Development of highly glyphosate-tolerant tobacco by coexpression of glyphosate acetyltransferase gat and EPSPS G2-aroA genes
Boqing Dun,Xujing Wng,Wei Lu,Ming Chen,Wei Zhng,Shuzhen Ping,Zhixing Wng,Boming Zhng,Min Lin,*
aInstitute of Crop Sciences,The National Key Facility for Crop Gene Resources and Genetic Improvement,Chinese Academy of Agricultural Sciences,Beijing 100081,China
bBiotechnology Research Institute,Chinese Academy of Agricultural Sciences,Beijing 100081,China
1.Introduction
Glyphosate(N-phosphonomethyl-glycine)is nonselective and the number-one selling herbicide in the world.It inhibits the enzyme enolpyruvylshikimate-3-phosphate synthase (EPSPS)in the plant chloroplast-localized pathway that leads to the biosynthesis of aromatic amino acids[1].The broad spectrum of weeds controlled by glyphosate and the safety and positive environmental profiles of the product have made its use for crop weed control attractive[2].
The commercialization of transgenic glyphosate-tolerant soybean in 1996 introduced a new pattern of use in which glyphosate can be applied to crops post-emergence to remove weeds without damage of crops.Since then,herbicide-tolerant crops have been quickly adopted by farmers.In 2012,herbicide tolerance,deployed in maize (Zea mays L.),Indian mustard(Brassica juncea L.),Anemone vitifolia Buch.-Ham.,soybean(Glycine max L.),sugar beet (Beta vulgaris L.),and erba medica(Medicago sativa L.) occupied 59% of 170.3 million hectares of transgenic crops planted globally[3].
Two basic strategies have been successfully used in glyphosate-tolerant crop development:expression of an insensitive form of the target enzyme EPSPS,and detoxification of the glyphosate molecule.The first strategy has been used in most existing commercial glyphosate-tolerant crops.They were obtained by employing a mutated (TIPS) or a microbial (CP4)form of EPSPS that is not inhibited by glyphosate [4,5].The theoretical disadvantage of this method is that glyphosate remains and accumulates in plant meristems,where it may hinder reproductive development and lower crop yield [6].The second approach avoids this limitation,because its functional mechanism is removal of herbicidal residue.N-acetylglyphosate is not herbicidal and does not inhibit EPSP synthase.Castle et al.[7,8]cloned glyphosate acetyltransferase (GLYAT) enzyme genes from Bacillus licheniformis.By DNA shuffling,a Glyat gene was obtained that had catalytic efficiency appropriate for commercial levels of resistance to glyphosate in crops.The first trait,in which GLYAT is deployed in soybean and canola(Brassica campestris L.),is in advanced stages of development(Pioneer Hi-Bred Technical Update)[1].
In China,a key problem in herbicide-tolerance gene engineering is the shortage of genes with higher glyphosate tolerance and independent intellectual property rights.Thus,it is of interest to seek new glyphosate-tolerance genes for developing glyphosate-tolerant crops that have high and stable heritability for glyphosate tolerance.Based on the biological diversity of microbial genetic resources in extremely polluted environments,a gat gene encoding N-acetyltransferase and a G2-aroA gene encoding EPSPS have been isolated by molecular biological methods [9,10].G2-aroA showed enhanced glyphosate tolerance in transgenic crops[11].
In the present study,we simultaneously introduced the G2-aroA and gat genes into tobacco,Nicotiana tabacum L.Glyphosate tolerance analysis indicated that transgenic tobacco coexpressing G2-aroA and gat displayed higher tolerance to glyphosate than transgenic tobacco containing G2-aroA or gat alone.These results showed that the combination of two approaches may enhance tolerance in transgenic crops and provide a new idea for development of glyphosate-tolerant crops.
2.Materials and methods
2.1.Materials
We previously isolated gat and G2-aroA from a glyphosate storage area with a long history of glyphosate pollution in Hebei Province,China.Transgenic tobacco G2 and GAT,N.tabacum var.NC89,Escherichia coli strain DH5α,Agrobacterium tumefaciens strain LBA4404,and vectors pSK,p4A,pGAT,and pG2 were maintained in our laboratory.
All products for restriction digests and ligations were purchased from New England Biolabs,Inc.and Promega,Inc.All other chemicals were analytical reagent grade.
2.2.Construction of plant expression vectors p2301G2-GAT
The polymerase chain reaction (PCR) was used to amplify gat gene from pGAT.The sequences of the primers along with underlined restriction enzyme sites were pGATF (5′-GCTCGAGATGATTGACGTGAACCCAAT-3′) and pGATR (5′-GGTTAACT TATGCGATCCTCTTGTACA-3′).The amplified product was inserted into the pMD18T-vector to produce pGAT-T.Gene gat was inserted into the Xho I/Hpa I site of p4Ato form intermediate vector pS4AGAT.The gat expression cassette was excised from pS4AGAT using Kpn I/Sma I and ligated into the plant expression vector pG2 to produce the plant expression vector p2301G2-GAT.
2.3.Transformation of tobacco
The plant expression vectors p2301G2-GAT were transferred into A.tumefaciens strain LBA4404 using the freeze-thaw method.LBA4404 was grown on YEB medium at 28°C and shaken at 150–250 r min-1overnight.Cultures were diluted 1:1 with YEB and allowed to grow to A550≈1.0.N.tabacum var.NC89 leaf discs from about 4-week-old tissue culture plantlets were used for A.tumefaciens-mediated transformation.After infection with A.tumefaciens,leaf discs were placed on cocultivation medium [MS (Murashige & Skoog) medium+3% sucrose+2.0 mg L-16-benzylaminopurine+0.1 mg L-1α-naphthaleneacetic acid] and incubated at 28°C in dark for 3–4 days.Leaf discs were cultured on differentiation medium (MS medium+3%sucrose+2.0 mg L-16-benzylaminopurine+0.1 mg L-1 α-naphthaleneacetic acid+500 mg L-1cephalosporin+100 mg L-1kanamycin)until plant regeneration.After regenerated seedlings had grown to 2–3 cm,they were placed in rooting medium (MS medium+3%sucrose+100 mg L-1kanamycin+500 mg L-1cephalosporin)in an Erlenmeyer flask for rooting.
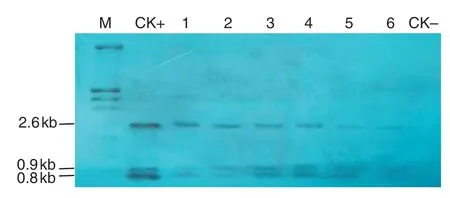
Fig.2-Southern blotting analysis of transgenic tobacco.M:molecular size marker;CK+:p2301G2-GAT plasmid DNA;1-6:transgenic plants.2,3,5,11,17,and 21;CK-:NC89.2.6 kb hybrid band containing full length G2-aroA gene;0.8 kb and 0.9 kb DNA band containing 320 bp and 121 bp gat gene,respectively.
2.4.Southern blotting detection of transgenic tobacco
Leaves of randomly chosen transgenic plants were collected for DNA isolation.Ten micrograms of genomic DNA of transgenic tobacco with gat/G2-aroA were fully digested with EcoR I/Kpn I and immobilized on a Hybond-N+membrane.The DNA samples of gat and G2-aroA were used for preparation of probes and Southern blotting analysis was performed using DIG-High Prime DNA Labeling and Detection Starter Kit II(Boehringer Mannheim Biochemicals).
2.5.RT-PCR detection of transgenic tobacco
Total RNA of transgenic tobacco was extracted with an RNA extraction kit (New England Biolabs,Inc.).RNA expression profiles of target genes in transgenic tobacco were assessed by RT-PCR using the ProtoScript First Strand cDNA Synthesis Kit(New England Biolabs,Inc.).Genes gat and G2-aroA were then amplified from transgenic tobacco cDNA using the following primers: gat gene 5′-ATGATTGACGTGAACCCAAT-3′ and 5′-TTA TGCGATCCTCTTGTACA-3′; and G2-aroA gene 5′-ATGGCGTGT TTGCCTGATGA-3′and 5′-TCAGTCGTTTAGGTGAACGCC-3′.
2.6.Western blotting analysis of transgenic tobacco
Protein was extracted from fresh tobacco leaves by homogenization in extraction buffer (200 mmol L-1Tris–HCl (pH 8.0),100 mmol L-1NaCl,400 mmol L-1sucrose,14 mmol L-1isoamyl alcohol,1 mmol L-1phenylmethylsulfonyl fluoride (PMSF) and 0.05% Tween-20).The extract was centrifuged at 12,500 r min-1for 20 min at 4 °C.The protein concentration of the supernatant was determined using the Bio-Rad protein assay.The protein samples were mixed with 50 μL of 3 × sodium dodecyl sulfate(SDS)loading buffer(Bio-Rad)and boiled for 10 min,and 8 μL of each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 12% Tris–glycine gels (Invitrogen).Protein bands were transferred to a Poly vinylidene fluoride(PVDF)membrane.After blocking with 5%BSA for 1 h at room temperature,the blots were incubated overnight at 4 °C with antiserum(1:10,000 dilution)in the presence of 1%BSA,washed three times(15 min each),and incubated with 1:30,000-diluted alkaline phosphate-conjugated anti-rabbit IgG for 1 h at room temperature.The reaction was visualized with a BCIP/NBT color development substrate(Promega,Inc.).The anti sera used were raised in rabbits.
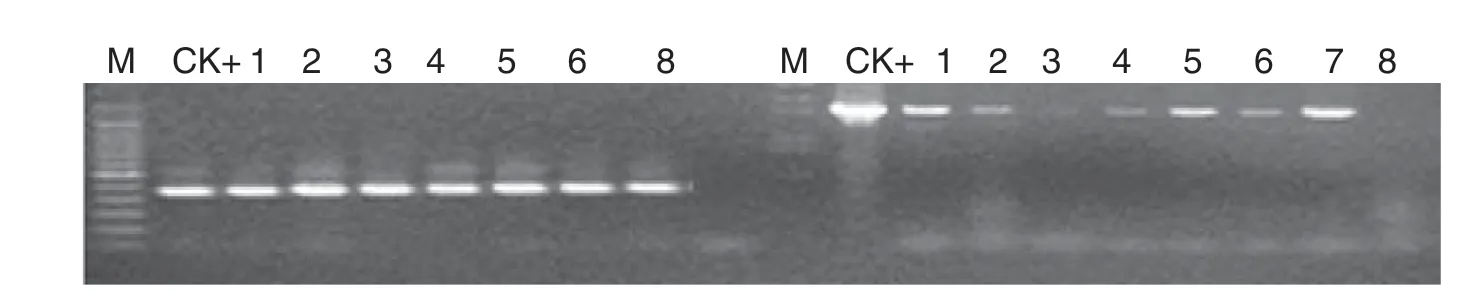
Fig.3-RT-PCR detection of gat and G2-aroA gene expression of transgenic tobacco.M:molecular size marker;CK+:p2301G2-GAT plasmid;1-7:transgenic plants 2,3,5,11,17,21,and 34;8:NC89.
2.7.Glyphosate tolerance analysis of transgenic tobacco
Two methods were used to analyze glyphosate tolerance in transgenic tobacco plants.For the leaf spraying experiment,6 to 8-leaf-stage transgenic plants grown in the green house were sprayed with the herbicide Roundup (isopropylamine salt of glyphosate as active ingredient),41.0%(w/v)at doses of 0.8–1.0 L ha-1.T1progeny seeds of transgenic tobacco containing gat,G2-aroA,or gat/G2-aroA were germinated on MS medium supplemented with 0,0.2,1.0,5.0,and 10.0 mmol L-1glyphosate.Seedlings were grown in growth chambers at 25 °C with 60%–70% relative humidity and a photosynthetic photon flux density of 24 μmol m-2s-1with a 10-h photoperiod.The growth status and viability of transgenic plants were evaluated after culturing for 4 weeks.
3.Results and discussion
3.1.Construction of plant expression vector
The gat gene was amplified by PCR using corresponding primers and template.After sequencing confirmation,the gene was inserted into pG2 to form plant expression vector p2301G2-GAT.In this vector,gat and G2-aroA genes were driven in tandem by a CaMV35S promoter with two enhancers and terminated with a NOS terminator at their 3′ ends.The T-regions in p2301G2-GAT also harbored 35SP::nptII::35SpolyA to provide kanamycin resistance.The structure of p2301G2-GAT is shown in Fig.1.
3.2.Molecular detection of transgenic tobacco
A total of 52 independent transgenic tobacco (N.tabacum cv.NC89) lines were generated by Agrobacterium-mediated gene transformation.The transgenic plants with G2-aroA and gat were named G2-GAT.Southern blotting,RT-PCR,and Western blotting analysis showed that the specific bands were present in tested samples (Figs.2–4),demonstrate that the target genes had been integrated into the tobacco genome and were expressed effectively at the RNA and protein levels.
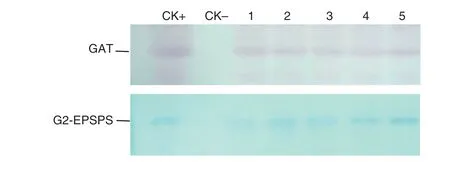
Fig.4-Western blotting analysis of transgenic tobacco.CK+:expression product of GAT or G2-EPSPS,which are from E.coli expressing gat or G2-aroA gene;CK-:NC89;1-5:Transgenic plants 2,3,5,11,and 17.
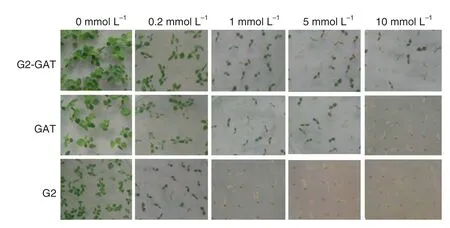
Fig.5-Seedling comparison of T1 seeds of transgenic tobacco on MS0 medium containing different concentrations of glyphosate.
3.3.Glyphosate tolerance analysis of transgenic tobacco
Glyphosate tolerance was compared among transgenic tobacco plants containing gat,G2-aroA,or both genes by assessing germination of T1transgenic tobacco seeds and by leaf spraying.T1seeds of transgenic tobacco G2,GAT,and G2-GAT(containing G2-aroA,gat,or G2-aroA/gat,respectively)were germinated after sterilization on MS medium containing different concentrations of glyphosate (Fig.5).Glyphosate tolerance was evaluated by seed germination and seedling growth on medium containing glyphosate after 4 weeks.On medium containing 0.2 mmol L-1glyphosate,no difference in seed germination was apparent among the 3 types of transgenic tobacco.All transgenic plants germinated anddeveloped normally,and there was little difference in seedling growth vigor compared with the control(plants growing on MS medium without glyphosate).On medium containing 1 mmol L-1glyphosate,all of the G2 transgenic plants died.No difference in viability was apparent among controls and GAT or G2-GAT transgenic plants,although the growth vigor of GAT and G2-GAT plants was obviously reduced.On media supplemented with 5 mmol L-1glyphosate,a difference in viability was apparent between GAT and G2-GAT transgenic plants,and their growth vigor was reduced compared with the control.On media supplemented with 10 mmol L-1glyphosate,all GAT transgenic plants died,but 14%of G2-GAT plants survived (Table 1).The segregation ratio of glyphosate resistant and sensitive plants was 3:1 in selection medium containing 0.2 mmol L-1glyphosate.We accordingly postulated that the genes introduced into these transgenic tobacco plants were inserted as single copies.
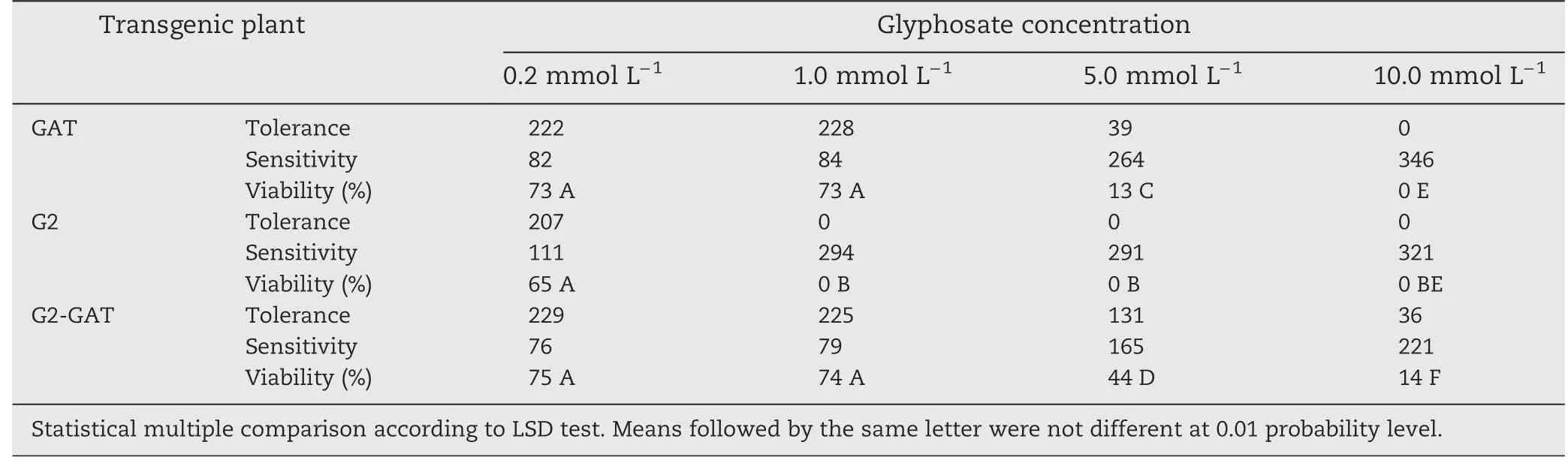
Table 1-Viability statistics analysis of transgenic tobacco T1 seedlings on MS medium containing different concentrations of glyphosate.
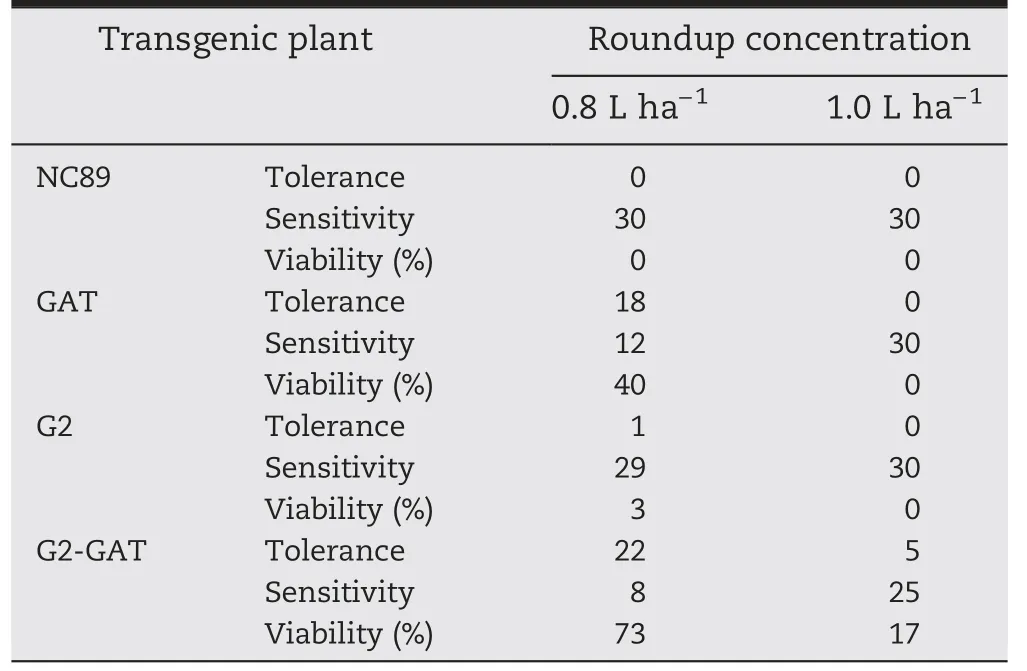
Table 2-Viability statistics of transgenic tobacco after leaf spraying with different concentrations of Roundup herbicide.
T1transgenic plants at 6 to 8-leaf-stage were sprayed with a 1.0% (v/v) solution of the herbicide Roundup (isopropylamine glyphosate salt as active ingredient,41.0%,w/v) at a dose of 0.8 L ha-1.In non-transgenic plants,the leaves and stem apex began to wilt 1–3 days after treatment.The non-transgenic control showed severe wilt and chlorosis on all leaves after 5 days and died 7 days after treatment.Twenty-four GAT plants grew well with normal morphology for 2 weeks after treatment,and 6 GAT plants begin to wilt 5 days after treatment and died after 2 weeks.Four G2 plants survived,but 3 showed partial leaf chlorosis and bleaching after 6 days.Twenty-six G2-GAT plants grew well with normal morphology for 2 weeks after treatment,and the remaining 4 plants exhibited wilting and bleaching 5 days after treatment and then died.All the three types of transgenic plants,except for 5 G2-GAT plants,died after glyphosate treatment at a dose of 1 L ha-1(Table 2 and Fig.6).
Glyphosate tolerance analysis indicated that the transgenic tobacco containing G2-aroA/gat showed higher tolerance to glyphosate than plants containing only G2-aroA or gat alone;however,a clear difference was observed between transgenic tobacco containing gat and plants containing G2-aroA.The G2-aroA-carrying plants were significantly more susceptible to glyphosate than those carrying gat.Either of the two explanations may account for this difference.The first is that G2-aroA was expressed at a low level,as confirmed by semi-quantitative RT-PCR analysis of the transgenic tobacco(data not shown).The second explanation is that the G2-aroA expression vector lacks a leader chloroplast signal peptide.In plants,the EPSPS protein is located and acts in the chloroplast,but EPSPS is expressed in the nucleolus and must enter the chloroplast via the chloroplast signal peptide.The transgenic plant carrying the bacterial EPSPS gene,which is expressed in the cytoplasm,may tolerate only a low concentration of glyphosate because it lacks the chloroplast signal peptide [12,13].The combination of the G2-aroA and gat genes was successfully used for construction of transgenic plants coexpressing glyphosate-tolerant EPSPS and glyphosatedetoxified GAT,and consequently conferred higher resistance to glyphosate.
There are increasing instances of evolved glyphosate tolerance in weed species following wide planting of glyphosate-tolerant crops,based mainly on EPSPS insensitive to the herbicide [2,14].In several cases,moderate tolerance is imparted by mutations of the target enzyme[15],but there is no documented case of a plant species having native or evolved tolerance to glyphosate by virtue of a metabolic enzyme [1].The combination of different strategies is thus a promising approach to the development of glyphosate-tolerant crops.Glyphosate oxidoreductase (GOX) and acetyltransferase (GAT)have the ability to detoxify glyphosate via the AMPA pathway(GOX-catalyzed oxidative cleavage of the carbon–nitrogen bond on thecarboxyl side,resulting in the formation of amino methylphosphonic acid (AMPA) and glyoxylate) and Nacetylation,respectively.Several agronomic crops transformed with both CP4 and GOX,including maize,A.vitifolia Buch.-Ham.,potato (Solanum tuberosum L.),Indian mustard,soybean,sugar beet,and tomato(Solanum lycopersicum L.),have been field tested and deregulated (http://www.nbiap.vt.edu/cfdocs/fieldtests1.cfm).However,in many crops carrying both genes,a chlorotic phenotype has been observed in response to glyphosate treatment.Growth of poplar transformed with CP4 alone was significantly better than that of poplar carrying both genes and exhibited less damage in response to glyphosate treatment[16].In the present study,we obtained high glyphosate-tolerant tobacco by coexpression of G2-aroA and gat genes,indicating the effectiveness of a combination of two strategies:expression of an insensitive form of the target enzyme EPSPS and metabolic detoxification of glyphosate.
This study was supported by the National Basic Research Program of China(2007CB707805),the National High Technology Research and Development Program of China (2006AA020101),and the National Natural Science Foundation of China(30470047 and 30200007).
[1] L.Pollegioni,E.Schonbrunn,D.Siehl,Molecular basis of glyphosate resistance-different approaches through protein engineering,FEBS J.278 (2011) 2753–2766.
[2] G.M.Dill,C.A.CaJacob,S.R.Padgette,Glyphosate-resistant crops: adoption,use and future considerations,Pest Manag.Sci.64(2008) 326–331.
[3] C.James,Global stutus of commercialized biotech/GM crops:2012,China Biotechnol.33(2013) 1–8.
[4] C.Herouet-Guicheney,D.Rouquié,M.Freyssinet,T.Currier,A.Martone,J.Zhou,E.E.Bates,J.M.Ferullo,K.Hendrickx,D.Rouan,Safety evaluation of the double mutant 5-enolpyruvylshikimate-3-phosphate synthase (2mEPSPS)from maize that confers tolerance to glyphosate herbicide in transgenic plants,Regul.Toxicol.Pharmacol.54 (2009)143–153.
[5] X.Xiao,H.Wu,X.Zhou,S.Xu,J.He,W.Shen,G.Zhou,M.Huang,The combination of quantitative PCR and western blot detecting CP4-EPSPS component in Roundup Ready soy plant tissues and commercial soy-related food stuffs,J.Food Sci.77(2012) C603–C608.
[6] W.A.Pline,J.W.Wilcut,S.O.Duke,K.L.Edmisten,R.Wells,Tolerance and accumulation of shikimic acid in response to glyphosate applications in glyphosate-resistant and nonglyphosate-resistant cotton (Gossypium hirsutum L.),J.Agric.Food Chem.50(2002) 506–512.
[7] L.A.Castle,D.L.Siehl,R.Gorton,P.A.Patten,Y.H.Chen,S.Bertain,H.J.Cho,N.Duck,J.Wong,D.Liu,M.W.Lassner,Discovery and directed evolution of a glyphosate tolerance gene,Science 304 (2004) 1151–1154.
[8] D.L.Siehl,L.A.Castle,R.Gorton,R.J.Keenan,The molecular basis of glyphosate resistance by an optimized microbial acetyltransferase,J.Biol.Chem.15(2007) 11446–11455.
[9] B.Q.Dun,W.Lu,S.Z.Ping,W.Zhang,M.Chen,Y.Q.Xu,D.Jin,Z.L.Zhao,M.Lin,Isolation of a novel glyphosate tolerance N-acetyltransferase gene and expression in E.coli,Chin.High Technol.Lett.16 (2006) 943–947.
[10] Y.Zhu,Z.L.Yu,M.Lin,Biotolerance or biodegradation of glyphosate and construction of transgenic plants,Mol.Plant Breed.1(2003) 435–441.
[11] G.R.Yu,W.-P.Du,J.Song,Q.Dou,Y.S.Liu,W.Lu,L.Y.Xu,Transformation of tolerant-gyphosate 2mG2-epsps gene by maize shoot tip and identification of resistance,Southwest China,J.Agric.Sci.23(2010) 1403–1408.
[12] S.R.Padgette,Q.K.Huynh,J.Borgmeyer,Bacterial expression and isolation of Petunia hybrida 5-enol-pyruvylshikimate-3-phosphate synthase,Arch.Biochem.Biophys.258 (1987) 564–573.
[13] H.Zhou,J.W.Arrowsmith,M.Fromm,Glyphosate tolerant CP4 and GOX genes as a selectable marker in wheat transformation,Plant Cell Rep.15(1995) 159–163.
[14] E.Waltz,Glyphosate resistance threatens Roundup hegemony,Nat.Biotechnol.28(2010)537–538.
[15] S.B.Powles,C.Preston,Evolved glyphosate resistance in plants: biochemical and genetic basis of resistance,Weed Technol.20(2006) 282–289.
[16] R.Meilan,K.-H.Han,C.Ma,S.P.DiFazio,J.A.Eaton,E.A.Hoien,B.J.Stanton,R.P.Crockett,M.L.Taylor,R.R.James,J.S.Skinner,L.Jouanin,G.Pilate,S.H.Strauss,The CP4 transgene provides high levels of tolerance to Roundup herbicide in field-grown hybrid poplars,Can.J.For.Res.32 (2002) 967–976
- The Crop Journal的其它文章
- Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton
- SSR genetic linkage map construction of pea (Pisum sativum L.) based on Chinese native varieties
- Rank correlation among different statistical models in ranking of winter wheat genotypes,
- Effects of exogenous ABA application on post-anthesis dry matter redistribution and grain starch accumulation of winter wheat with different staygreen characteristics
- Genetic characterization and linkage disequilibrium mapping of resistance to gray leaf spot in maize(Zea mays L.)
- Overexpression of GmDREB1 improves salt tolerance in transgenic wheat and leaf protein response to high salinity

