糖尿病血管病变中的表观遗传现象
楼旭丹(综述) 汪海东△(审校) 夏世金Sven Skog
(1复旦大学附属华东医院内分泌科 上海 200040;2复旦大学附属华东医院上海市老年医学研究所 上海 200040;3华瑞分子生物医学研究院 深圳 518057)
糖尿病血管病变中的表观遗传现象
楼旭丹1(综述) 汪海东1△(审校) 夏世金2Sven Skog3
(1复旦大学附属华东医院内分泌科 上海 200040;2复旦大学附属华东医院上海市老年医学研究所 上海 200040;3华瑞分子生物医学研究院 深圳 518057)
糖尿病及相关的微血管和大血管并发症对公众健康构成了极大威胁,其高发病率和死亡率是造成社会经济负担的主要原因。最近研究指出表观遗传学可能在糖尿病血管病变中发挥重要作用,这种表型的变化并不涉及DNA序列的改变,然而可影响基因的表达并遗传给下一代。此外,大型临床试验UKPDS和DCCT-EDIC均证实代谢记忆的存在,即使血糖控制在理想状态,血管炎症仍能持续发生。尽管表观遗传现象尚未完全阐明,但为糖尿病的发病机制提供了新的思路,并有望成为难治性疾病的治疗靶点。
DNA甲基化; 组蛋白修饰; 糖尿病血管病变; 代谢记忆
IntroductionThe incidence of diabetes mellitus has increased at an unprecedented speed and caused widespread concern.Both type 1 and type 2 diabetes have accelerated rates of developing into vascularcomplications,including microvascular complications such as diabetic nephropathy,retinopathy,neuropathy,and macrovascular complications such as cardiovascular diseases,hypertension and stroke[1].Studies have demonstrated that diabetic vascular diseases can be triggered at the level of epigenetics[2],which includes DNA methylation,histone modifications,chromatin remodeling,genetic imprinting,random chromosome(X) inactivation, RNA interference,etc[3].Hyperglycemia,dyslipidemia,increasing age or pharmacological intervention can lead to epigenetic changes,which in turn affect genomic stability and up-regulate or down-regulate genes expression.That is to say,epigenetics provide a link between phenotype and genotype,thus has a significant impact on diverse biological processes and are correlated with corresponding changes in pathophysiology[4-6].In this review,we mainly elaborate the epigenetic phenomenon,particularly DNA methylation and histone modifications in diabetic vascular diseases.There are few reviews giving a comprehensive and systematic description of the pathogenesis of diabetes-related vascular complications from the epigenetic filed.Here we provide a more focused review which not only presents our own experiments data and the latest research results to improve the integrity and authority of our article,but also includes some clinical studies using inhibitors of epigenetic modifications to highlight the article values in guiding therapy.
DNA methylationAs the first epigenetic marker,DNA methylation has been extensively studied.DNA methylation is mediated by DNA methyltransferases (DNMTs)which transfer a methyl group from S-adenosyl-methionine(SAM)to the 5'position of the cytosine ring,generating 5-methyl-cytosine (5-mC)[7].In mammals,this specific activation mainly occurs at the cytosine phosphate guanosine (CpG) di-nucleotides.However,parts of Cp G di-nucleotides sequences,which contain high quantity of GCs,called Cp G islands,mostly remain non-methylated[8].Located at the promoters of regulatory genes,the abnormal methylation of Cp G islands,generally hypermethylation,can lead to transcriptional repression or make the gene silencing[9].The following three ways may be the underlying mechanisms either direct or indirect affecting genes expression:First,preventing transcription factors from binding to cognate DNA sequences.Second,recruiting additional inhibitory molecules,such as methyl-Cp G-binding proteins(MBPs)and histone deacetylases (HDACs).Third,agglutination of chromatin structures may hinder the binding of transcription factors to its regulatory sequences[10].Therefore,DNA methylation is associated with gene silencing,while non-methylation is associated with gene activating,and demethylation often correlates with reactivating of a silent gene[11].
Histone modificationsN-terminal amino acid residues exposed to the surface of the nucleosome are the major sites of post-translational modifications (PTM ),including methylation,acetylation,phosphorylation,ubiquitination,ADP ribosylation and other covalent modifications.Different modified types of histones with different amino acid residues,often seen in lysine (K)or arginine (R),suggest the existence of “histone codes”[12],which determine the state of genes transcription either by affecting the affinity of histones and DNA duplexes to make chromatin structures loose or condensed,or by playing a gene regulatory role to affect transcription factors binding to promoter or enhancer regions.Histone lysine methylation is considered to be more stable and long lasting,it can be mediated mono-,di-,or trimethylation by specific histone lysine methyltransferases(HMTs).Lysine residues are usually methylated in H3 or H4,studies have shown that the methylation of H3 in lysine 4,36,79 sites activates the transcription of chromatin,while methylation in 9,27 and H4 lysine 20 sites presents repressive marks[13].Histone acetyltransferases (HATs) and histone deacetylases(HDACs)are both involved in the transference of an acetyl group from acetyl Co A.HATs have been considered activating marks,which prevent the formation of highly condensed chromatin and promote genes transcription,while HDACs remove acetyl groups from lysine residues,generating counteractions[14].The dynamic genes expression processes are regulated by a balance between the opposing activities of HMTs and histone lysine demethylases(HDMs),as well as HATs and HDACs[15].
DNA methylation under diahetic conditionsIncreasing evidence shows that the level of DNA methylation varies under diabetic conditions.Recent reports manifested thatβ-cell dysfunction and insulin genes expression could be regulated by DNA methylation[16-17].In another case,progressive epigenetic silencing through DNA methylation at the promoter of Pdx1(a transcription factor that has an effect onβcell differentiation and gene activation)would lead to the development of diabetes[18].In damaged diabetic islet,peroxisome proliferator-activated receptor-γ (PPARγ)was highly methylated at DNA Cp G islands[19].Several inflammatory genes relevant to atherosclerosis showed DNA methylation alterations in vascular smooth muscle cells(VSMCs)and endothelial cells (ECs).In addition,DNA methylation was considered to be linked with diabetic-associated cardiovascular diseases (CVDs)and chronic kidney diseases(CKDs).There was an increased level of homocysteine being identified as a risk factor for both CVDs and CKDs,activated mainly by inhibiting DNA methyltransferases,as reported in some studies[20-22].DNA hypomethylation was generally seen in diabetic nephropathy(DN),while hypermethylation at the promoter of UNC13B(a gene known as an indicator of DN)was found in type 1 diabetes with DN[23].Thus the role of DNA methylation in diabetic complications was still full of confusion and needed more investigations.
Resultsof our preliminary studies confirmed that high expression of interleukin-17 (IL-17)significantly accelerated the development of metastatic diabetes in NOD/SCID mice compared with siRNA-IL-17 and control groups,along with notably elevated levels of tumor necrosis factor-α(TNF-α),interferon-γ (IFN-γ),interleukin-6 (IL-6)in peripheral blood[24-25].In our follow-up studies,we found that diabetic rats'aortas showed reduced DNA methylation levels at the promoters of IL-17,IL-6,TNF-αand IFN-γcompared with normal groups,in parallel with increased protein and mRNA levels of each cytokine,proved by Western blotting and real-time PCR,indicating that DNA methylation may be one of the potential mechanisms regulating diabetic vascular diseases.A new study identified that type 1 diabetesassociated DNA methylation variable positions arose early in the etiological process before diagnosis[26],suggesting it may be a useful clinical biomarker for early diagnosis of diabetes and its management.
Histone methylation-associated diahetic vascular diseasesNF-κB is one of the major transcription factors activated in inflammatory pathways under diabetic conditions,which allows monocytes and macrophages to be assembled in the blood vessels and leads to macrovascular atherosclerosis[27].Evidence indicated that the activating events of NF-κB P65 gene may be modulated by epigenetic modifications.Based on the crystal structure,Set7 acted as an unique mono-methyltransferase exactly targeting on H3K4,generating H3K4m1.Chromatin immunoprecipitation purification (ChIP)of bovine aortic endothelial cells which were previously exposed to hyperglycemia,found Set7 enriching at the NF-κB p65 gene promoter.However,when Set7 was silenced or knocked down by siRNA,NF-κB p65 gene failed to be induced under the same condition[28].As a result,NF-κB p65 gene expression induced by hyperglycemia may be achieved through the pathway of H3K4 m1 catalyzed by Set7.
It has also been indicated that cytokines and chemokines such as IL-6,monocyte chemoattractant protein-1(MCP-1)and macrophage colony stimulating factor-1(MCSF-1)are detected in vascular smooth muscle cells (VSMCs)in both diabetes and atherosclerosis[29].Until recently,researchers come up with the idea that aberrant changes of histonemodifications may influence inflammatory factors expression.H3K9m3 is a particular lysine residue trimethylated by Suv39h1,they may recruit other inhibitory complexes such as heterochromatin protein 1α (HP1α)and HDACs to further propagate suppressive marks[30].Compared with db/+control mice,IL-6,MCP-1 and MCSF mRNA levels were significantly increased in VSMCs of db/db mice(db/db refers to a model established for type 2 diabetes with insulin resistance),while H3K9m3 levels decreased at the promoters of inflammatory genes in accordance with reduced Suv39h1[31].When db/db VSMCs were transfected with human Suv39H1,the over-expression and hypersensitivity could be reversed,suggesting that the enhanced inflammatory factors expression may be associated with loss of inhibitory histone modifications.
Lysine-specific demethylase 1(LSD1)is a nuclear amine oxidase that removes the methyl from H3K4m1 and H3K4m2,it can not only make loss of activated gene transcription signals,but can also demethylate H3K9m1 and H3K9m2,resulting in silent genes to be re-activated[32-33].In high glucose,LSD1 enriched in the NF-κB P65 gene promoters,however,the occupancies of LSD1 were dramatically decreased at IL-6 and MCP-1 genes promoters in db/db VSMCs[34].As discussed above,it is certain that LSD1 is linked to inflammatory genes expression events regardless of which one(H3K4 or H3K9)does LSD1 exactly act on.In summary,mutual suppression or mutual antagonism takes place in the process of histone methylation,respective methyltransferases and demethylases coexist a system which eventually leads to deleterious diabetic vascular diseases(Fig 1).
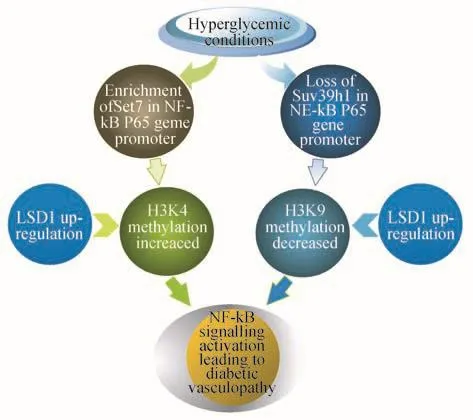
Fig 1 Proposed histone methylation mechanisms of hyperglycemia on vascular cells
Histone acetylation-associated diahetic vascular diseasesIn genome-wide analysis of primary human aortic endothelial cells,gene activation events were identified to be strongly correlated with hyperacetylation[35].Islet specific transcription factor Pdx1 was found to promoteβcell expression by recruiting HAT p300,while p300 inhibitor curcumin could prevent glucose-induced gene transcription and insulin secretion[36-37].Kaur,et al found that p300 may regulate fibronectin expression via NF-κB and poly ADP-ribose polymerase (PARP)activation in diabetes,leading to structural alterations such as basement membrane thickening and extracellular matrix(ECM)proteins like fibronectin and collagen deposition in tissues in chronic diabetic complications[38].Studies further confirmed that the upregulation of glucose-induced histone methylation and acetylation were important up-stream epigenetic mechanisms,regulating genes expression of vasoactive factors and ECM proteins in endothelial cells,and were potential therapeutic targets for diabetic complications[39-40].Moreover,IFN-γcould disrupt the expression of genes key to cellular metabolism and energy expenditure by repressing the activity of silent mating type information regulation 2 homolog 1(SIRT1)[41].The NAD+dependent deacetylase SIRT1 has emerged as a critical coordinator of inner environment primarily by impacting deacetylation levels and thus fine-tuning the activation of key transcription factors involved in the inflammatory processes.
Interestingly,in a recent study by Zhong et al[42],hyperglycemia was mentioned to enhance advanced glycation end products (AGEs)and reactive oxygen species(ROS),which stimulated HDACs(mainly HDAC1,HDAC2 and HDAC8)while inhibited HATs activity,resulting in an accelerated development of diabetic retinopathy.The results are quite distinct,not only due to multiple cross-talk modifications in response to cellular “language”,but also because regulatory determinants are segregated into intricate pathways[43].Nevertheless,HDAC inhibitors such as trichostatin A (TSA)are currently being tested as anticancer agents in clinical trials.Recent studies even demonstrated antifibrotic and renoprotective effects of HDAC inhibitors in diabetic kidneys,suggesting that HDAC inhibitors may prove to be a novel class of multitarget agents in the treatment of DN[44].Crosson,et al provided the first evidence that suppressing HDAC activity could protect the retina from ischemic injury[45].Although our knowledge of HDAC biology is very limited,the progress in understanding the role of HDAC inhibitors identifies epigenetic mechanism as a powerful tool for therapeutic intervention in tissue pathological lesion including diabetic vasculopathy.
Epigenetic role in metaholic memoryEarly or transient exposure to high glucose levels will induce long-lasting deleterious changes in the vasculature even after the patients have achieved normal glycemic control,which refers to “metabolic memory”[4]or“legacy effect”[46].The Diabetes Control and Complications Trial (DCCT),and the subsequent Epidemiology of Diabetic Complications and Interventions Trial (EDIC)demonstrated that the progression of diabetic vascular diseases occurred in both conventional and intensive glycemic control groups,and this effect could continue to operate for more than 5 years though the glycosylated hemoglobin had normalized for a period of time[47].The United Kingdom Prospective Diabetes Study(UKPDS)found that in type 2 diabetes lower fasting blood glucose at the time of diagnosis correlated with decreased risk of macrovascular diseases[48],but there was no statistically significance whether tight glycemic control would lead to better outcomes.The Action in Diabetes and Vascular Disease Preterax and Diamicron Modified Release Controlled Evaluation (ADVANCE)and Action to Control Cardiovascular Risk in Diabetes(ACCORD)suggested that intensive glycemic control helped delay both micro-and macrovascular complications,but raised the question whether this strict treatment was beneficial to all diabetics[49-50].
In fact,metabolic memory phenomenon was first observed in vitro models more than 20 years ago.When the retina of diabetic dogs were placed in high glucose environment for 2.5 years and then switched to normal glucose level for another 2.5 years,the incidence of retinopathy was similar to the control group which in poor glucose all over the 5 years[51].Endothelial cells cultured in high glucose could continuously express ECM proteins in spite of returning to subsequent normal glucose[52].In addition,reinstitution of normoglycemia after previous short episodes of hyperglycemia still had good effects on retina,whereas achieving glucose control after persistent exposure to hyperglycemia failed to appear protection[53].In vivo studies on rats showed that exposure from hyperglycemia to normoglycemia possibly led to irreparable damages of diabetic neuropathy[54].
Epigenetics is reported to be the underlying mechanism contributing to metabolic memory.When endothelial cells were incubated in high glucose(30 mmol/L)for a while then replaced in normal level (5 mmol/L),H3K4m1 and Set7 continued to increase at the NF-κB p65 gene promoter,and LSD1 was recruited along with the above changes,on the contrary,H3K9m3 presented sustained reduction[28].This effect lasted for six days,during the short period of time,NF-κB dependent proteins MCP-1 and VCAM-1 also showed elevated expression levels[55].When streptozotocin (STZ)-induced rats were either in poor glycemic control(PC,GHb>11%)or in good glycemic control(GC,GHb<6%)for 12 months,or in PC for 6 months and then in GC for 6 months,termination of hyperglycemia provided no benefits to retinal capillary cells,HDACs remained active and HATs kept compromised,revealing histone acetylation was important in this legacy effect,too[42].Besides,epigenetic pathways of AGEs and ROS were manifested to play a key role in the persistent genes expression[56].Although none of the experiments can solely clarify the phenomenon,when takentogether,the evidence strongly suggests that epigenetics are associated with metabolic memory,at least can partly explain it.
ConclusionsHyperglycemia can incur epigenetic changes through provoking variousbiological pathways to dysregulate genes expression in vessels,and the persistent changes imply that epigenetics may be a contributor to metabolic memory.Epigenetics not only reveals the pathogenesis of diabetes in the levels of etiology and pathology,but also represents a new treatment to interfere or overcome diabetes and its severe complications because of the reversible modifications.In fact,some epigenetic drugs are being studied for cancer and other diseases,such as DNA methylation inhibitors and HDACs inhibitors.We have every reason to believe that epigenetics will become the most powerful and reliable tool to conquer diabetes and vascular complications.
[1] Villeneuve LM,Natarajan R.The role of epigenetics in the pathology of diabetic complications[J].Am J Physiol Renal Physiol,2010,299(1):F14-F25.
[2] Reddy MA,Natarajan R.Epigenetic mechanisms in diabetic vascular complications[J].Cardiovasc Res,2011,90(3):421-429.
[3] Krause B,Sobrevia L,Casanello P.Epigeneticsnew concepts of old phenomena in vascular physiology[J].Curr Vasc Pharmacol,2009,7(4):513-520.
[4] Siebel AL,Fernandez AZ,El-Osta A.Glycemic memory associated epigenetic changes[J].Biochem Pharmacol,2010,80(12):1853-1859.
[5] McQuown SC,Wood MA.Epigenetic regulation in substance use disorders[J].Curr Psychiatry Rep,2010,12(2):145-153.
[6] Portela A,Esteller M.Epigenetic modiflcations and human disease[J].Nat Biotechnol,2010,28(10):1057-1068.
[7] Chen ZX,Riggs AD.DNA methylation and demethylation in mammals[J].J Biol Chem,2011,286(21):18347-18353.
[8] Szyf M.DNA methylation,the early-life social environment and behavioral disorders[J].J Neurodev Disord,2011,3(3):238-249.
[9] Miranda TB,Jones PA.DNA methylation The nuts and bolts of repression[J].J Cell Physiol,2007,213(2):384-390.
[10] Klose RJ,Bird AP.Genomic DNA methylationthe mark and its mediators[J].Trends Biochem Sci,2006,31(2):89-97.
[11] Fuks F.DNA methylation and histone modificationsteaming up to silence genes[J].Curr Opin Genet Dev,2005,15(5):490-495.
[12] Sims RR,Reinberg D.Is there a code embedded in proteins that is based on post-translational modifications?[J].Nat Rev Mol Cell Biol,2008,9(10):815-820.
[13] Berger SL.The complex language of chromatin regulation during transcription[J].Nature,2007,447(7143):407-412.
[14] Wang GG,Allis CD,Chi P.Chromatin remodeling and cancer,Part ICovalent histone modifications[J].Trends Mol Med,2007,13(9):363-372.
[15] Villeneuve LM, Reddy MA, Natarajan R.Epigeneticsdeciphering its role in diabetes and its chronic complications[J].Clin Exp Pharmacol Physiol,2011,38(7):401-409.
[16] Kuroda A,Rauch TA,Todorov I,et al.Insulin gene expression is regulated by DNA methylation[J].PLoS One,2009,4(9):e6953.
[17] Gilbert ER,Liu D.Epigeneticsthe missing link to understandingβ-cell dysfunction in the pathogenesis of type 2 diabetes[J].Epigenetics,2012,7(8):841-852.
[18] Park JH,Stoffers DA,Nicholls RD,et al.Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1[J].J Clin Invest,2008,118(6):2316-2324.
[19] Ling C,Del Guerra S,Lupi R,et al.Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion[J].Diabetologia,2008,51(4):615-622.
[20] Zhang D,Jiang X,Fang P,et al.Hyperhomocysteinemia promotes inflammatory monocyte generation and accelerates atherosclerosis in transgenic cystathionine beta-synthasedeflcient mice[J].Circulation,2009,120(19):1893-1902.
[21] Ekstrom TJ,Stenvinkel P.The epigenetic conductora genomic orchestrator in chronic kidney disease complications? [J].J Nephrol,2009,22(4):442-449.
[22] Ingrosso D,Perna AF.Epigenetics in hyperhomocysteinemic states.A special focus on uremia[J].Biochim Biophys Acta,2009,1790(9):892-899.
[23] Bell CG,Teschendorff AE,Rakyan VK,et al.Genomewide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus[J].BMC Med Genomics,2010,3:33.
[24] Wang HD,Sun J,Xia SJ.Interleukin-17 mediates the pathogenesis of diabetes induced by diabetogenic T lymphocytes in NOD mice[J].Chin J Endocrinol Metab,2009,25(5):539-543.
[25] Wang HD,Sun J,Xia SJ.Construction of the retrovirus vectors carrying the IL-17 or siRNA-IL-17 genes and expression of IL-17 gene in the transgenic diabetogenic T cells[J].Chin Gerontol J,2009,29(22):2873-2877.
[26] Rakyan VK,Beyan H,Down TA,et al.Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis[J].PLoS Genet,2011,7(9):e1002300.
[27] Keating ST,El-Osta A.Chromatin modificationsassociated with diabetes[J].J Cardiovasc Transl Res,2012,5(4):399-412.
[28] Brasacchio D,Okabe J,Tikellis C,et al.Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail[J].Diabetes,2009,58(5):1229-1236.
[29] Devaraj S,Glaser N,Griffen S,et al.Increased monocytic activity and biomarkers of inflammation in patients with type 1 diabetes[J].Diabetes,2006,55(3):774-779.
[30] Shilatifard A.Chromatin modifications by methylation and ubiquitinationimplications in the regulation of gene expression[J].Annu Rev Biochem,2006,75:243-269.
[31] Villeneuve LM,Reddy MA,Lanting LL,et al.Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes[J].Proc Natl Acad Sci USA,2008,105(26):9047-9052.
[32] Metzger E,Wissmann M,Yin N,et al.LSD1 demethylates repressive histone marks to promote androgen-receptordependent transcription[J].Nature,2005,437(7057):436-439.
[33] Lan F,Zaratiegui M,Villen J,et al.S.pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription[J].Mol Cell,2007,26(1):89-101.
[34] Reddy MA,Villeneuve LM,Wang M,et al.Role of the lysine-specific demethylase 1 in the proinflammatory phenotype of vascular smooth muscle cells of diabetic mice[J].Circ Res,2008,103(6):615-623.
[35] Pirola L,Balcerczyk A,Tothill RW,et al.Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells[J].Genome Res,2011,21(10):1601-1615.
[36] Ma J,Phillips L,Wang Y,et al.Curcumin activates the p38MPAK-HSP25 pathway in vitro but fails to attenuate diabetic nephropathy in DBA2J mice despite urinary clearance documented by HPLC[J].BMC Complement Altern Med,2010,10:67.
[37] Yun JM,Jialal I,Devaraj S.Epigenetic regulation of high glucose-induced proinflammatory cytokine production in monocytes by curcumin[J].J Nutr Biochem,2011,22(5):450-458.
[38] Kaur H,Chen S,Xin X,et al.Diabetes-induced extracellular matrix protein expression is mediated by transcription coactivator p300[J].Diabetes,2006,55(11):3104-3111.
[39] Sun G,Reddy MA,Yuan H,et al.Epigenetic histone methylation modulates fibrotic gene expression[J].J Am Soc Nephrol,2010,21(12):2069-2080.
[40] Chen S,Feng B,George B,et al.Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells[J].Am J Physiol Endocrinol Metab,2010,298(1):E127-E137.
[41] Li P,Zhao Y,Wu X,et al.Interferon gamma (IFN-γ)disrupts energy expenditure and metabolic homeostasis by suppressing SIRT1 transcription[J].Nucleic Acids Res,2012,40(4):1609-1620.
[42] Zhong Q,Kowluru RA.Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon[J].J Cell Biochem,2010,110(6):1306-1313.
[43] Cooper ME,El-Osta A.Epigeneticsmechanisms and implications for diabetic complications[J].Circ Res,2010,107(12):1403-1413.
[44] Lee HB,Noh H,Seo JY,et al.Histone deacetylase inhibitorsa novel class of therapeutic agents in diabetic nephropathy[J].Kidney Int,2007,72(106):S61-S66.
[45] Crosson CE,Mani SK,Husain S,et al.Inhibition of histone deacetylase protects the retina from ischemic injury[J].Invest Ophthalmol Vis Sci,2010,51(7):3639-3645.
[46] Chalmers J,Cooper ME.UKPDS and the legacy effect[J].N Engl J Med,2008,359(15):1618-1620.
[47] Pop-Busui R,Low PA,Waberski BH,et al.Effects of prior intensive insulin therapy on cardiac autonomic nervous system function in type 1 diabetes mellitusthe Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications study(DCCT/EDIC)[J].Circulation,2009,119(22):2886-2893.
[48] Holman RR,Paul SK,Bethel MA,et al.10-year follow-up of intensive glucose control in type 2 diabetes[J].N Engl J Med,2008,359(15):1577-1589.
[49] Patel A,Mac Mahon S,Chalmers J,et al.Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes[J].N Engl J Med,2008,358(24):2560-2572.
[50] Gerstein HC,Miller ME,Byington RP,et al.Effects of intensive glucose lowering in type 2 diabetes[J].N Engl J Med,2008,358(24):2545-2559.
[51] Engerman RL,Kern TS.Progression of incipient diabetic retinopathy during good glycemic control[J].Diabetes,1987,36(7):808-812.
[52] Roy S,Sala R,Cagliero E,et al.Overexpression of flbronectin induced by diabetes or high glucosephenomenon with a memory[J].Proc Natl Acad Sci USA,1990,87(1):404-408.
[53] Chan PS,Kanwar M,Kowluru RA.Resistance of retinal inflammatory mediators to suppress after reinstitution of good glycemic controlnovel mechanism for metabolic memory[J].J Diabetes Complications,2010,24(1):55-63.
[54] Kennedy JM,Zochodne DW.Experimental diabetic neuropathy with spontaneous recoveryis there irreparable damage? [J].Diabetes,2005,54(3):830-837.
[55] El-Osta A,VBrasacchio D,Yao D,et al.Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia[J].J Exp Med,2008,205(10):2409-2417.
[56] Tonna S,El-Osta A,Cooper ME,et al.Metabolic memory and diabetic nephropathypotential role for epigenetic mechanisms[J].Nat Rev Nephrol,2010,6(6):332-341.
Epigenetic phenomenon in diahetic vascular diseases
LOU Xu-dan1,WANG Hai-dong1△,XIA Shi-jin2,Sven Skog3
(1Department of Endocrinology,Huadong Hospital,Fudan University,Shanghai200040,China;2Shanghai Institute of Geriatrics,Huadong Hospital,Fudan University,Shanghai200040,China;3Sino-Swed Molecular Bio-Medicine Research Institute,Shenzhen518057,Guangdong Province,China)
DNA methylation; histone modification; diabetic vascular diseases; metabolic memory
R 587.1
B
10.3969/j.issn.1672-8467.2014.02.021
2013-01-11;编辑:张秀峰)
上海市科委科技发展基金基础研究重点课题(10JC1404800);国家自然科学基金面上项目(31171129)
△Corresponding author E-mail:wanghaidong@medmail.com.cn
【Ahstract】Diabetes mellitus and its associated micro-and macrovascular complications have posed a great threat to public health and become the main cause of morbidity and mortality burden to state economy.Recent studies have raised that epigenetics may play an important role in the pathology of diabetic vascular diseases,which refers to phenotype changes that are not related to the underlying DNA sequence,yet the alterations in gene expression can possibly be inherited into next generation.Furthermore,UKPDS and DCCT-EDIC demonstrate a phenomenon called metabolic memory,even if the blood glucose level is controlled in an ideal state,vascular inflammation still persists.Although epigenetics has not been fully elucidated,it provides a new insight into the pathogenesis of diabetes and is expected to become the targeted therapy for severe diseases.
*This work was supported hy Shanghai Committee of Science and Technology Development Funds for Basic Research under grant(10JC1404800),and the General Program of National Natural Science Foundation of China(31171129).

