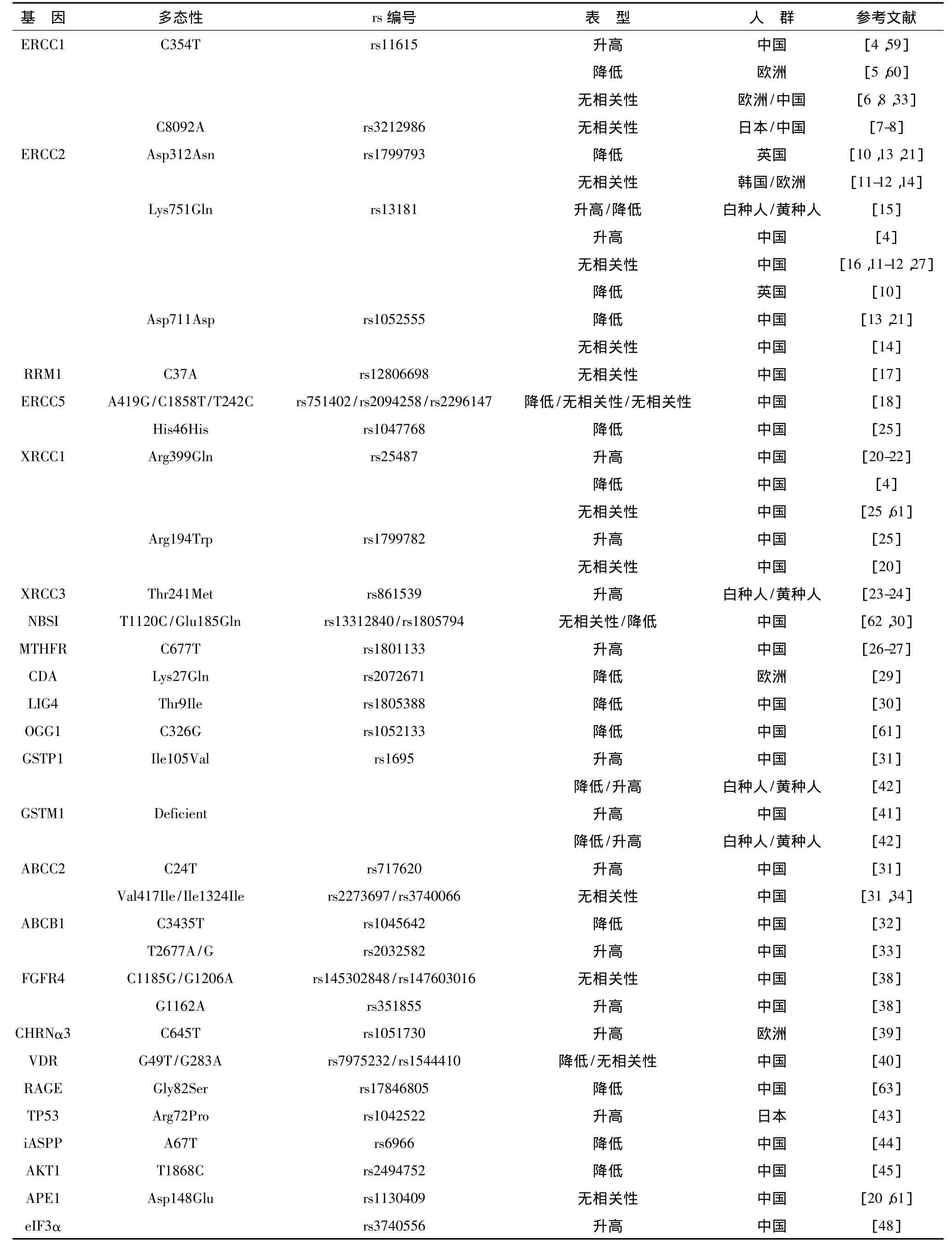
表1 与铂类药物化疗敏感性相关的主要遗传变异
2 转运体与药物受体
转运体和受体在药物的吸收、分布和消除过程中发挥了重要作用,当前针对铂类药物研究较为充分的主要是三磷酸腺苷结合盒转运体(ATP-binding cassette transporters,ABC)。Sun 等[31]在113 例 NSCLC患者中研究了ABCC2的突变与铂类化疗敏感性的关系,发现C-24T突变型等位基因携带者的铂类化疗敏感性(63.3%)要显著高于野生型患者;而Val417Ile的GA+AA基因型患者敏感性显著低于GG基因型患者,Ile1324Ile的CT+TT基因型患者敏感性显著低于CC 基因型。Yan 等[32]和 Chen 等[33]则研究了 ABCB1,他们发现C3435T野生型患者的铂类化疗敏感性(21.4%)要高于CT(19.0%)、TT(12.5%)基因型患者,而T2677A/G的T等位基因携带者(36.2%)敏感性高于G(51.0%)、A(69.6%)等位基因携带者。Yoh等[34]对72例NSCLC 患者的 ABCC1、ABCC3 进行了研究,未发现它们的多态性与铂类化疗敏感性相关。除了ABC 家族转运体,Wang 等[35-36]和 Li等[37]也对其他转运体与NSCLC患者铂类化疗敏感性进行了系统全面的研究,发现TMEM205 rs896412、OCT2 rs1869641和rs195854、AQP9 rs1516400、AQP2 rs7314734 及ATP7B rs9535826和rs9535828多态性可以影响铂类的化疗敏感性。
除了转运体,受体也得到了广泛研究。如Fang等[38]在629例NSCLC患者中研究了成纤维细胞生长因子受体4(fibroblast growth factor receptor 4,FGFR4)的多态性,发现rs351855 AA基因型患者的铂类化疗敏感性要显著高于GG基因型患者。Carcereny等[39]则在305例NSCLC患者中发现,尼古丁乙酰胆碱受体 α3(cholinergic receptor nicotinic alpha 3,CHRNα3)rs1051730的CT基因型患者铂类化疗敏感率高于CC、TT基因型。Xiong等[40]则针对755例中国患者研究了维生素D受体(vitamin D receptor,VDR),发现ApaI G>T基因rs7975232的携带GG基因型的患者比携带TT基因型的患者更有可能对铂类化疗敏感。
3 药物代谢与解毒通路
代谢和解毒是药物在体内的重要过程,药物经过体内一些酶的转化后其药理活性发生变化,因此该通路的基因多态性可以影响药物的疗效,当前针对铂类药物研究较多的是GST。
Sun等[31]研究了113例NSCLC患者,发现GSTP1 Ile105Val突变G等位基因携带者的铂类化疗敏感性(40.5%)要高于AA基因型(18.3%)患者。而Li等[41]对217例NSCLC患者的研究表明,GSTM1缺失型多态性在敏感患者中出现的频率(69.8%)与耐药患者(45.8%)不同。Yang 和Xian[42]对GSTP1 Ile105Val和GSTM1进行了荟萃分析,结果显示在东亚人中,GSTP1 Ile105Val的G等位基因携带者对铂类化疗敏感性较好,而白种人中无相关性。同样的情况也存在于GSTM1插入缺失型突变,东亚人群缺失型患者比插入型患者对铂类的敏感性要高,这些结果与Yin等[9]研究一致。此外,Li等[41]也对细胞色素 P4501A1的T264C进行了研究,发现TT基因型在敏感患者中的频率(47.7%)与耐药患者(25.5%)有显著差异,TC基因型患者的铂类化疗敏感率要高于其他基因型。
4 细胞凋亡通路
铂类药物最终通过诱导肿瘤细胞凋亡来发挥疗效,因此该通路基因变异可以影响其疗效,也得到了广泛研究。Shiraishi等[43]对640例日本NSCLC患者的研究表明,肿瘤抑制蛋白p53(tumor protein p53,TP53)Arg72Pro的非同义突变 TP53-72Pro基因型患者的铂类化疗敏感性(54.3%)要显著高于p53-72Arg基因型(29.1%)患者。Su等[44]对 230例NSCLC患者的研究表明,iASPP A67T的A等位基因携带者对化疗更敏感。Xu 等[45]和 Wang等[46]则研究了AKT1 rs2494752和Bcl-2结合抗凋亡基因1(Bcl-2 associated athanogene 1,BAG-1)C324T,发现AKT1 rs2494752突变型的铂类化疗敏感性降低,而BAG-1 C324T的CC基因型患者铂类化疗敏感率高于CT基因型患者。另外,也有人研究了基质金属蛋白酶-2、脱嘌呤/脱嘧啶核酸内切酶1等基因,但均未发现其与化疗疗效相关[44,47]。
5 其他
除了以上通路的基因多态性,其他基因也被广泛研究,如Xu等[48]在771例不同地区中国NSCLC患者中发现真核翻译起始因子3α(eukaryotic translation initiation factor 3α,eIF3α)的rs3740556多态性与铂类药物化疗敏感性相关,携带A等位基因的患者铂类化疗敏感性显著高于GG基因型的患者;并且Yin等[49-50]进一步阐明了 eIF3α rs3740556 改变铂类化疗敏感性的具体机制。此外对pre-miRNA-27α 的 rs895819、NQO1 C609T、galectin-3 A191C 和A292C都有相关报道,但尚需更多研究进行重复[51-53]。全基因组关联分析(Genome Wide Association Studies,GWAS)是近年在药物基因组学领域兴起的研究方法,有研究人员利用HapMap数据库结合体外药物敏感性实验对铂类药物的敏感性进行GWAS研究,取得了一些结果,但是尚缺乏体内研究数据[54-58]。
随着最近这些年的研究,NSCLC铂类化疗药物基因组学吸引了越来越多研究人员的兴趣,同时也发现了大量相关遗传变异。但是该领域的研究结果往往不一致,其主要原因有以下几个方面,同时也是以后研究需要解决的问题。①当前所开展研究样本量较少,也未经多中心验证,因此导致结果难以被重复;②当前所发现的单个遗传变异均对NSCLC铂类化疗贡献较小,因此依靠单个位点难以准确预测化疗敏感性;③除了遗传因素,环境因素也对铂类化疗敏感性有影响,但目前的研究均缺乏环境因素的考虑。相信随着以上问题的解决,最终可以建立准确预测NSCLC铂类化疗疗效的药物基因组学模型,从而实现个体化治疗。
[1]Siegel R,Ma J,Zou Z,et al.Cancer statistics,2014[J].CA Cancer J Clin,2014,64(1):9-29.
[2]周宏灏.遗传药理学[M].北京:人民军医出版社,2003:1-14.
[3]Kelland L.The resurgence of platinum-based cancer chemotherapy[J].Nat Rev Cancer,2007,7(8):573-584.
[4]Li D,Zhou Q,Liu Y,et al.DNA repair gene polymorphism associated with sensitivity of lung cancer to therapy[J].Med Oncol,2012,29(3):1622-1628.
[5]Kalikaki A,Kanaki M,Vassalou H,et al.DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer[J].Clin Lung Cancer,2009,10(2):118-123.
[6]Isla D,Sarries C,Rosell R,et al.Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer[J].Ann Oncol,2004,15(8):1194-1203.
[7]Takenaka T,Yano T,Kiyohara C,et al.Effects of excision repair cross-complementation group 1(ERCC1)single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients[J].Lung Cancer,2010,67(1):101-107.
[8]Yu D,Shi J,Sun T,et al.Pharmacogenetic role of ERCC1 genetic variants in treatment response of platinum-based chemotherapy among advanced non-small cell lung cancer patients[J].Tumour Biol,2012,33(3):877-884.
[9]Yin JY,Huang Q,Zhao YC,et al.Meta-analysis on pharmacogenetics of platinum-based chemotherapy in non small cell lung cancer(NSCLC)patients[J].PLoS One,2012,7(6):e38150.
[10]Booton R,Ward T,Heighway J.Xeroderma pigmentosum group D haplotype predicts for response,survival,and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer[J].Cancer,2006,106(11):2421-2427.
[11]Kim SH,Lee GW,Lee MJ,et al.Clinical significance of ERCC2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy[J].Lung Cancer,2012,77(3):578-584.
[12]Provencio M,Camps C,Cobo M.Prospective assessment of XRCC3,XPD and Aurora kinase A single-nucleotide polymorphisms in advanced lung cancer[J].Cancer Chemother Pharmacol,2012,70(6):883-890.
[13]Li XD,Han JC,Zhang YJ,et al.Common variations of DNA repair genes are associated with response to platinum-based chemotherapy in NSCLCs[J].Asian Pac J Cancer Prev,2013,14(1):145-148.
[14]Wu W,Li H,Wang H,et al.Effect of polymorphisms in XPD on clinical outcomes of platinum-based chemotherapy for Chinese non-small cell lung cancer patients[J].PLoS One,2012,7(3):e33200.
[15]Qin Q,Zhang C,Yang X,et al.Polymorphisms in XPD gene could predict clinical outcome of platinum-based chemotherapy for non-small cell lung cancer patients:a meta-analysis of 24 studies[J].PLoS One,2013,8(11):e79864.
[16]Qiu M,Yang X,Hu J,et al.Predictive value of XPD polymorphisms on platinum-based chemotherapy in non-small cell lung cancer:a systematic review and meta-analysis[J].PLoS One,2013,8(8):e72251.
[17]Feng JF,Wu JZ,Hu SN,et al.Polymorphisms of the ribonucleotide reductase M1 gene and sensitivity to platinbased chemotherapy in non-small cell lung cancer[J].Lung Cancer,2009,66(3):344-349.
[18]He C,Duan Z,Li P,et al.Role of ERCC5 promoter polymorphisms in response to platinum-based chemotherapy in patients with advanced non-small-cell lung cancer[J].Anticancer Drugs,2013,24(3):300-305.
[19]Zhu XL,Sun XC,Chen BA,et al.XPC Lys939Gln polymorphism is associated with the decreased response to platinum based chemotherapy in advanced non-small-cell lung cancer[J].Chin Med J(Engl),2010,123(23):3427-3432.
[20]Zhao W,Hu L,Xu J,et al.Polymorphisms in the base excision repair pathway modulate prognosis of platinumbased chemotherapy in advanced non-small cell lung cancer[J].Cancer Chemother Pharmacol,2013,71(5):1287-1295.
[21]Zhang L,Ma W,Li Y,et al.Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy[J].Genet Mol Res,2014,13(1):228-236.
[22]Chen J,Zhao QW,Shi GM,et al.XRCC1 Arg399Gln and clinical outcome of platinum-based treatment for advanced non-small cell lung cancer:a meta-analysis in 17 studies[J].J Zhejiang Univ Sci B,2012,3(11):875-883.
[23]Qiu M,Xu L,Yang X,et al.XRCC3 Thr241Met is associated with response to platinum-based chemotherapy but not survival in advanced non-small cell lung cancer[J].PLoS One,2013,8(10):e77005.
[24]Shen XY,Lu FZ,Wu Y,et al.XRCC3 Thr241Met polymorphism and clinical outcomes of NSCLC patients receiving platinum-based chemotherapy:a systematic review and meta-analysis[J].PLoS One,2013,8(8):e69553.
[25]Sun X,Li F,Sun N,et al.Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients[J].Lung Cancer,2009,65(2):230-236.
[26]Cui LH,Yu Z,Zhang TT,et al.Influence of polymorphisms in MTHFR 677 C→T,TYMS 3R→2R and MTR 2756 A→G on NSCLC risk and response to platinum-based chemotherapy in advanced NSCLC[J].Pharmacogenomics,2011,12(6):797-808.
[27]Zhu N,Gong Y,He J,et al.Influence of methylenetetrahydrofolate reductase C677T polymorphism on the risk of lung cancer and the clinical response to platinum-based chemotherapy for advanced non-small cell lung cancer:an updated meta-analysis[J].Yonsei Med J,2013,54(6):1384-1393.
[28]Cheng H,Sun N,Sun X,et al.Polymorphisms in hMSH2 and hMLH1 and response to platinum-based chemotherapy in advanced non-small-cell lung cancer patients[J].Acta Biochim Biophys Sin(Shanghai),2010,42(5):311-317.
[29]Tibaldi C,Giovannetti E,Vasile E,et al.Correlation of CDA,ERCC1,and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced nonsmall cell lung cancer patients[J].Clin Cancer Res,2008,14(6):1797-803.
[30]Jiang YH,Xu XL,Ruan HH,et al.The impact of functional LIG4 polymorphism on platinum-based chemotherapy response and survival in non-small cell lung cancer[J].Med Oncol,2014,31(5):959.
[31]Sun N,Sun X,Chen B,et al.MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer[J].Cancer Chemother Pharmacol,2010,65(3):437-446.
[32]Yan PW,Huang XE,Yan F,et al.Influence of MDR1 gene codon 3435 polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancer[J].Asian Pac J Cancer Prev,2011,12(9):2291-2294.
[33]Chen S,Huo X,Lin Y,et al.Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients[J].Int J Hyg Environ Health,2010,213(2):140-145.
[34]Yoh K,Ishii G,Yokose T,et al.Breast cancer resistance protein impacts clinical outcome in platinum-based chemotherapy for advanced non-small cell lung cancer[J].Clin Cancer Res,2004,10(5):1691-1697.
[35]Wang Y,Yin JY,Li XP,et al.The association of transporter genes polymorphisms and lung cancer chemotherapy response[J].PLoS One,2014,9(3):e91967.
[36]Wang Y,Li XP,Yin JY,et al.Association of HMGB1 and HMGB2 genetic polymorphisms with lung cancer chemotherapy response[J].Clin Exp Pharmacol Physiol,2014,41(6):408-415.
[37]Li XP,Yin JY,Wang Y,et al.The ATP7B genetic polymorphisms predict clinical outcome to platinum-based chemotherapy in lung cancer patients[J].Tumour Biol,2014[Epub ahead of print].
[38]Fang HM,Tian G,Zhou LJ,et al.FGFR4 genetic polymorphisms determine the chemotherapy response of Chinese patients with non-small cell lung cancer[J].Acta Pharmacol Sin,2013,34(4):549-554.
[39]Carcereny E,Ramirez JL,Sanchez-Ronco M,et al.Bloodbased CHRNA3 single nucleotide polymorphism and outcome in advanced non-small-cell lung cancer patients[J].Lung Cancer,2010,68(3):491-497.
[40]Xiong L,Cheng J,Gao J,et al.Vitamin D receptor genetic variants are associated with chemotherapy response and prognosis in patients with advanced non-small-cell lung cancer[J].Clin Lung Cancer,2013,14(4):433-439.
[41]Li W,Yue W,Zhang L,et al.Polymorphisms in GSTM1,CYP1A1,CYP2E1,and CYP2D6 are associated with susceptibility and chemotherapy response in non-small-cell lung cancer patients[J].Lung,2012,190(1):91-98.
[42]Yang Y,Xian L.The association between the GSTP1 A313G and GSTM1 null/present polymorphisms and the treatment response of the platinum-based chemotherapy in non-small cell lung cancer(NSCLC)patients:a metaanalysis[J].Tumour Biol,2014,35(7):6791-6799.
[43]Shiraishi K,Kohno T,Tanai C,et al.Association of DNA repair gene polymorphisms with response to platinumbased doublet chemotherapy in patients with non-smallcell lung cancer[J].J Clin Oncol,2010,28(33):4945-4952.
[44]Su D,Ma S,Liu P,et al.Genetic polymorphisms and treatment response in advanced non-small cell lung cancer[J].Lung Cancer,2007,56(2):281-288.
[45]Xu JL,Wang ZW,Hu LM,et al.Genetic variants in the PI3K/PTEN/AKT/mTOR pathway predict platinum-based chemotherapy response of advanced non-small cell lung cancers in a Chinese population[J].Asian Pac J Cancer Prev,2012,13(5):2157-2162.
[46]Wang YD,Ha MW,Cheng J,et al.The role of expression and polymorphism of the BAG-1 gene in response to platinum-based chemotherapeutics in NSCLC[J].Oncol Rep,2012,27(4):979-986.
[47]Zhao X,Wang X,Wu W,et al.Matrix metalloproteinase-2 polymorphisms and clinical outcome of Chinese patients with nonsmall cell lung cancer treated with first-line,platinum-based chemotherapy[J].Cancer,2012,118(14):3587-3598.
[48]Xu X,Han L,Yang H,et al.The A/G allele of eIF3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients[J].Lung Cancer,2013,79(1):65-72.
[49]Yin JY,Shen J,Dong ZZ,et al.Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair[J].Clin Cancer Res,2011,17(13):4600-4609.
[50]Yin JY,Dong ZZ,Liu RY.Translational regulation of RPA2 via internal ribosomal entry site and by eIF3a[J].Carcinogenesis,2013,34(6):1224-1231.
[51]Xu J,Yin Z,Shen H.A genetic polymorphism in pre-miR-27a confers clinical outcome of non-small cell lung cancer in a Chinese population[J].PLoS One,2013,8(11):e79135.
[52]Tian G,Wang M,Xu X.The role of NQO1 polymorphisms in the susceptibility and chemotherapy response of Chinese NSCLC patients[J].Cell Biochem Biophys,2014,69(3):475-479.
[53]Wu F,Hu N,Li Y,et al.Galectin-3 genetic variants are associated with platinum-based chemotherapy response and prognosis in patients with NSCLC[J].Cell Oncol(Dordr),2012,35(3):175-180.
[54]Wheeler HE,Gamazon ER,Stark AL,et al.Genome-wide meta-analysis identifies variants associated with platinating agent susceptibility across populations[J].Pharmacogenomics J,2013,13(1):35-43.
[55]Ziliak D,O'Donnell PH,Im HK,et al.Germline polymorphisms discovered via a cell-based,genome-wide approach predict platinum response in head and neck cancers[J].Transl Res,2011,157(5):265-272.
[56]Huang RS,Duan S,Shukla SJ,et al.Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach[J].Am J Hum Genet,2007,81(3):427-437.
[57]Huang RS,Johnatty SE,Gamazon ER,et al.Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients[J].Clin Cancer Res,2011,17(16):5490-5500.
[58]O'Donnell PH,Gamazon E,Zhang W,et al.Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants[J].Pharmacogenet Genomics,2010,20(5):327-337.
[59]Li F,Sun X,Sun N,et al.Association between polymorphisms of ERCC1 and XPD and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer[J].Am J Clin Oncol,2010,33(5):489-494.
[60]Krawczyk P,Wojas-Krawczyk K,Mlak R,et al.Predictive value of ERCC1 single-nucleotide polymorphism in patients receiving platinum-based chemotherapy for locallyadvanced and advanced non-small cell lung cancer--a pilot study[J].Folia Histochem Cytobiol,2012,50(1):80-86.
[61]Peng Y,Li Z,Zhang S,et al.Association of DNA base excision repair genes(OGG1,APE1 and XRCC1)polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients[J].Int J Cancer,2014[Epub ahead of print].
[62]Xu JL,Hu LM,Huang MD,et al.Genetic variants of NBS1 predict clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer in China[J].Asian Pac J Cancer Prev,2012,13(3):851-856.
[63]Wang X,Cui E,Zeng H,et al.RAGE genetic polymorphisms are associated with risk,chemotherapy response and prognosis in patients with advanced NSCLC[J].PLoS One,2012,7(10):e43734.

